Abstract
Polycomb group genes (PcG) encode a group of about 16 proteins that were first identified in Drosophila as repressors of homeotic genes. PcG proteins are present in all metazoans and are best characterized as transcriptional repressors. In Drosophila, these proteins are known as epigenetic regulators because they remember, but do not establish, the patterned expression state of homeotic genes throughout development. PcG proteins, in general, are not DNA binding proteins, but act in protein complexes to repress transcription at specific target genes. How are PcG proteins recruited to the DNA? In Drosophila, there are specific regulatory DNA elements called Polycomb group response elements (PREs) that bring PcG protein complexes to the DNA. Drosophila PREs are made up of binding sites for a complex array of DNA binding proteins. Functional PRE assays in transgenes have shown that PREs act in the context of other regulatory DNA and PRE activity is highly dependent on genomic context. Drosophila PREs tend to regulate genes with a complex array of regulatory DNA in a cell or tissue-specific fashion and it is the interplay between regulatory DNA that dictates PRE function. In mammals, PcG proteins are more diverse and there are multiple ways to recruit PcG complexes, including RNA-mediated recruitment. In this review, we discuss evidence for PREs in vertebrates and explore similarities and differences between Drosophila and vertebrate PREs.
1. INTRODUCTION
Polycomb group proteins (PcG) are important regulators of developmental genes in all metazoans (two reviews: Beisel & Paro, 2011; Simon & Kingston, 2009). First discovered in genetic studies as regulators of homeotic genes in Drosophila, genome-wide chromatin-immunoprecipitation (ChIP) studies using antibodies against various PcG proteins show that there are likely hundreds of PcG targets in Drosophila (Négre et al., 2006; Schwartz et al., 2006, 2010; Tolhuis et al., 2006). In vertebrate embryonic stem (ES) cells, PcG proteins are important regulators of developmental genes and are important in both maintenance of pluripotency and in differentiation. Because they regulate developmental and cell cycle genes, altered expression of PcG genes has been associated with many cancers (reviews: Geini & Hendzel, 2009; Sparmann & van Lohuizen, 2006).
There are at least 16 PcG genes in Drosophila, identified by mutations that derepress homeotic gene expression. Many PcG genes encode proteins that act in protein complexes to regulate transcription via alterations to chromatin structure. The two best studied PcG protein complexes are PRC1 and PRC2 (Polycomb repressive complexes 1 and 2), which are conserved from flies to mammals (for reviews, see Kerppola, 2009; Müller & Verrijzer, 2009; Schuettengruber & Cavalli, 2009; Simon & Kingston, 2009). PRC2 contains the PcG proteins Enhancer of zeste (E(z)), Extra sex combs (Esc), Suppressor of zeste 12 (Su(z)12), as well as the protein p55 (Cao et al., 2002; Czermin et al., 2002; Müller et al., 2002), and in some tissues Polycomb-like (Pcl), which modifies PRC2 function (Nekrasov et al., 2007; Savla, Benes, Zhang, & Jones, 2008). E(z), the catalytic component of PRC2, trimethylates histone H3 lysine 27 creating the H3K27me3 mark characteristic of Polycomb-regulated genes. The PRC1 core complex is comprised of Polycomb (Pc), Polyhomeotic (Ph), Posterior sex combs (Psc), and Sex combs extra/dRing1 (Sce/dRing1; Fritsch, Beuchle, & Müller, 2003) and acts to inhibit chromatin remodeling and compact chromatin (Saurin, Shao, Erdjument-Bromage, Tempst, & Kingston, 2001; Shao et al., 1999). Psc and Sce/dRing1 are also in another complex called dRAF that contains a histone demethylase, dKDM2 (Lagarou et al., 2008). Another PcG complex, PR-DUB (Polycomb repressive deubiquitinase) consists of two other PcG proteins, Calypso and Additional sex combs (Asx), which deubiquitinates H2A118 (Scheuermann et al., 2010; Schuettengruber & Cavalli, 2010). Additional PcG proteins required for homeotic gene silencing not yet assigned to a protein complex include Sex combs on middle leg (Scm), which loosely associates with PRC1, and Super sex combs (Sxc/Ogt) that encodes O-GlcNAc transferase and modifies Ph (Gambetta, Oktaba, & Müller, 2009). Finally, Drosophila has the protein complex Pho-repressive complex (PhoRC), consisting of the DNA binding PcG protein Pleiohomeotic (Pho) and the methyl-lysine-binding protein, dSfmbt (Klymenko et al., 2006). Both Pho and dSfmbt have homologues in vertebrates, but no vertebrate PhoRC complex has been described.
Genome-wide studies have shown that PcG proteins colocalize to many or most PcG target genes in Drosophila. Despite this, mutations in various PcG genes give very different phenotypes (Breen & Duncan, 1986). This suggests that different target genes have different requirements for specific PcG proteins. For example, at the PcG target gene engrailed (en), a gene required for segmentation, mutations in ph cause a massive en derepression in embryos, while mutations in Pc cause very little en misexpression (Moazed & O’Farrell, 1992). Since Ph and Pc are both components of PRC1, and both are bound to en PREs in Drosophila embryos (Négre et al., 2006), why en is so sensitive to the loss of Ph but not Pc is unclear. In a recent study, Gutiérrez et al. (2012) began to classify different PcG targets based on their response to Sce mutations. They studied targets that bind the PcG proteins Pho, Sce, Pc, Ph, and Psc, and found that expression of only a subset of the targets was altered in Sce mutants. Clearly, there is still a lot we need to learn about the role of different PcG proteins in transcriptional repression of different target genes.
In this review, we focus on the fundamental question of how PcG proteins are recruited to their target genes. This topic has been comprehensively covered in a number of recent reviews (Beisel & Paro, 2011; Müller & Kassis, 2006; Ringrose & Paro, 2007; Schuettengruber & Cavalli, 2009); here, we bring our own perspective and discuss unresolved issues. In Drosophila, we concentrate on target genes that are bound by both PRC1 and PRC2 and describe what is known about Polycomb group response element (PREs), DNA fragments that can recruit PRC1 and PRC2 to target genes via a complex array of DNA binding proteins. In vertebrates, recruitment of PcG proteins is more complicated, given that there are many more PcG proteins and varieties of PcG protein complexes (Beisel & Paro, 2011; Kerppola, 2009). While it has been demonstrated that RNA is involved in recruitment of PcG protein complexes at some vertebrate PcG targets, a role for RNA in PcG protein binding in Drosophila has not been demonstrated. Thus, for the vertebrate system, we limit our discussion to presumptive PREs.
2. PREs IN DROSOPHILA
2.1. PRE maintenance assays
PREs were first discovered in transgenes of regulatory DNA from the Bithorax complex (BX-C). In 1991, Müller and Bienz, while studying regulatory DNA from the Ubx gene, identified a DNA fragment that they called BXD, which, in combination with another regulatory fragment that initiated correct Ubx expression, could maintain β-galactosidase expression throughout embryonic development in a pattern similar to Ubx. Notably, in a Polycomb mutant, β-galactosidase expression was derepressed late in embryonic development, suggesting that Polycomb directly repressed the expression of the transgene (Müller & Bienz, 1991). In 1993, Simon et al. identified three discrete fragments of regulatory DNA from the BX-C that rendered transgenes responsive to PcG repression and coined the term Polycomb group response elements (PREs). Several important principles came out of this and other work on PREs from the BX-C complex. First, PREs act with other regulatory DNA to remember boundaries set up directly by gap and pair-rule regulators. Second, PREs respond to mutations in many different PcG genes, suggesting PcG proteins act together to repress gene expression. Third, discrete BX-C PREs have similar activities and can be interchangeable within transgenes (Simon, Chiang, Bender, Shimell, & O’Connor, 1993).
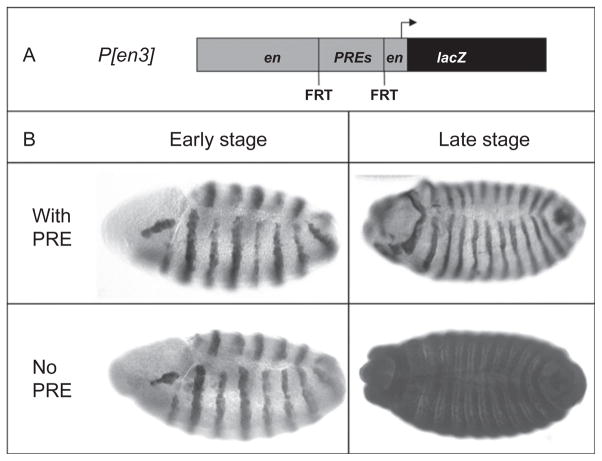
The principles of PREs learned from the early experiments on BX-C PREs apply to most if not all of the PREs studied to date. We study PREs from the Drosophila engrailed (en) and invected (inv) genes. en and inv encode coregulated, highly related homeodomain proteins which function together during development to control embryonic segmentation and formation of the posterior compartment in imaginal disks (Gustavson, Goldsborough, Ali, & Kornberg, 1996). en and in exist in a 115-kb chromatin domain, marked by the distinctive H3K27me3 modification put on by the PRC2 PcG protein complex (Kharchenko et al., 2011). There are four major PREs present in the en/inv domain (Cunningham, Brown, & Kassis, 2010; DeVido, Kwon, Brown, & Kassis, 2008) and their activity can be demonstrated in the functional assay shown in Fig. 3.1. A transgene containing 8 kb of upstream en regulatory DNA fused to the reporter gene lacZ expresses β-galactosidase in en-like stripes throughout embryonic development. However, deletion of a 2-kb fragment, that includes two PREs, while still giving en-like stripes early in development, gives expression throughout the embryo in late development. Thus, the 2-kb fragment, containing two PREs, is required for maintenance of en stripes. In this construct, the activity of both PREs is required for full maintenance (DeVido et al., 2008). Interestingly, the en PREs are also able to act as PREs in an embryonic Ubx-reporter construct (Americo et al., 2002) and PRED, a PRE from the Ubx gene, can act with en regulatory DNA to maintain en-expression in stripes (Cunningham et al., 2010). These data emphasize the functional similarity between en and Ubx PREs.
Figure 3.1.
Embryonic maintenance PRE assay. (A) Structure of a vector used to test for PRE activity of the en PREs. Eight kilobases of en sequence is fused to the reporter gene, lacZ. This leads to the expression of β-galactosidase in en-like stripes throughout embryonic development. This 8-kb of en includes a 2-kb region that encompasses the en PREs. Here, the 2-kb fragment containing the PREs is flanked by FRT sites allowing for removal of the PRE with FLP recombinase (Cunningham et al., 2010; DeVido et al., 2008). (B) Early and late stage embryos stained for β-galactosidase with a line that carries the 2-kb en PRE and a line that has the 2 kb PRE excised. With the PRE fragment, lacZ expression is maintained in en-like stripes throughout embryonic development. When the en PRE is excised, lacZ is expressed throughout the embryo. Early stage embryos: lateral view, anterior left, dorsal up, stage 11. Late stage embryos: dorsal view, anterior left, stage 16.
In addition to embryonic reporter constructs, there are at least two imaginal disk Ubx-lacZ-reporter constructs for testing PRE activity (Fritsch, Brown, Kassis, & Müller, 1999; Poux, Kostic, & Pirrotta, 1996); both constructs contain combinations of Ubx-regulatory DNA, including an imaginal disk enhancer. Without a PRE, β-galactosidase is ubiquitously expressed in the wing disk from these constructs, where Ubx is not normally expressed; addition of a PRE represses wing disk expression of the Ubx-lacZ-reporter. An Ubx-lacZ imaginal disk reporter construct was used to refine the size of the Ubx bxd PRE, PRED to a 567-bp fragment and to demonstrate the importance of the PcG-DNA-binding protein Pho to PRED activity in larvae (Fritsch et al., 1999). Recently, an imaginal Ubx-lacZ-reporter was used to test the activity of presumptive PREs from the Psc–Su(z)2 gene complex (Park, Schwartz, Kahn, Asker, & Pirrotta, 2012). These results again demonstrated that DNA from different genes can act as PREs with Ubx-regulatory DNA.
Only a few of the hundreds of PREs identified in genome-wide studies have been tested for activity in either embryonic or imaginal disk PRE reporter constructs and even fewer have been tested in both (see Table 3.1). It might be that some PREs are stage or tissue specific (see below). The development of a series of PRE vectors, with regulatory DNA from different genes, designed to assess activity at different developmental stages using the phi-C31 system (Groth, Fish, Nusse, & Calos, 2004) to insert transgenes at the same chromosomal location, would be key to assessing the functional equivalency between PREs.
Table 3.1.
Summary of Drosophila genes with PREs that have been characterized by the PSS assay or a maintenance assay in embryos (E) or larvae (L)
| PRE | Maintenance assay | PSS mini-white |
|---|---|---|
| en | Yes, E | Yes |
| inv | Yes, E | Yes |
| eve | Yes, E | Yes |
| Psc/Su(z)2 | Yes, L | Yes |
| Scr | Yes, E | Yes |
| pb | Yes, E | Yes |
| vg | ND | Yes |
| ph | ND | Yes |
| esg | ND | Yes |
| hh | ND | Yes |
| cad | ND | Yes |
| aPKC | ND | Yes |
| prod | ND | Yes |
| BX-C | ||
| Mcp | Yes, E | Yes |
| bxd(PRED) | Yes, E, L | Yes |
| iab-2 | Yes, E* | Yes |
| iab-6 | ND | Yes |
| iab7 (Fab7) | Yes E | Yes |
| iab-8 | Yes E | Yes |
iab-2 fragment used required additional DNA to work in the embryonic maintenance assay.
ND is for not done. Some of these genes have more than one PRE that have been tested in these assays (see text for discussion of this). Scr, Sex combs reduced; aPKC, atypical protein kinase C; prod, proliferation disruptor; hh, hedgehog; vg, vestigal; cad, caudal; esg, escargot; pb, probosipedia; BX-C, Bithorax complex. Mcp, bxd (PRED), iab fragments are regulatory regions within the BX-C.
References: en: Kassis et al. (1991), Kassis (1994), Brown, Mucci, Whiteley, Dirksen, and Kassis (1998), Americo et al. (2002), and DeVido et al. (2008). inv: Cunningham et al. (2010). eve: Fujioka, Emi-Sarker, Yusibova, Goto, and Jaynes (1999) and Fujioka, Yusibova, Zhou, and Jaynes (2008). Psc/Su(z)2: Park et al. (2012). Scr: Gindhart and Kaufman (1995). pb: Kapoun and Kaufman (1995). vg: Okulski, Druck, Bhalerao, and Ringrose (2011). ph: Bloyer et al. (2003). esg: Kassis (1994). hh: Rank, Prestel, and Paro (2002), Maurange and Paro (2002) and Chanas and Machat (2005). cad, aPKC, and prod: Ringrose, Rehmsmeier, Dura, and Paro (2003). BX-C: Chan, Rastelli, and Pirrotta (1994), Gruzdeva, Kyrchanova, Parshikov, Kullyev, and Georgiev (2005), Vazquez, Muller, Pirrotta, and Sedat (2006), Pérez-Lluch, Cuartero, Azorin, and Espinås (2008), Mishra et al. (2001), Shimell, Peterson, Burr, Simon, and O’Connor (2000), and Barges et al. (2000).
2.2. Mini-white silencing
The discovery of silencing of the mini-white gene in transgenic Drosophila coincided with the discovery of PREs (Kassis, Vansickle, & Sensabaugh, 1991). In 1988, Pirrotta developed pCaSpeR, a vector for P-element transformation using the mini-white gene as a reporter for transgenic flies (Pirrotta, 1988). white encodes a member of the ABC family of transporter superfamily and is necessary for the transport of the precursors of eye color pigment into the eye. Flies with a null mutation in white have a white-eye color. To make transgenic flies with the pCaSpeR vector, embryos mutant for the white gene are injected, mated, and screened for transgenic progeny that have colored eyes. The amount of white gene product produced determines the eye color; that is, a little White product gives a light yellow eye, more White product gives an orange eye all the way to the wild-type red eye color. The mini-white gene lacks most of the enhancers that cause white to be expressed in the eye; eyes from pCaSpeR-containing transgenic flies have an eye color ranging from yellow to red. This is because the expression of mini-white is highly susceptible to regulation by flanking genomic DNA. The pCaSpeR vector soon became the vector of choice because of two properties: (1) it was easier to clone into than other P-based vectors and (2) transgenic flies with independent insertion sites could be recognized by their different eye colors.
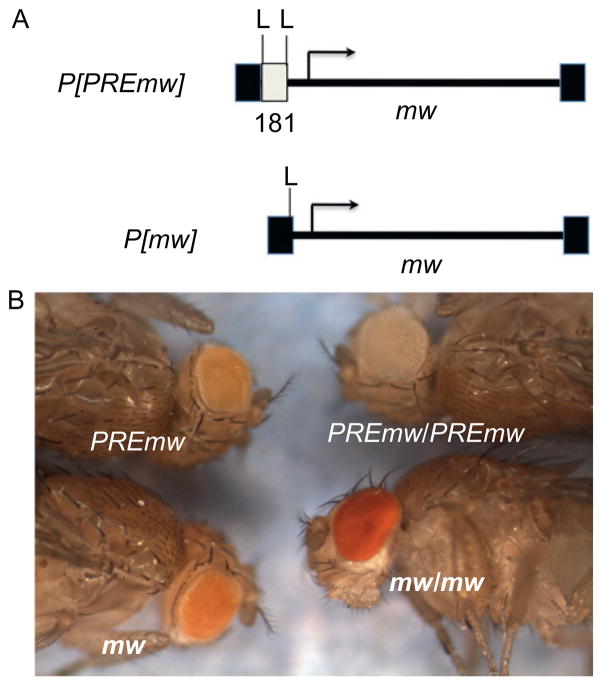
In 1991, Kassis et al. reported that a fragment of regulatory DNA from the Drosophila en gene could mediate mini-white silencing in flies homozygous for the en-mini-white containing transgenes (P[en-mw]). Normally, when using a vector with mini-white, flies homozygous for the transgene have a darker eye color than flies heterozygous for it (P[mw] in Fig. 3.2). However, when a 2.4-kb fragment of en regulatory DNA was included in the mini-white vector, the eye color of homozygous flies was often lighter than that of heterozygous flies, sometimes even white, suggesting that the mini-white expression was completely silenced in the eye. This mini-white silencing was dependent on the two transgenes being inserted near each other in the genome, either in cis (a duplicated P[en-mw] element would also silence mini-white) or in trans, that is, on homologous chromosomes (Kassis et al., 1991). Somatic chromosomes are paired in Drosophila, and this mini-white silencing was dependent on the pairing of the two P[en-mw] transgenes. This phenomenon has been called pairing-dependent repression or pairing-sensitive silencing (PSS) (Kassis, 1994). The 2.4-kb en fragment contained within the P[en-mw] transgenes contains two PREs (Americo et al., 2002; DeVido et al., 2008; Kassis, 1994).
Figure 3.2.
PSS of mini-white expression in transgenic flies. (A) Diagrammatic representation of the two reporter constructs used to test for PSS of an en PRE. P[mw] is the P-element vector control without any PRE insert. P[PREmw] is the same transformation vector carrying a 181-bp PRE from en cloned upstream of the mini-white promoter. The filled boxes represent the P-element ends. The open box represents the PRE. L is loxP sites that allow excision of the PRE. (B) Shows the eye color of transformants carrying the above constructs. mw and PREmw are heterozygous for the construct. mw/mw and PREmw/PREmw are homozygous for the constructs. PSS can be seen by comparing heterozygous versus homozygous eye colors for the two different constructs. Figure adapted from Noyes, Stefaniuk, Cheng, Kennison, and Kassis (2011).
Mini-white silencing is a property of PREs (reviewed in Kassis, 2002). In 1994, Chan, Rastelli, and Pirrotta reported that a 2.2-kb PRE from Ubx caused silencing of the mini-white gene in the pCaSpeR vector. In that case, an Ubx PRE repressed mini-white expression even in the heterozygous state; many Ubx PRE-mini-white transformants had variegated eyes, some with the eye color only present in a few ommatidia. Using a different vector that contained both mini-white and hs-neo genes and selecting flies using G418 resistance, many of the lines obtained showed no detectable eye color and thus would not have been obtained if eye color had been used as the selectable marker. The 2.2-kb Ubx bxd PRE fragment they used was subsequently dissected into subfragments, six of which, when multimerized, mediated mini-white suppression (Horard, Tatout, Poux, & Pirrotta, 2000) in slightly different ways. Some subfragments gave transgenic flies with variegated eyes; one gave flies with patterned eyes (eye color in only part of the eye, but reproducibly in the same part of each eye). Of three multimerized subfragments tested for PREs in a maintenance assay in embryos, only one acted as a PRE. The data suggest that the 2.2-kb Ubx bxd PRE fragment contains multiple elements that function together to regulate Ubx expression throughout development.
Use of the FRT-Flp and LoxP-Cre recombinase systems to remove PREs from pCaSpeR-containing transgenes has shown that all PREs tested repress eye color to some extent in the heterozygous state. Figure 3.2 shows an example of this. In P[PREmw], the 181-bp en PRE is flanked by LoxP sites. At this chromosomal insertion site, P[PREmw] flies have a light orange eye color. Removal of the PRE via Cre recombinase yields flies with a slightly darker eye color. The effect of the 181-bp en PRE is much more dramatic in the homozygous flies; with the PRE, the eye color is white; without the PRE, the eye color is red (Noyes et al., 2011).
Silencing of mini-white expression either in the heterozygous state or via PSS is the most common assay used to test the function of a presumptive PRE (Table 3.1). Is this a legitimate assay for PRE activity? There are a few reported cases where PREs that work in maintenance assays in embryos or larvae do not repress mini-white expression and vice versa (reviewed in Kassis, 2002), but, in general, there is an excellent correlation between PRE activity and PSS. Some minimal PREs may be stage or tissue specific and may not work in both assays. Results from our lab show a perfect correlation between fragments of en/inv DNA that mediate PcG repression in embryos and PSS (Americo et al., 2002; Brown, Grau, DeVido, & Kassis, 2005; Cunningham et al., 2010; DeVido et al., 2008). Thus, PSS is a good indication of PRE activity.
2.3. Effect of flanking regulatory DNA on PRE activity
In vivo, PREs can be located tens of kilobases away from the promoter they regulate. How are they able to act over such large distances? In vitro, PRC1 inhibits chromatin remodeling and transcription (reviewed in Simon & Kingston, 2009). Interestingly, PRC1 bound to a polynucleosome template in vitro can recruit a second template and inhibit chromatin remodeling and transcription of that template as well (Lavigne, Francis, King, & Kingston, 2004). Further, the Drosophila PRC1 subunit Psc can bind to and compact chromatin, an activity conserved in the M33 Polycomb homolog in mice (Grau et al., 2011).
The PSS assay suggests that PREs act together to facilitate gene silencing. The physical interaction between the PREs and promoters of the BX-C has been well documented by chromatin chromosome capture (3C) and fluorescent in situ hybridization (Lanzuolo, Roure, Dekker, Bantignes, & Orlando, 2007). Genome-wide chromatin conformation studies suggest that PcG proteins are a major organizer of the three-dimensional structure of the genome (recently reviewed in Delest, Sexton, & Cavalli, 2012; Pirrotta & Li, 2012). PcG proteins are present in “Polycomb bodies” where concentrations of PcG proteins appear as “dots” in the cell. Interestingly, PcG target genes colocalize with PcG bodies only in those cells where they are repressed (see Delest et al., 2012; Pirrotta & Li, 2012).
In a PRE-transgene assay, a PRE is removed from its normal chromatin context, often combined with regulatory DNA from a gene it normally does not regulate, and then inserted into a region of the genome where it normally does not reside. Given that PREs are designed to interact with other regulatory DNA, it is not surprising that the activity of a PRE in a transgene is dependent on where it is inserted in the genome. For example, PSS mediated by en PREs occurs in only about 50% of transgenic lines, and the extent of silencing also depends on the chromosomal insertion site (Americo et al., 2002; Noyes et al., 2011). Various models can be proposed to explain this. First, since PREs often work together, perhaps PSS only occurs in regions of the genome where there is another PRE nearby; there is some evidence to suggest that PRE-containing transgenes have a tendency to insert in the genome near PcG-regulated genes (reviewed in Kassis, 2002 and see below). Second, if the transgene inserts downstream of a promoter, it might be transcribed; transcription through a PRE has been shown to inactivate it (Schmitt, Prestel, & Paro, 2005). Finally, genomic enhancers flanking the PRE-transgene insertion site might activate mini-white expression via a mechanism not subject to PcG repression. A mammalian enhancer that can evict PcG proteins has recently been described (Vernimmen et al., 2011).
Recent work from our lab shows the effect of chromatin context (or flanking DNA) on PRE function. We carried out a genetic screen for dominant suppressors of PSS of an en PRE-mini-white transgene (P[enPRE-mw]), hoping to recover mutations in PcG genes or other genes necessary for the PSS activity of the en PRE (Noyes et al., 2011). We screened for suppressors of mini-white silencing of a P[enPRE-mw] transgene inserted on chromosome 2R between the genes CG30456 and GstS1. We reasoned that if the mutations we recovered affected the activity of the en PRE directly, then they should suppress mini-white silencing by P[enPRE-mw] regardless of where it was inserted in the genome. Mutations that suppressed the mini-white silencing of a specific P[enPRE-mw] insertion must act on flanking regulatory DNA. Interestingly, most of the mutations we recovered suppressed the mini-white silencing at only one or a subset of P[enPRE-mw] insertion sites, again showing the profound effect of genomic location on PRE activity.
We have characterized two of the dominant suppressors of PSS by P[enPRE-mw] (Cunningham et al., 2012; Noyes et al., 2011). One was a dominant mutation in the gene encoding the transcriptional activator Without children (WocD) (Noyes et al., 2011). WocD is caused by a single amino acid change that we hypothesize increases the ability of Woc to activate transcription. WocD suppresses P[enPRE-mw]-mediated PSS at only two chromosomal insertion sites (out of 16 tested). This suggests that WocD does not act at the PRE directly but rather acts on flanking genomic DNA. WocD may change the local chromatin conformation, perhaps by increasing the transcription of genes flanking the P[enPRE-mw] insertion site, inactivating PRE function at that chromosomal location.
The other PSS suppressor mutation we characterized was a dominant mutation in the cohesin-associated regulatory protein wings apart-like (wapl) (Cunningham et al., 2012), waplAG. Interestingly, waplAG mutants have a classic PcG phenotype, sex comb teeth on the second and third legs (Kennison, 1995), suggesting that Wapl is also a regulator of the homeotic gene Scr. Wapl acts in a protein complex with Pds5 to remove cohesin from chromosomes (Shintomi & Hirano, 2009). Our results suggest that increasing cohesin-binding stability antagonizes PcG silencing (Cunningham et al., 2012). The relationship between cohesin binding and PcG activity has been recently reviewed (Dorsett, 2011).
2.4. PREs can facilitate transcriptional activation or repression
The patterned expression of the HOX genes in Drosophila is initially set up by the gap and pair-rule transcription factors that directly bind HOX-regulatory DNA. Soon after, these direct regulators disappear, and the PcG and trithorax-group (trxG) proteins take over to maintain the repressed or active state, respectively. It is well established that PcG proteins act through PREs, and there is evidence that trxG proteins act through the same or closely linked DNA elements (trxG response elements, TREs, reviewed in Ringrose & Paro, 2004). Interestingly, in genome-wide studies, the C-terminus of Trithorax localizes to the same locations as PcG proteins, the presumptive PREs (Schuettengruber et al., 2009; Schwartz et al., 2010).
The first indication that PREs could maintain either the repressed or active chromatin state was for a DNA fragment from the BX-C, Fab-7 (Cavalli & Paro, 1998). Remarkably, Fab-7 was able to maintain either transcriptional repression or activation of two reporter genes throughout both mitosis and meiosis depending on the transcriptional state of the transgenes in the embryo. The authors designed a vector with a GAL4-inducible lacZ gene followed by the mini-white gene. Inclusion of the Fab-7 fragment (that contains a PRE) repressed the expression of both lacZ and mini-white. After transient activation of the lacZ gene by GAL4 in embryos, both lacZ and mini-white expression were maintained throughout development. This fragment of DNA was subsequently called a “cellular memory module” or CMM. Three PRE-containing fragments from the BX-C have been shown to act as CMMs in this assay: Fab7, Mcp, bxd, and also a PRE from the hedgehog (hh) gene (Maurange & Paro, 2002; Rank et al., 2002). The en PRE did not act as a CMM in this assay (DeVido et al., 2008). Further experiments indicated that transcription through a PRE could turn it from a PRE to a TRE (Schmitt et al., 2005). One model states that it is the transcriptional state of PREs that determines whether they will maintain the active or repressed chromatin state. This may be true for some genes, but it is unlikely that this simple model will explain how the activity state is set for all PREs.
The ability of two closely linked engrailed PREs to mediate either transcriptional activation or repression was revealed in an enhancer-trap assay. An en-lacZ transgene containing 2.4 kb of upstream en upstream sequences (that includes the two PREs) driving lacZ expression had no intrinsic patterning activity on its own, but lacZ was expressed in patterns driven by genomic enhancers near the transgene insertion site. By flanking the en PREs with loxP or FRT sites, the expression of the en-lacZ transgene with and without the PREs in the same location could be examined. Remarkably, each PRE could act with flanking regulatory DNA as either a positive or negative regulatory element depending on the genomic context and the tissue examined. These data suggest that PREs, or closely associated sequences, mediate long-range interactions with enhancers or silencers around the PRE-transgene insertion site.
An excellent example of a fragment of DNA that acts as both a PRE and a presumptive TRE is provided by studies on a PRE from the eve locus (Fujioka et al., 2008). A 300-bp eve DNA fragment was shown to (1) mediate PSS of mini-white; (2) act as a PRE in an Ubx-reporter construct in embryos; and (3) maintain the continued expression of an eve-lacZ reporter construct in cells of the larval CNS; that is, without the PRE, the eve-lacZ gene was not expressed in the larval CNS. Remarkably, all three activities were dependent on a single binding site for the PcG-DNA binding protein Pho (Fujioka et al., 2008). Pho has been shown to interact with both PcG proteins and the trxG proteins Brahma, Moira, and Osa in Drosophila embryo extracts (Mohd-Sarip, Venturini, Chalkley, & Verrijzer, 2002). It would be interesting to know whether PREs from other genes could maintain the expression of the eve-lacZ reporter in the larval CNS, or whether this activity is specific for the eve PRE.
2.5. DNA binding proteins and sequence motifs associated with PREs
PREs are made up of multiple binding sites for many different factors. Despite the overwhelming amount of literature investigating the nature of Drosophila PREs, we still do not have a clear understanding of what precise combination of DNA binding sites/proteins are required for PRE activity. Studies of a number of different Drosophila PREs from en (Americo et al., 2002; DeVido et al., 2008; Kassis, 1994), inv (Cunningham et al., 2010), eve (Fujioka et al., 2008), Ph (Bloyer, Cavalli, Brock, & Dura, 2003), hh (Chanas & Machat, 2005; Maurange & Paro, 2002), vestigal (vg) (Okulski et al., 2011), and PREs of the BX-C: Fab7, Mcp, bxd (also called PRED), and iab-2 (Busturia et al., 2001, Hagstrom, Müller, & Schedl, 1997, Mishra et al., 2001, Strutt, Cavalli, & Paro, 1997) have identified several different factors/binding sites as being important for PRE function. These include Pho/Phol, Sp1/KLF factors, GAGA/Psq, Zeste/Fs(1)h, Dsp1, Grainyhead (Grh), and other as yet unidentified factors (reviewed in Müller & Kassis, 2006; Ringrose & Paro, 2007) that we will discuss below.
Genome-wide ChIP studies are yielding specific information about the distribution of these proteins at PREs in different cell types and at different developmental stages (Kwong et al., 2008; Négre et al., 2006; Schuettengruber et al., 2009; Schwartz et al., 2006, 2010; Tolhuis et al., 2006). These studies show that the identified PRE-associated DNA binding proteins are not present at all PREs and bind to many sites that are not within PREs. The phenotypes of mutants of some of these DNA binding factors argue for a role of some of these proteins in trxG gene activation rather than PcG repression. This may reflect the close association of PREs and TREs.
In addition, genome-wide studies have shown that PcG repression is not the all or nothing system that was originally thought and is in fact much more dynamic. A study of PcG distribution in different tissue culture cells reflecting different activity states of some genes shows that PcG proteins can be bound to PREs in both the on and the off transcriptional state raising interesting mechanistic questions (Schwartz et al., 2010). Interestingly, Papp and Müller (2006) found binding of PcG proteins in both the on and off transcriptional states at Ubx PREs in imaginal disks. Comparison of embryo versus imaginal tissue PcG protein binding suggests the presence of stage-specific PREs (Kwong et al., 2008; Schuettengruber et al., 2009). The general consensus seems to be that no one factor alone is responsible for PRE activity and that PcG recruitment to PREs involves the cooperative contribution of a number of different DNA binding factors. The exact combination of factors that constitute a PRE may be different at different genes.
2.5.1 Pho and Phol
The only known bonafide member of the PcG genes that encodes a DNA binding protein is pho. pho zygotic mutants die as pharate adults with sex combs on the second and third legs (the “classic” PcG phenotype), while embryos derived from germ line clones that lack both maternal and zygotic pho die at the embryonic stage with segmentation defects (Breen & Duncan, 1986). The Drosophila genome encodes a protein highly related to Pho in the DNA binding domain, Pho-like (Phol) (Brown, Fritsch, Müller, & Kassis, 2003). phol mutants are homozygous viable with no homeotic phenotypes. Genetic experiments suggest that Pho and Phol act together to repress homeotic gene expression since pho; phol double mutants have more derepression of the homeotic gene Ubx in wing disks than do pho mutants. Phol also works independently of Pho; eggs derived from Phol females are fertilized but do not develop, a phenotype not observed for eggs that lack Pho (Breen & Duncan, 1986).
Pho and Phol are the Drosophila homologs of the mammalian proteins YY1 and YY2, respectively (Brown et al., 1998, 2003 Drews, Klar, Dame, & Bräuer, 2009; Nguyen, Zhang, Olashaw, & Seto, 2004). YY1 is a very dynamic protein that can be involved with repression, activation, and transcriptional initiation in a context-dependent way (Gordon, Akopyan, Garban, & Bonavida, 2006; Wang, Chen, & Yang, 2006). Pho has 96% amino acid identity with YY1 over four zinc fingers that are used for DNA binding and protein–protein interactions. There is also conservation of another region of the protein (18/22 amino acid identity between YY1 and Pho), a small spacer region implicated in protein–protein interactions. Phol has 80% amino acid identity to Pho over the four zinc fingers, and 10/22 and 11/22 amino acid identity to YY1 and Pho, respectively, in the spacer region. YY1 has been postulated to be involved in PcG repression in vertebrates (discussed in more detail below), and mammalian YY1 can partially rescue a pho zygotic mutant (Atchison, Ghias, Wilkinson, Bonini, & Atchinson, 2003).
Although Pho and Phol both bind to Ubx PRED and Phol can silence Ubx transcription in the absence of Pho (Wang et al., 2004), genome-wide studies show that, in wild-type embryos, the distribution of Pho and Phol is different (Schuettengruber et al., 2009). Consistent with the PcG phenotype of zygotic pho mutants, Pho binding is highly correlated with the binding of Ph and Pc. Of genomic positions that share Ph and Pc binding, 96% correspond to Pho peaks but only 21% correspond to Phol peaks (Schuettengruber et al., 2009). The data suggest that Phol preferentially binds to a slightly different sequence than Pho and may be involved in transcriptional activation rather than repression (Schuettengruber et al., 2009). The 96% overlap between regions that bind Pc/Ph and Pho suggest that Pho could be absolutely required for PRC1 recruitment. However, in another study, also in embryos, there was only a 50% overlap of Pc binding sites with Pho (Kwong et al., 2008). It should also be pointed out that there are many Pho sites that do not overlap with either Pc or Ph binding.
What does Pho do at the PRE? In embryos, Pho was found in two protein complexes, Pho-dINO80, a nucleosome-remodeling complex, and PhoRC where it is in a complex with dSfmbt, a protein that has methyl-lysine-binding activity (Grimm et al., 2009; Klymenko et al., 2006). Both Pho and Phol form stable complexes with dSfmbt in Sf9 cells. In contrast, Phol is not found associated with dINO80. dSfmbt is also bound to PREs, and mutations in the gene encoding dSfmbt cause homeotic derepression (Klymenko et al., 2006; Oktaba et al., 2008) showing it is also important for PcG repression. Therefore, one function of Pho and Phol may be to bring dSfmbt to the PRE. Biochemically, Pho has been reported to interact with components of both PRC1 and PRC2 in coimmunoprecipitation and GST pulldown assays (Mohd-Sarip, Cleard, Mishra, Karch, & Verrijzer, 2005; Wang et al., 2004). Further, both Pho and dSfmbt copurified with a Pc protein labeled in vivo by biotinylation (Strübbe et al., 2011). Pho and Phol are thought to play a key role in the binding of both PRC1 and PRC2 to chromatin (Wang et al., 2004). Interestingly, the PcG protein Scm can bind to an Ubx PRE in the absence of Pho/Phol (Wang et al., 2010).
There is no doubt that Pho plays a key role at PREs. To our knowledge, all PREs tested in either the mini-white silencing or the embryonic or larval maintenance assays bind Pho. Mutation of Pho binding sites within PREs abrogated the function of all PREs where it has been tested (reviewed in Kassis, 2002; Fujioka et al., 2008). There is some data that other PRE-binding proteins may be necessary to facilitate the binding of Pho to the PRE. For example, Grh, GAF, and Dsp1 have all been postulated to cooperatively assist Pho binding to chromatinized substrates (Blastyák, Mishra, Karch, & Gyurkovics, 2006; Dejardin et al., 2005; Mahmoudi, Zuijderduijn, Mohd-Sarip, & Verrijzer, 2003). Pho is generally thought to have a repressive role but, as discussed above, Fujioka et al. (2008) have shown that Pho can act as an activator in a specific subset of cells in the Drosophila nervous system. It remains to be seen whether this activating role of Pho is specific for this case or can be generalized to many PRE/TREs.
2.5.2 Sp/KLF proteins
An analysis of a PRE from the en gene identified a Sp1/KLF binding site as being required for PSS and PRE activity of a 181-bp PRE (Americo et al., 2002; Brown et al., 2005). There are nine members of the Sp1/KLF family of proteins in Drosophila and in vitro transcription/translation of the DNA binding domains showed that 8/9 of the proteins were capable of binding to the site in the en PRE (Brown et al., 2005). As seen in the extensive family of mammalian Sp1/KLF proteins (reviewed in Kaczynski, Cook, & Urrutia, 2003), there were differences in the binding site preferences of the Drosophila family members. Further analysis identified a member of this group, Spps (Sp1 factor for PSS) as being involved in PRE activity of the en PRE (Brown & Kassis, 2010). Flies mutant for Spps die as late pharate adults and Spps enhances the pho mutant phenotype. This indicates that Pho and Spps either work together or in parallel parts of the same pathway to silence PcG target genes. PSS of mini-white in constructs carrying either an en PRE or a bxd PRE is lost in Spps mutants, showing that Spps is required for PSS. ChIP-qPCR showed that Spps binds to both the en and bxd PREs in larvae. ChIP experiments showing the genome-wide distribution of Spps have not yet been published. However, a general role for Spps in PRE activity is predicted given that Spps colocalizes almost perfectly with Psc in polytene chromosomes (Brown & Kassis, 2010). Mammalian YY1 and Sp1 have been shown to be able to interact (Lee, Calvin, & Shi, 1993; Seto, Lewis, & Shenk, 1993) which raises the possibility for interaction here. The involvement of other members of the Drosophila Sp1/KLF factors in PcG repression has yet to be determined. The mammalian Sp1/KLF proteins are a dynamic set of proteins involved in many aspects of gene expression, differentiation, and cancer (Kaczynski et al., 2003; McConnell & Yang, 2010).
2.5.3 GAGAG sites
The sequence GAGAG is required for the activity of many PREs (reviewed in Kassis, 2002; Fujioka et al., 2008). GAGAG elements are binding sites for two proteins, GAGA factor (GAF) and Pipsqueak (Psq) (Lehmann, Siegmund, Lintermann, & Korge, 1998). GAF recruits a chromatin-remodeling complex and the role of GAF in PRE activity may be to remove nucleosomes at the PRE, allowing the binding of other transcription factors (Mahmoudi et al., 2003). Genome-wide studies show that GAF is present at about 50% of sites bound by both Pc and Ph (Négre et al., 2006; Schuettengruber et al., 2009). Psq also binds to the GAGAG sequence although it is reported to have a preference for longer GAGA sequences in keeping with the fact that it has a different DNA binding domain (HLH; Siegmund & Lehmann, 2002) from GAF (a single zinc finger; Lehmann et al., 1998). Psq has been isolated from S2 cells in a protein complex (CRASCH; Huang & Chang, 2004; Huang, Chang, Yang, Pan, & King, 2002) with PcG proteins. The genome-wide distribution of Psq binding is not known but GAF and Psq proteins colocalize on polytene chromosomes. GAF and Psq both have the same type of protein–protein interaction domains, BTB domain, and can interact with each other (Schwendemann & Lehman, 2002) and with some other members of this class of proteins, for example, Tramtrack (Pagans, Ortiz-Lombardia, Esinas, Bernues, & Azorin, 2002). GAF is not a stable component of any of the PcG complexes isolated to date. Recently, a vertebrate homolog of GAF was identified that also binds to GAGAG sequences and shows extensive homology with GAF (Matharu, Hussain, Sankaranarayanan, & Mishra, 2010). It will be interesting to know whether this protein plays a role in PcG silencing in mammals.
GAF is encoded by the gene trithorax-like (trl; Farkas et al., 1994). As suggested by the name, trl mutants have a trxG phenotype. GAF is required for many aspects of gene expression and also for nuclear division in Drosophila embryos (Bhat et al., 1996). GAF is bound to GAGA satellite sequences. Because of its multifunctional role in controlling gene expression and cell division, it is impossible to directly test the role of GAF at PREs in embryos. Psq acts during oogenesis in the establishment of the anterior/posterior axis of the oocyte (Siegel, Jongens, Jan, & Jan, 1993). A role for Psq in PcG silencing is suggested by a genetic interaction between a psq and Pc. A mutation in psq strongly enhances the derepression of Ubx in wing and leg disks of larvae heterozygous for a Pc allele, strongly suggesting Psq plays a role in PcG silencing (Huang et al., 2002). The exact role of GAF versus Psq at the PREs needs further exploration.
2.5.4 GTGT sequence
This sequence has shown up in a number of studies to be enriched in PRE sequences (Ringrose et al., 2003; Schuettengruber et al., 2009). Deletion of the GTGT sequences in the vg PRE led to reduction in silencing activity of this fragment suggesting that this sequence may play a role in PcG repression (Okulski et al., 2011). Interestingly, in mammals, the Sp1/KLF factor Sp2 was shown to bind to a GTGT box (GGTGTGGGG) implicated in regulating T-cell receptor gene expression (Kaczynski et al., 2003; Kingsley & Winoto, 1992; Suske, Bruford, & Philipsen, 2005). It would be interesting to know whether the GTGT sequences deleted in the vg PRE bind to Spps or other Drosophila Sp1/KLF family members.
2.5.5 Dsp1
Dsp1 was identified as a PRE DNA binding protein important for the recruitment of PcG complexes to polytene chromosomes (Dejardin et al., 2005). These authors reported that Dsp1 binds to the consensus sequence GAAAA and that mutation of GAAAA within the en and Fab7 PREs not only abrogated PSS of mini-white but also changed the PREs into constitute TREs. These data suggest that Dsp1 is required for PRE repression activity.
Genome-wide studies with 4–12 h embryo extracts have shown that Dsp1 binds to a subset of PREs (about 50% of the Ph/Pc sites identified (225/439); Schuettengruber et al., 2009). Curiously, the GAAAA consensus sequence is not enriched within these PREs. Clearly, the DNA binding site of Dsp1 requires further definition; however, Dsp1 might recognize DNA structure instead of sequence. Dsp1 is a member of the class of HMG proteins that have two HMG domains; these proteins bind to the minor groove of DNA without sequence specificity, instead recognizing DNA structural features. This type of HMG protein has been reported to be able to introduce a significant distortion or bend into the DNA (for reviews, see Agresti & Bianchi, 2003; Stros, 2010). Dsp1 could facilitate PcG protein binding or long-range interactions by changing the structure of the DNA.
Mutants of Dsp1 exhibit features more typical of a trxG gene than a PcG gene (Decoville, Giacomello, Leng, & Locker, 2001; Rappailles, Decoville, & Locker, 2005). Dsp1 has been shown to coimmunoprecipitate with the chromo domain containing protein, Corto (Salvaing et al., 2006). These authors suggest that Dsp1 binds to the Scr PRE only when Scr is being transcribed, thus Dsp1 plays a critical role in TRE activity. Clearly, more work needs to be done to understand the role of Dsp1 in PcG and trxG activity.
2.5.6 Grh
Grh was found to bind to the iab-7 PRE and to cooperatively interact with Pho both in vitro and genetically (Blastyák et al., 2006). The binding site for Grh identified in the BX-C iab-7 PRE was reported to be TGTTTTTT. However, other groups report a Grh consensus sequence as WCHGGTT (where W is A or T and H is not G) (Almeida & Bray, 2005; Venkatesan, McManus, Mello, Smith, & Hansen, 2003). To date, a genome-wide study of Grh binding in relationship to PcG protein binding has not been reported. A presumptive Grh binding site is present in an eve PRE (Fujioka et al., 2008) but not in the 181-bp en PRE (J. Lesley Brown and Judith A. Kassis, unpublished observation), suggesting it may be important for only a subset of PREs.
Other evidence that Grh may be involved in PcG repression comes from work on the mammalian Grh-family member CP2. CP2 was shown to interact with a mammalian Ring protein, DinG. The CP2–DinG interaction was necessary for transcriptional repression (Tuckfield et al., 2002). The authors also showed that Drosophila Grh could interact with dRing in Gst-pull down experiments in vitro.
Drosophila Grh regulates the expression of many genes and can act as either a transcriptional repressor or activator dependent on the context (see references cited in McQuilton, Pierre, Thurmon, & The FlyBase Consortium, 2012). Of particular interest for this review, Grh, Zeste, and GAGA binding sites activate transcription of an Ubx promoter reporter construct through intermingled clusters of binding sites (Hur, Laney, Jeon, Ali, & Biggin, 2002 and references therein). Interestingly, Hur et al. (2002) found that Zeste and GAGA promoter-proximal binding sites were able to maintain PcG-mediated repression of a 22-kb Ubx-lacZ reporter construct in embryos, whereas Grh binding sites did not have this activity. The role of Grh in PcG repression requires further study.
2.5.7 Zeste binding sites
The role of Zeste in PcG repression is also unclear. Zeste has been reported to be an activator or repressor of transcription dependent on the context and has been reported to be a stoichiometric component of PRC1 (Saurin et al., 2001). Schuettengruber et al. (2009) found only a 25% overlap of Ph with Zeste in genome-wide binding data. Oktaba et al. (2008) report very little overlap between Zeste and Pho in genome-wide binding data. Zeste mutants are homozygous viable and fertile and do not show PcG phenotypes (Goldberg, Colvin, & Mellin, 1989). female sterile (1) homeotic (fs(1)h) is a member of the trxG genes. fs(1)h encodes a double bromodomain-containing protein that binds to Zeste sites within the Ubx promoter and activates transcription of the Ubx gene (Chang, King, Lin, Kennison, & Huang, 2007). It is not known if Fs(1)h binds to Zeste sites in PREs.
2.6. Are all PREs alike?
Algorithms based on clustered consensus binding sites for PRE DNA binding proteins have been marginally successful in identifying PREs, giving both false positive and false negative results (Fiedler & Rehmsmeier, 2006; Ringrose et al., 2003; Zeng, Kirk, Gou, Wang, & Ma, 2012). In one study, of 167 potential PREs that Ringrose et al. (2003) identified, only 16% of them were PcG binding sites in PcG ChIP genome-wide studies of Drosophila embryos (Schuettengruber et al., 2009). A weakness of this approach is the unreliability of the consensus binding sequences that are used in the search. A consensus sequence is our best guess but it does not take into account cooperativity between weak sites, the influence of nearby binding proteins, or the fact that a protein may be able to bind to sites that deviate from the consensus. For example, in a study on PREs at the invected locus (Cunningham et al., 2010), a fragment that had both PSS and PRE activity did not contain any matches to the core consensus binding sequence for Pho (GCCAT); thus this fragment was not predicted to be a PRE by the jPREdictor algorithm; however, this fragment clearly bound Pc and Ph in ChIP studies, suggesting it was a PRE. There were several CCAT sequences in this PRE, and 5/6 bound Pho in vitro. In addition, some PREs require Pho binding sites that contain sequences beyond the minimal Pho consensus. Two extended Pho binding sites were required to interact and recruit Pc complexes to chromatin in the iab-7 PRE (Mohd-Sarip et al., 2002; reported to be a PcG target Mohd-Sarip et al., 2005, 2006). In studies examining the genome-wide distribution of Pho, Oktaba et al. (2008) noted that an extended Pho consensus sequence was enriched in regions that bound both Pho and dSfmbt. They also examined the number of Pho consensus sites within a Pho-bound region and found that, while often there was more than one Pho site, the spacing of these sites was variable, and many fragments had only one Pho binding site (Oktaba et al., 2008). Further, the number and order of Pho and other motifs varies greatly between orthologous PREs in different Drosophila species (Hauenschild, Ringrose, Altmutter, Paro, & Rehmsmeier, 2008); clearly there is great flexibility in PRE-binding site configuration.
PRE prediction algorithms make the assumption that all PREs have the same requirements for activity. This seems unlikely given the complexity of the different PcG target loci and the indication that different PREs function at different times in development. There is also a difference in the arrangement of PREs within a gene that may reflect underlying differences in the way the PRE works. A subset of PREs are located immediately upstream of the promoter, while others are many kilobases away. The available data suggest that some DNA binding proteins are present at most PREs (Pho), for example; however, it is likely that locus specific factors influence the activity of at least a subset of PREs.
A limited number of PREs have been shown to work with regulatory DNA from other genes (discussed above). For example, PREs from the genes en, eve, and Abd-B maintain silenced expression of an Ubx-lacZ reporter construct in the correct pattern in embryos. Further, the iab-7 and iab-8 core PREs completely replaced the activity of a core bxd PRE in the endogenous Ubx gene (Kozma, Bender, & Sipos, 2008). In general, these studies used minimal PRE fragments of only a few hundred base pairs or less that contained binding sites for Pho and GAF. Larger PRE fragments are more likely to contain other regulatory DNA and may not work in these assays. For example, while a 181-bp minimal en PRE maintained an Ubx-like expression pattern of an Ubx-lacZ transgene in embryos, a larger fragment (2.6 kb) that contained the 181-bp PRE as well as an additional PRE and flanking DNA totally silenced the Ubx-lacZ reporter gene in a Pc-dependent way (Americo et al., 2002). Recently, Okulski et al. (2011) used the phi-C31 system to integrate two different PRE-mini-white transgenes (vg and Fab7) at four different chromosomal insertion sites and quantitated the amount of mini-white repression by the PREs both in flies heterozygous and homozygous for the transgene. They noted that the vg and Fab7 PREs gave quantitatively different amounts of mini-white silencing and behaved differently at the different insertions sites. Although this was a very careful study, one problem with it was that the PRE fragments they used were 1.6 kb and were likely to have other regulatory DNA for their respective loci. In this regard, the 181-bp en PRE also has promoter-tethering activity (Kwon et al., 2009). It is important to keep in mind that PREs act in the context of other regulatory DNA and that other regulatory elements may be closely linked or even intertwined.
Strübbe et al. (2011) recently purified Pc under physiological conditions designed to maintain the weakest associations and then analyzed copurifying products by mass spectrometry. This identified all of the PRC1 core components Pc, Psc, Sce/dRing, Ph and Su(z)2 and PhoRC (dSfmbt and Pho) but components of PRC2 were not found. Fs(1)h and Grh also copurified. In addition to known PcG complexes, the data also showed unexpected interactions with the cohesin complex and with proteins related to the Moz/Morf histone acetyltransferase complex. This type of approach, combined with a rigorous analysis of individual PREs will be necessary to understand how PcG proteins are recruited.
3. CHARACTERISTICS OF PcG TARGET GENES IN DROSOPHILA
3.1. Many PcG targets have multiple PREs
PcG target genes tend to have a large amount of regulatory DNA reflecting their complex and often dynamic expression patterns throughout development (Kharchenko et al., 2011; Négre et al., 2006). PcG target genes often occur in gene clusters. The best known of these are the HOX gene clusters including the BX-C that contains the homeotic genes Ubx, Abd-B, and Abd-A. The arrangement and function of PREs within the BX-C have been extensively studied (reviewed in Maeda & Karch, 2006, 2011); in fact, much of what we know about PREs comes from studies on BX-C PREs. Here, we describe the PREs of three PcG target loci, eve, an example of a single gene PcG target and two PcG targets with two coregulated genes, en/inv and Psc/Su(z)2.
3.1.1 eve
The eve gene is a pair-rule gene required for segmentation and the development of specific lineages in the mesoderm and nervous system. A single eve 1.5-kb transcription unit is flanked by about 9 kb of regulatory DNA downstream and 6-kb upstream, containing many eve enhancers (Fujioka et al., 1999). PcG genes regulate eve expression in the nervous system (Fujioka et al., 2008; Smouse, Goodman, Mahowald, & Perrimon, 1988). In S2 cells, a 20-kb “eve domain” is covered with the characteristic H3K27me3 PcG mark generated by PRC2 (Kharchenko et al., 2011). There are two fragments of eve DNA that mediate PSS of mini-white, a 300-bp fragment located 8.9-kb downstream of the eve transcription unit (eve PRE300), and another 441-bp fragment that includes the eve promoter. eve PRE300 acts as a PRE in a Ubx-lacZ reporter gene and also as a positive element to maintain expression of an eve-reporter gene in specific cells of the larval nervous system (discussed above). The eve proximal-promoter PRE has not been tested in these assays. Both eve PREs contain Pho, GAF, and Zeste binding sites (Fujioka et al., 2008). Interestingly, eve PRE300 is not required for correct eve expression in an eve rescue transgene (Fujioka et al., 1999). This suggests that the promoter-proximal eve PRE is sufficient for maintaining eve expression.
3.1.2 en/inv
The en and inv genes are coexpressed genes that share regulatory DNA (Gustavson et al., 1996) and form a 115-kb H3K27me3 domain in S2 cells, embryos, and larvae (Kharchenko et al., 2011). There are four well-characterized PREs in the inv/en locus; two closely linked ones located between −400 bp and −2 kb upstream of the en transcription start site (DeVido et al., 2008) and two upstream of inv, one coincident with the major inv transcription start site and another about 6-kb upstream of that (Cunningham et al., 2010). In S2 cells and adults, PcG proteins are bound to a single 2-kb peak just upstream of the en transcription start site (encompassing the two PREs) and to the two inv PREs, however, in embryos and larvae, PcG proteins bind to additional regions upstream of the en gene (Négre et al., 2006; Oktaba et al., 2008). It is not known whether these other PcG binding regions represent additional stage-specific PREs or are the result of the three-dimensional structure of the locus causing cross-linking to fragments because of chromatin compaction. PRE activity assays will need to be done on these other fragments to distinguish between these two possibilities. Interestingly, deletion of a 1.5-kb fragment (enΔ1.5, that includes the two PREs upstream of the en transcription start site) from the endogenous en/inv locus does not alter the pattern of en or inv expression (Cheng, Kwon, Arai, Mucci, & Kassis, 2012). These data suggest that PREs might act redundantly in the en/inv domain; however, it is not know whether deletion of the PREs in enΔ1.5 causes quantitative difference in en/inv expression.
3.1.3 Psc/Su(z)2
The PcG group genes Psc and Su(z)2 exist in an H3K27me3 domain that extends over about 91 kb in S2 cells; insulator elements may limit the spread of H3K27me3 into flanking genes (Park et al., 2012). Psc and Su(z)2 are ubiquitously expressed genes, so, obviously, PcG proteins do not silence gene expression in this case. It is thought that PcG proteins regulate the level of Psc and Su(z)2 expression. ChIP experiments in S2 cells show four large discrete Psc binding sites as well as additional smaller peaks (Schwartz et al., 2006). Park et al. (2012) showed that two of these peaks had PRE activity in both a mini-white silencing assay and in a larval Ubx-reporter gene functional assay. Another Psc binding fragment behaved as a downregulatory module rather than a typical PRE. Deletion of one of the Psc/Su(z)2 PREs from the endogenous locus caused a twofold increase in the level of Psc and Su(z)2 transcription suggesting that this element downregulates the level of transcription. Why do PREs from the Psc/Su(z)2 gene act as silencers in transgenes, but not silence expression of the endogenous gene? One hypothesis is that the type of enhancers associated with this gene are not subject to PcG silencing. More work needs to be done on this interesting locus.
3.2. Homing and PcG target genes
P-element based vectors have been used to make transgenic Drosophila for about 30 years (Rubin & Spradling, 1982). Aside from some hotspots and a preference to insert near the 5′ end of transcription units, P-based vectors insert in the genome in a relatively nonselective manner and as such, have been used as a tool to generate thousands of transposon-induced mutations in Drosophila (Bellen et al., 2011). In 1990, Hama, Ali, and Kornberg reported that P-based constructs that contain en regulatory DNA (P[en]) insert near or into the en/inv domain at a very high frequency. They called this phenomenon “homing” (Hama, Ali, & Kornberg, 1990). Further studies on P[en] homing by en regulatory DNA showed that a 2-kb fragment, extending from −2.4- to −0.4-kb upstream of the en transcription start site was sufficient for homing, and that the en PREs contributed to the homing activity of this fragment (Cheng et al., 2012). Further, P[en] inserted near PcG targets at an increased frequency compared with a P-based vector used to generate insertions for the Drosophila gene disruption project (Cheng et al., 2012). These data suggest that either the chromatin structure or PcG proteins themselves bring P[en] to the region of PcG-regulated loci. This is consistent with the view that PcG targets colocalize in the nucleus in PcG bodies (Delest et al., 2012; Pirrotta & Li, 2012) and suggest that these bodies also occur in germ cells where homing occurs. Transgenes containing polyhomeotic PREs and BX-C PREs have also been reported to insert at a high frequency in or near PcG target genes (reviewed in Kassis, 2002).
Fragments of DNA from the BX-C and eve PcG targets have been shown to mediate homing (Bender & Hudson, 2000; Fujioka, Wu, & Jaynes, 2009). Like P[en], insertions mediated by the BX-C and eve homing fragments occurred over a large region, causing insertions throughout and near the parent locus. For these two cases, the fragments that mediate homing are most likely insulators, not PREs (Bender & Hudson, 2000; Fujioka et al., 2009). Anecdotally, 1/5 insertions of a P-based vector that contained one of the Psc/Su(z)2 PREs was inserted near the promoter of the Su(z)12 gene, strongly suggesting that this fragment mediates homing. Note that one reported case of homing, that of the linotte gene (now called drl; Taillenbourg & Dura, 1999), is not reported to be a PcG target gene in embryos, larvae, or S2 cells; however, it is possible it could be a PcG-regulated gene in germ cells where P-element homing occurs.
4. PREs IN VERTEBRATES
PcG proteins in vertebrates are much more diverse and play a role in gene regulation in many different tissues and stages of development (two recent reviews: Beisel & Paro, 2011; Kerppola, 2009), including X-inactivation (one recent review: Jeon, Sarma, & Lee, 2012). The mechanism of PcG protein recruitment varies dependent on the target and can be either RNA or protein mediated. In vertebrates, there are gene-specific DNA binding proteins that recruit specific PcG proteins to specific genes (reviewed in Beisel & Paro, 2011; Kerppola, 2009; Schuettengruber & Cavalli, 2009). Here, we focus on vertebrate DNA fragments that have been shown to have properties similar to Drosophila PREs.
In ES cells, CpG islands have been shown to be required for PRC2 recruitment (for review, see Deaton & Bird, 2011). In one elegant study, Mendenhall et al. (2010) showed that a CpG island was sufficient for recruitment of PRC2 to a BAC transgene integrated in ES cells. Binding sites for transcriptional activators in or next to the GC-rich sequence abrogated its ability to recruit PRC2 pointing to a competition between transcriptional activation and repression. Remarkably, even a GC-rich region from E. coli could recruit PRC2 when integrated into the transgene. Interestingly, PRC1 was not recruited to the DNA in their experiments. Lynch et al. (2012) provide a definitive PRE assay in mouse ES cells at the α-globin gene locus. They used recombination-mediated cassette exchange to study PcG protein binding and histone modifications to a single-copy human α-globin locus integrated into a mouse ES cell line. They also showed the importance of CpG islands in PcG recruitment and their experiments also pointed to a competition between transcriptional activation versus PcG repression. In addition, a key role for unmethylated CpG dinucleotides in PcG recruitment was established as they found de novo sites of PcG recruitment at CpG-rich sequences in methyltransferase-deficient ES cells. It is not known if PRC2 recruitment to CpG-rich sequences is protein or RNA mediated or occurs by default in the absence of transcriptional activators.
Are there any PREs in mammals that have binding sites for proteins important for PRE activity in Drosophila? Pho/Phol (YY1-homologues), Spps (a Sp1/KLF family member), Dsp1 (a HMG protein), and GAF all have mammalian homologues. In embryonic and neural stem cells, YY1 and PRC2 show almost no colocalization but, instead, YY1 binding is correlated with transcribed genes (Mendenhall et al., 2010). However, there is some genetic and biochemical evidence that YY1 might recruit PcG proteins at some loci (Garcia, Marcos-Gutierrez, del Mar Lorente, Moreno, & Vidal, 1999; Lorente et al., 2006). Below, we describe three examples of vertebrate PREs, studied in tissues other than ES cells that bear some similarities to Drosophila PREs.
Sing et al. (2009) identified a putative PRE (PRE-kr) that regulates expression of the Maf/Kreisler gene in rhomdomeres in mice. Endogenous PRE-kr binds PRC1 and PRC2 components. A reporter construct containing PRE-kr recruits PcG proteins. Like Drosophila PREs, PRE-kr contains binding sites for YY1 and GAF. The role of the YY1 and GAF binding sites in PRE-kr function has not been tested. Interestingly, PRE-kr mediates mini-white silencing and recruits PcG proteins to polytene chromosomes in Drosophila showing that this 3-kb fragment also acts as a PRE in Drosophila. Further studies would be necessary to determine if the same sequence motifs are required for PRE activity in the mouse and fly.
A vertebrate PRE from a HOX gene also shows characteristics of a Drosophila PRE. Woo, Kharchenko, Daheron, Park, and Kingston (2010) identified a 1.8-kb region between the human HOXD11 and HOXD12 genes (D11.12) that is bound by PcG proteins at the endogenous locus and functions as a PcG-dependent repressor in reporter constructs. Analysis of this element showed that there is a cluster of conserved YY1 sites important for the recruitment of BMI1 (a PRC 1 component) but less so for the recruitment of the PRC2 component SUZ12. A 237-bp region that is highly conserved across vertebrates was essential for the recruitment of members of both PRC1 and PRC2. The D11.12 region also contains a CpG island but the functional importance of this has not been tested. These results imply that PRC1 and PRC2 have different methods of recruitment.
A potential human PRE was identified in resting T cells downstream of the SLCA17 gene (Cuddapah et al., 2012). In resting T cells, the SLCA17 gene is not transcribed and PcG proteins are bound to the presumptive PRE. Treatment of HeLa cells with RNAi to SUZ12 caused a decrease in SUZ12 levels and to the levels of Bmi1 and Ring1B binding to the presumptive SLCA17 PRE. A 3-kb putative SLCA17 PRE contains YY1 and GAF binding sites and acted as a pairing-sensitive silencer in Drosophila, again suggesting that PcG proteins in Drosophila and vertebrates can be recruited by similar mechanisms (Cuddapah et al., 2012).
5. OUTLOOK
Much has been learned about Drosophila PREs since their discovery almost 20 years ago. First discovered as silencers of homeotic gene expression, genome-wide studies have revealed hundreds (or, in some studies, thousands) of potential PREs in Drosophila. Available data suggest that not all PREs are alike, but the differences between PREs are not well defined. The situation in mammals is even more complex with the increased diversity of mammalian PcG proteins and the involvement of PcG proteins in so many diverse tissues, stages of development, and processes. It is still too early to conclude whether there is a class of mammalian PREs that closely resemble Drosophila PREs. There are many fundamental questions about Drosophila PREs that need to be addressed to understand PRE composition and function; some of the questions we have discussed in this review are summarized below.
Are all PREs alike? Functional assays on hundreds of potential PREs using the phi-C31 integration system and reporter assays for different developmental stages will be necessary to assess whether there are different classes of PREs. In these assays, it will be important to use minimal PRE fragments (hundreds of base pairs) as larger DNA fragments will most likely have gene-specific activities including enhancers and silencers that could interfere with PRE assays.
Mutations in different PcG genes in Drosophila (and mammals) lead to different phenotypes suggesting that not all the PcG proteins act together all the time or at all targets. Do different PREs recruit different PcG complexes or does the nature of the gene activators determine which PcG proteins are required at a particular target gene?
At some targets, PcG proteins are bound to transcribed genes. What do PcG proteins do at actively transcribed genes? How can PcG proteins repress transcription at some targets and silence it at others?
Many PcG target genes are expressed in many different tissues at many different times in development; only a subset of the expression patterns is subject to PcG regulation. Are there some types of enhancers (or promoters for that matter) that cannot be regulated by PcG proteins?
How do PRE-binding proteins recruit or anchor PcG protein complexes? The mechanisms are far from known.
Despite all these questions, it is quite clear that PREs play a major role in regulating the expression of many genes in the Drosophila genome. It is incumbent on us to figure out how they work.
Acknowledgments
We thank Yuzhong Cheng and Payal Ray for comments on this chapter. We thank the authors of many recent reviews on this topic; their comprehensive reviews made it easier for us to highlight only a subset of the literature on this important topic. We apologize for any omissions or misstatements of data. J. A. K. and J. L. B. are supported by the intramural research program of the National Institutes of Health and Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- Agresti A, Bianchi M. HMGβ proteins and gene expression. Current Opinion in Genetics and Development. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mechanisms of Development. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Americo J, Whiteley M, Brown JL, Fujioka M, Jaynes JB, Kassis JA. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a Polycomb group response element from the Drosophila engrailed gene. Genetics. 2002;160:1561–1571. doi: 10.1093/genetics/160.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchinson ML. Transcription factor YY1 functions as a PcG protein in vivo. The EMBO Journal. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, et al. The fab-8 boundary defines the distal limit of the Bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R. Silencing chromatin: Comparing modes and mechanisms. Nature Reviews Genetics. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, et al. The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- Blastyák A, Mishra RK, Karch F, Gyurkovics H. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Molecular and Cellular Biology. 2006;26:1434–1444. doi: 10.1128/MCB.26.4.1434-1444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloyer S, Cavalli G, Brock H, Dura JM. Identification and characterization of polyhomeotic PREs and TREs. Developmental Biology. 2003;261:426–442. doi: 10.1016/s0012-1606(03)00314-2. [DOI] [PubMed] [Google Scholar]
- Breen TR, Duncan IM. Maternal expression of genes that regulate the Bithorax complex of Drosophila melanogaster. Developmental Biology. 1986;118:442–456. doi: 10.1016/0012-1606(86)90015-1. [DOI] [PubMed] [Google Scholar]
- Brown JL, Fritsch C, Müller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Brown JL, Grau DJ, DeVido SK, Kassis JA. An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Research. 2005;33:5181–5189. doi: 10.1093/nar/gki827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Kassis JA. Spps, a Drosophila Sp1/KLF family member binds to PREs and is required for PRE activity late in development. Development. 2010;137:2597–2602. doi: 10.1242/dev.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Molecular Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element coveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. The EMBO Journal. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas G, Machat F. Tissue specificity of hedgehog repression by the Polycomb group during Drosophila melanogaster development. Mechanisms of Development. 2005;122:975–987. doi: 10.1016/j.mod.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Chang YL, King B, Lin SC, Kennison JA, Huang DH. A double-bromodomain protein, FSH-S, activates the homeotic gene Ultrabithorax through a critical promoter-proximal region. Molecular and Cellular Biology. 2007;15:5486–5489. doi: 10.1128/MCB.00692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Kwon DY, Arai AL, Mucci D, Kassis JA. P-element homing is facilitated by engrailed polycomb-group response elements in Drosophila melanogaster. PLoS One. 2012;7:e30437. doi: 10.1371/journal.pone.0030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, et al. A novel human polycomb binding site acts as a functional polycomb response element in Drosophila. PLoS One. 2012;7:e36365. doi: 10.1371/journal.pone.0036365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MD, Brown JL, Kassis JA. Characterization of the Polycomb group response elements of the Drosophila melanogaster invected locus. Molecular and Cellular Biology. 2010;30:820–828. doi: 10.1128/MCB.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, et al. Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development. 2012;139:4172–4179. doi: 10.1242/dev.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Deaton A, Bird A. CpG islands and the regulation of transcription. Genes & Development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoville M, Giacomello E, Leng M, Locker D. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics. 2001;157:237–244. doi: 10.1093/genetics/157.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, Locker D, et al. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2005;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- Delest A, Sexton T, Cavalli G. Polycomb: A paradigm for genome organization from one to three dimensions. Current Opinion in Cell Biology. 2012;24:405–414. doi: 10.1016/j.ceb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- DeVido SK, Kwon D, Brown JL, Kassis JA. The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development. 2008;135:669–676. doi: 10.1242/dev.014779. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Cohesin: Genomic insights into controlling gene transcription and development. Current Opinion in Genetics and Development. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews D, Klar M, Dame C, Bräuer AU. Developmental expression profile of the yy2 gene in mice. BMC Developmental Biology. 2009;9:45. doi: 10.1186/1471-213X-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Fiedler T, Rehmsmeier M. jPREdictor: A versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Research. 2006;34:W546–W550. doi: 10.1093/nar/gkl250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Beuchle D, Müller J. Molecular and genetic analysis of the Polycomb group gene sex combs extra/Ring in Drosophila. Mechanisms of Development. 2003;120:949–954. doi: 10.1016/s0925-4773(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Müller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Yusibova GL, Zhou J, Jaynes JB. The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development. 2008;135:4131–4139. doi: 10.1242/dev.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase Sxc/Ogt in Polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex and with the transcription factor YY1. The EMBO Journal. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geini RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells and cancer. Biochemistry and Cell Biology. 2009;87:283–306. doi: 10.1139/O09-057. [DOI] [PubMed] [Google Scholar]
- Gindhart JG, Kaufman TC. Identification of Polycomb and Trithorax Group responsive elements in the regulatory region of the Drosophila Homeotic Gene Sex combs reduced. Genetics. 1995;139:797–814. doi: 10.1093/genetics/139.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg ML, Colvin RA, Mellin AF. The Drosophila zeste locus is non-essential. Genetics. 1989;123:145–155. doi: 10.1093/genetics/123.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes and Development. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Stuerwald U, Lindner D, Rybin V, et al. Molecular recognition of histone lysine methylation by the polycomb group repressor dSfmbt. The EMBO Journal. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phi-C31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pair-wise interactions and can facilitate enhancer-promoter communication. Molecular and Cellular Biology. 2005;25:3682–3789. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavson E, Goldsborough AS, Ali Z, Kornberg TB. The Drosophila engrailed and invected genes: Partners in regulation, expression, and function. Genetics. 1996;142:893–906. doi: 10.1093/genetics/142.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, Müller J. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–127. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Müller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes and Development. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- Hauenschild A, Ringrose L, Altmutter C, Paro R, Rehmsmeier M. Evolutionary plasticity of polycomb/trithorax response elements in Drosophila species. PLoS Biology. 2008;28:e261. doi: 10.1371/journal.pbio.0060261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B, Tatout C, Poux S, Pirrotta V. Structure of a Polycomb response element and in vitro binding of Polycomb group complexes containing GAGA factor. Molecular and Cellular Biology. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Chang YL. Isolation and characterization of CHRASCH, a Polycomb-containing silencing complex. Methods in Enzymology. 2004;377:267–282. doi: 10.1016/S0076-6879(03)77016-5. [DOI] [PubMed] [Google Scholar]
- Huang DW, Chang YL, Yang CC, Pan IC, King B. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Molecular and Cellular Biology. 2002;22:6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur MW, Laney JD, Jeon SH, Ali J, Biggin MD. Zeste maintains repression of Ubx transgenes: Support for a new model of Polycomb repression. Development. 2002;129:1339–1344. doi: 10.1242/dev.129.6.1339. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X inactivation. Current Opinion in Genetics and Development. 2012;22:62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biology. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA. Pairing-sensitive silencing, Polycomb group response elements, and transposon homing in Drosophila. Advances in Genetics. 2002;46:421–438. doi: 10.1016/s0065-2660(02)46015-4. [DOI] [PubMed] [Google Scholar]
- Kassis JA, Vansickle EP, Sensabaugh SM. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoun AM, Kaufman TC. Regulatory regions of the homeotic gene proboscipedia are sensitive to chromosomal pairing. Genetics. 1995;140:643–658. doi: 10.1093/genetics/140.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annual Review of Genetics. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. Polycomb group complexes—Many combinations, many functions. Trends in Cell Biology. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley C, Winoto A. Cloning of GT box-binding proteins: A novel Sp1 multigene family regulating T-cell receptor gene expression. Molecular and Cellular Biology. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes and Development. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma G, Bender W, Sipos L. Replacement of a Drosophila Polycomb response element core, and in situ analysis of its motifs. Molecular Genetics and Genomics. 2008;9:595–603. doi: 10.1007/s00438-008-0336-3. [DOI] [PubMed] [Google Scholar]
- Kwon D, Mucci D, Langlais K, Americo JL, DeVido SK, Cheng Y, et al. Enhancer-promoter communication at the Drosophila engrailed locus. Development. 2009;136:3067–3075. doi: 10.1242/dev.036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C, Adryan B, Bell I, Meadows L, Russell S, Manak JR, et al. Stability and dynamics of Polycomb target sites in Drosophila development. PLoS Genetics. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-sarip A, Moshkin YM, Chalkley GE, Bezatarosti K, Demmers JA, et al. dKDM2 couples histone H2A ubiquitination to histone H3 demethylation during Polycomb group silencing. Genes and Development. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignes F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nature Cell Biology. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lavigne M, Francis NJ, King IF, Kingston RE. Propagation of silencing: Recruitment and repression of naïve chromatin in trans by polycomb repressed chromatin. Molecular Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- Lee JS, Calvin KM, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proceedings of the National Academy of Sciences. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Siegmund T, Lintermann K, Korge G. The Pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. The Journal of Biological Chemistry. 1998;273:28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
- Lorente M, Pérez C, Sánchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mechanisms of Development. 2006;123:312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lynch MD, Smith AJH, De Gobbi M, Fienley M, Hughes JR, Vernimmen D, et al. An interspecies analysis reveals a key role for undermethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. The EMBO Journal. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Gene expression in time and space: Additive vs hierarchical organization of cis-regulatory regions. Current Opinion in Genetics and Development. 2011;21:187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Zuijderduijn LMP, Mohd-Sarip A, Verrijzer CP. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Research. 2003;31:4147–4156. doi: 10.1093/nar/gkg479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate Homologue of Drosophila GAGA Factor. Journal of Molecular Biology. 2010;400:434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Maurange C, Paro R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes and Development. 2002;16:2672–2683. doi: 10.1101/gad.242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiological Reviews. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmon J The FlyBase Consortium. FlyBase 101—The basics of navigating FlyBase. Nucleic Acids Research. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, et al. GC-Rich Sequence Elements recruit PRC2 in Mammalian ES Cells. PLOS Genetics. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, et al. The iab-7 Polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and Pleiohomeotic for silencing activity. Molecular and Cellular Biology. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, O’Farrell PH. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development. 1992;116:805–810. doi: 10.1242/dev.116.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes and Development. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a polycomb nucleoprotein complex. Molecular Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Molecular and Cellular Biology. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. The EMBO Journal. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyl transferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Müller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Current Opinion in Genetics and Development. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Current Opinion in Genetics and Development. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Négre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biology. 2006;4:170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. The EMBO Journal. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. The Journal of Biological Chemistry. 2004;279:25927–25934. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- Noyes A, Stefaniuk C, Cheng Y, Kennison JA, Kassis JA. Modulation of the activity of a Polycomb-group response element in Drosophila by a mutation in the transcriptional activator Woc. G3 (Bethesda) 2011;1:471–478. doi: 10.1534/g3.111.001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K, Gutiérrez L, Gagneur J, Girardot C, Sengupta A, Furlong EEM, et al. Dynamic regulation of Polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Developmental Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Okulski H, Druck B, Bhalerao S, Ringrose L. Quantitative analysis of Polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics and Chromatin. 2011;4:1–16. doi: 10.1186/1756-8935-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S, Ortiz-Lombardia M, Esinas ML, Bernues J, Azorin F. The Drosophila transcription factor tramtrack (TTK) Interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Research. 2002;30:4406–4413. doi: 10.1093/nar/gkf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes and Development. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Schwartz YB, Kahn TG, Asker D, Pirrotta V. Regulation of Polycomb group genes Psc and Su(z)2 in Drosophila melanogaster. Mechanisms of Development. 2012;128:536–547. doi: 10.1016/j.mod.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Pérez-Lluch S, Cuartero S, Azorin F, Espinås ML. Characterization of new regulatory elements within the Drosophila bithorax complex. Nucleic Acids Research. 2008;36:6926–6933. doi: 10.1093/nar/gkn818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Current Opinion in Genetics and Development. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, Kostic C, Pirrotta V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb Group Response Element. The EMBO Journal. 1996;15:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Molecular and Cellular Biology. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappailles A, Decoville M, Locker D. DSP1, a Drosophila HMG protein, is involved in spatiotemporal expression of the homeotic gene Sex combs reduced. Biology of the Cell. 2005;97:779–785. doi: 10.1042/BC20040508. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annual Review of Genetics. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Developmental Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Salvaing J, Decoville M, Mouchel-Vielh E, Bussiere M, Daulny A, Boldyreva L, et al. Corto and DSP1 interact and bind to a maintenance element of the Scr Hox gene: Understanding the role of Enhancers of trithorax and Polycomb. BMC Biology. 2006;4:9. doi: 10.1186/1741-7007-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes and Development. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. The DUBle life of polycomb complexes. Developmental Cell. 2010;18:878–880. doi: 10.1016/j.devcel.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, et al. Functional anatomy of Polycomb and Trithorax chromatin landscapes in Drosophila embryos. PLoS Biology. 2009;7:0001–0018. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature Genetics. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genetics. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendemann A, Lehman M. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12883–12888. doi: 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Peterson AJ, Burr J, Simon JA, O’Connor MB. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Developmental Biology. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- Shintomi D, Hirano T. Releasing cohesin from chromosome arms in early mitosis: Opposing actions of Wapl-Pds5 and Sgo1. Genes and Development. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Jongens TA, Jan LY, Jan YN. Pipsqueak, an early acting member of the posterior group of genes, affects vasa level and germ cell-somatic cell interaction in the developing egg chamber. Development. 1993;119:1187–1202. doi: 10.1242/dev.119.4.1187. [DOI] [PubMed] [Google Scholar]
- Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA binding proteins. Development Genes and Evolution. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Developmental Biology. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nature Reviews Molecular Cell Biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiakakis A, Sturgeon K, Djabali M, Ellis J, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Smouse D, Goodman C, Mahowald A, Perrimon N. Polyhomeotic: A gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes and Development. 1988;2:830–842. doi: 10.1101/gad.2.7.830. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Reviews Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stros M. HMGβ proteins: Interactions with DNA and chromosomes. Biochimica et Biophysica Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Strübbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, et al. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. PNAS. 2011;108:5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. The EMBO Journal. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Taillenbourg E, Dura JM. A novel mechanism for P element homing in Drosophila. Proceedings of the National Academy of Sciences. 1999;96:6856–6861. doi: 10.1073/pnas.96.12.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nature Genetics. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tuckfield A, Clouston DR, Wilanowski TM, Zhao LL, Cunningham JM, Jane SM. Binding of the RING polycomb proteins to specific target genes in a complex with the grainyhead-like family of developmental transcription factors. Molecular and Cellular Biology. 2002;22:1936–1946. doi: 10.1128/MCB.22.6.1936-1946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp Element Mediates Stable Long-Range Chromosome-Chromosome Interactions in Drosophila. Molecular Biology of the Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U. Functional conservation between family members of an ancient duplicated transcription factor family. Nucleic Acids Research. 2003;31:4304–4316. doi: 10.1093/nar/gkg644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, Lynch MD, De Gobbi M, Garrick D, Sharpe JA, Sloane-Stanley JA, et al. Polycomb eviction as a new distant enhancer function. Genes and Development. 2011;25:1583–1588. doi: 10.1101/gad.16985411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Molecular Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wang CC, Chen JJW, Yang PC. Multifunctional transcription factor YY1: A therapeutic target in human cancer. Expert Opinion on Therapeutic Targets. 2006;10:253–266. doi: 10.1517/14728222.10.2.253. [DOI] [PubMed] [Google Scholar]
- Wang L, Jahren N, Miller EL, Ketel CS, Mallin DR, Simon JA. Comparative analysis of chromatin binding by sex Comb on Midleg (SCM) and other Polycomb group repressors at a Drosophila Hox gene. Molecular and Cellular Biology. 2010;30:2584–2593. doi: 10.1128/MCB.01451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD Cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Kirk BD, Gou Y, Wang Q, Ma J. Genome-wide polycomb target prediction in Drosophila melanogaster. Nucleic Acids Research. 2012;40:5848–5863. doi: 10.1093/nar/gks209. [DOI] [PMC free article] [PubMed] [Google Scholar]