Abstract
The nucleosome remodeling and deacetylase (NuRD; also known as Mi-2) complex regulates gene expression at the level of chromatin. The NuRD complex has been identified – using both genetic and molecular analyses – as a key determinant of differentiation in mouse embryonic stem cells and during development in various model systems. Similar to other chromatin remodelers, such as SWI/SNF and polycomb complexes, NuRD has also been implicated in the regulation of transcriptional events integral to oncogenesis and cancer progression. Emerging molecular details regarding recruitment of NuRD to specific loci during development and modulation of these events in cancer are used to illustrate how inappropriate localization of the complex could contribute to tumor biology.
Introduction
The nucleosome remodeling and deacetylase (NuRD; also known as Mi-2) complex is one of four major types of ATP-dependent chromatin remodeling complexes1. Like other classes of chromatin remodeling complexes, the NuRD complex has important roles in processes such as transcription, chromatin assembly, cell cycle progression and genomic stability. The NuRD complex is highly conserved in plants and animals and is broadly expressed in most tissues2. The NuRD complex consists of different protein subunits and combinatorial assembly of these subunits determines the function of NuRD in genomic targeting and mediating cell-type specific functions. Recent progress in understanding the mechanisms of transcriptional regulation by the NuRD complex in cancer biology, where it has dual roles in promoting and suppressing tumorigenesis, form the focus of this Review. Emerging non-transcriptional roles of this complex in processes such as chromatin assembly and the DNA damage/repair response, and their implications in maintaining the genome integrity are also discussed.
Biology and function of the NuRD complex
The NuRD complex was first purified about a decade ago in cells from different species3-6, and contains 6 core subunits2 (Figure 1). This complex was unique on discovery in that it contained at least two subunits with enzymatic functions: the Mi-2α and Mi-2β subunits (also known as CHD3 and CHD4 - chromodomain/helicase/DNA-binding), which have ATP-dependent chromatin remodeling activity, and histone deacetylases 1 and 2 (HDAC1, HDAC2) that catalyze protein deacetylation. More recently, it has been shown that the histone demethylase LSD1 can also be associated with the NuRD complex in certain cell types7, although this association has not been confirmed independently (http://www.cell.com/comments/S0092-8674(09)00710-7). Other non-enzymatic subunits include methyl CpG binding domain 2 (MBD2) and MDB3, metastasis associated 1 (MTA1), MTA2 and MTA3, and retinoblastoma binding protein 4 (RBBP4) and RBBP7 (also known as RBAp46 and RBAp48). Several laboratories also report the association of GATAD2A and GATAD2B (also known as p66α and p66β) to the NuRD complex8-10. p66α, p66β RBAp46 and RBAp48 subunits are thought to be structural components of the NuRD complex and have been shown to directly associate with histone tails11-13. The MBD and MTA subunits, on the other hand, are implicated in targeting the complex to different genomic locations by associating with methylated DNA14 or with transcription factors15, respectively.
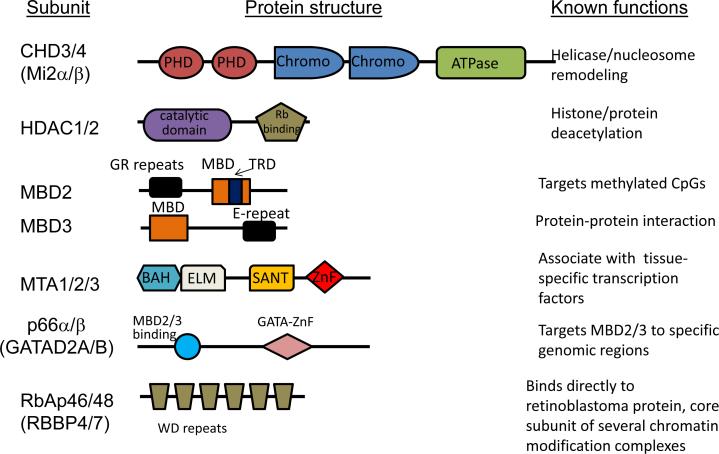
Figure 1. Core components of the NuRD complex.
Conserved protein domains and known functions within the complex are shown for each NuRD subunit and their variants. The CHD3/CHD4 subunit consists of 2 PHD fingers, 2 chromodomains, and an ATPase domain. The PHD fingers are required for interaction with HDAC16 and for modified histone tails96,97. The chromodomains display DNA binding activity and are indispensable for ATPase, nucleosome mobilization, and nucleosome binding98. The ATPase domain carries out ATP hydrolysis which provides the energy necessary for remodeling nucleosome by either histone displacement99,100 or histone octamer sliding101. HDAC1/2 are class I HDACS that share homology to the yeast RPD3 gene and consist of a zinc containing deacetylase catalytic domain. HDAC1 and HDAC2 uniquely contain an additional C terminal Rb binding motif102. MBD2 and MBD3 contain a conserved MBD motif. The MBD domain of MBD2 but not MBD3 binds methylated DNA17,18. MBD2 also contains a glycine and arginine (GR) rich region and a transcriptional repression domain (TRD) involved in recruiting HDAC103,104. MBD3 contains a glutamate (G) repeat region near the C-terminus. MTA1/2/3 share four highly conserved functional domains including the bromo-adjacent homology (BAH) domain, the Egl27/MTA1 (ELM) domain, the SW13, ADA2, N-CoR and TFIII B B” (SANT) domain, and a zinc finger DNA binding domain. The BAH domain is thought to be involved in protein-protein interaction105, while the SANT domain seems to contribute to MTA2-HDAC1 interaction106. The zinc finger domain has been shown to be necessary for interaction with transcription factors or transcriptional co-regulators such as FOG-2107. Function of the ELM domain of MTA proteins remains undefined. p66a/b contain 2 conserved regions. The amino terminal conserved region directly interact with MBD2 or MBD3, while the carboxyl terminal conserved region can interact with histone tails and is important for targeting to specific genomic loci10,13. Both RbAp46 and RbAp48 contain 6 WD-40 repeats which fold into a seven-bladed b propeller structure and binds to histone H4108.
Combinatorial assembly of the non-enzymatic subunits is proposed to be a fundamental mechanism to confer functional specificity of the NuRD complex. For example, MBD2 and MBD3 are found in mutually exclusive NuRD complexes16. While MBD2 can recognize and bind to methylated DNA, a function that has been conserved throughout evolution, mammalian MBD3 contains an amino acid change at the MBD–DNA interface and cannot bind methylated DNA17,18. Instead, the MBD domain in MBD3 may function as a protein-protein interaction domain and has been shown to bind the oncoprotein JUN19. Analysis of Mbd2 and Mbd3 knockout (KO) mice confirmed the functional difference between the two MBD protein family members – Mbd3 KO mice are embryonic lethal whereas Mbd2 KO mice are viable and have only mild defects20. Similar to the MBD subunit, MTA family proteins also form exclusive alternative NuRD complexes associating with different transcription factors and targeting distinct gene loci. For example, only MTA3 can directly interact with the transcriptional repressor BCL6 to maintain a germinal center B cell identity in activated B cells15. These examples highlight the functional differences between the various family members of NuRD complex and suggest roles in promoting specialized functions of the complex in different cell types and biological systems.
In addition to their functions within the NuRD complex, some NuRD subunits can also associate with other protein complexes. For example, RBAp46 and RBAp48 are found in several other multi-subunit chromatin modification complexes12. They presumably act to provide structural support and to promote protein-protein interactions, rather than to provide functional specificity of protein complexes12. HDAC1 and HDAC2 are also the core enzymatic subunits of CoREST and Sin3 complexes21. Like NuRD, these complexes are also associated with transcriptional repression22,23. It remains unclear whether the different HDAC1/2 complexes can act synergistically to repress common downstream targets, or the different complexes are specifically targeted to different regions of the genome.
KO and transgenic animal models of NuRD complex components reveal it has functions in normal developmental processes as well as in tumorigenesis24. The NuRD complex is required at various stages of hematopoietic differentiation, including hematopoietic stem cell maintenance and differentiation into lymphoid and myeloid lineage cells25. A NuRD complex containing MTA3 has been shown to be important in initiating hematopoiesis in zebrafish embryos26. The NuRD complex is also involved in transcriptional regulation of key genes that promote the progression of T and B lymphocyte development27-29. Transcriptional repression of multiple lineage-specific genes by the NuRD complex during hematopoiesis was found to be mediated through FOG-1, which binds MTA family proteins and recruits NuRD to GATA family transcription factors30,31. Other hematopoietic lineage-specific transcription factors are also associated with the NuRD complex includes Ikaros and BCL11B32-34. The MBD3 containing form of the NuRD complex is required for the maintainence of pluripotency in embryonic stem cells and for the elaboration of normal differentiation programs35,36. In the context of cancer, the NuRD complex has been associated with processes such as metastasis and epithelial-to-mesechymal transition (EMT). The remainder of this Review will focus on the recent progress in understanding transcriptional regulation by the NuRD complex in promoting tumorigenesis, as well as its involvement in physiological cellular processes that maintain genome stability to prevent the development of cancer.
Transcriptional regulation by the NuRD complex in cancer
Biology of MTA family subunits
Of all the NuRD complex subunits, the MTA family members are the best studied in the context of cancer development. MTA1 was first cloned and characterized as a candidate metastasis-associated gene from a differential cDNA hybridization screen comparing nonmetastatic and highly metastatic rat mammary adenocarcinomas37. Increased levels of MTA1 were subsequently observed in tumors derived from various tissue origins, including breast, colorectal, gastric, esophageal, endometrial, pancreatic, ovarian, non-small cell lung and prostate cancer, hepatocellular carcinoma, and diffuse large B cell lymphoma (DLBCL) in humans38. MTA1 overexpression correlates with higher tumor grade, microvascular invasion and poor prognosis in many cancer types38. The broad nature of MTA1 overexpression in different types of advanced malignancies is likely due to MTA1 being a downstream target of the MYC oncoprotein39. Silencing of MTA1 was found to abrogate the ability of MYC to transform mammalian cells39. Mechanisms of MTA1-mediated oncogenesis are discussed in detail below.
In the context of breast cancer, MTA1 and MTA2, but not MTA3, have been shown to repress estrogen functions40. Although MTA1 promotes breast tumor progression, MTA3 has an opposing role by inhibiting epithelial-to-mesechymal transition (EMT)41. EMT is characterized by loss of cell adhesion and increased cellular motility, a process thought to be critical to the initiation of cancer metastasis42. Activation of the ERBB2 (also known as HER2) pathway results in upregulation of MTA1 levels, which in turn suppresses estrogen-receptor (ER) element (ERE)-driven transcription by physically interacting with ER43. Overexpression of MTA1 in ERα positive (ERα+) breast cancer cells is sufficient to reduce levels of ER target genes including BRCA1, resulting in enhanced invasive growth in an anchorage-independent manner43,44. The initial description of MTA3 revealed further intertwining of the biology of the MTA gene family with ER41-45. Removal of estrogen leads to loss of MTA3 expression, and MTA3 expression positively correlates with ER expression in human primary breast tumors41. A MTA3 containing NuRD complex has been shown to repress transcription of SNAIL, a critical transcription factor promoting EMT41. MTA1 and MTA3 exhibit opposing patterns during tumor progression in a transgenic mouse strain that develops spontaneous breast cancer46. MTA3 is highly expressed in epithelial cells in normal ducts, and its expression decreases in the early stages of tumorigenesis and becomes silenced in late stage invasive carcinoma. In contrast, MTA1 expression progressively increases during breast cancer progression. This opposing pattern of MTA1 and MTA3 expression is in agreement with the molecular connection between MTA1, ER, and MTA3, and further supporting the model that different MTA family members promote target specificity of the NuRD complex. The biology of MTA family in breast cancer typifies the current characterization of NuRD complex – combinatorial assembly of subunits underlie seemingly contradictory biological outcomes. In this sense, NuRD complex, and its roles in cancer, are considerably different than those documented for other chromatin remodelers like SWI/SNF and polycomb47,48.
Recruitment of the complex by oncogenes and tumor suppressors
Multiple lines of evidence converge on the conclusion that NuRD complex associates with oncogenic transcription factors to promote transcriptional repression of downstream targets. Several examples of this mechanism have been observed in different types of malignancies. In B cell lymphomas of germinal center or post-germinal center origin, such as DLBCL, increased expression of MTA3 is commonly observed49. As mentioned above, MTA3 can directly interact with BCL615, a transcriptional repressor and oncogene that has a causal role in a substantial proportion of DLBCL50. In this system, MTA3 is required for the BCL6-dependent repression of the transcriptional program associated with plasma cell differentiation15.
In three cases of aggressive B-cell chronic lymphocytic leukemia, chromosomal translocations involving the immunoglobulin heavy chain locus resulted in deregulated expression of BCL11A, a Kruppel-like zinc-finger transcriptional repressor51. As MTA proteins within the NuRD complex directly interact with a closely related protein BCL11B in T-cell leukemia and lymphoma cell lines32,33, it is likely that BCL11A also recruits the NuRD complex to promote B-lineage lymphoid malignancies. BCL11A and BCL11B are transcriptional repressors and KO mice indicated that BCL11A and BCL11B are indispensable for early B and T cell development, respectively, affecting differentiation as well as cell survival programs in these cells52,53.
TWIST, a basic helix loop helix transcription factor, can act as a master regulator of cancer metastasis and EMT in a similar fashion to SNAIL54. Increased expression of TWIST is observed in several types of cancers, including breast, gastric, hepatocellular, prostate, uterine and bladder cancers, and correlates with a poor prognosis55. In breast cancer cells, an MTA2 containing NuRD complex was found to associate with TWIST56.In this case, TWIST recruits the NuRD complex to the promoter of a target gene, CDH1 which encodes E-cadherin), to mediate transcriptional repression and to promote the EMT. This finding places NuRD complex as integral to prevention41 and promotion56 of EMT, depending on the cellular context.
The chimeric protein promyelocytic leukaemia (PML)–retinoic acid receptor α (RARα), a well characterized oncogenic transcription factor resulting from chromosomal translocation in human acute promyelocytic leukemias, also recruits NuRD complex through direct protein interaction57. The NuRD complex is recruited by PML–RARα to target genes including the tumor suppressor retinoic acid receptor β2 (RARB2). The NuRD complex in turn facilitates the recruitment of other epigenetic modifiers including the Polycomb complex and DNA methyltransferases to establish the repressive marks H3K27 methylation and DNA methylation, respectively, and to promote gene silencing events that result in the blockade of cellular differentiation57.
In addition to the association with oncoproteins by the MTA subunits, other components of the NuRD complex can also directly interact with transcription factors. For example, NAB2, a co-repressor of the early growth response (EGR) family of transcriptional transactivators, preferentially binds the C-terminal domain of either CHD3 or CHD4 to co-repress EGR activities involved in the progression of prostate cancer58. The functions of EGR1 are broad in that it regulates cell growth, differentiation and apoptotic programs59. In prostate cancer, EGR1 targets include insulin like growth factor 2 (IGF2), transforming growth factor β1 (TGFβ1), and platelet derived growth factor α (PDGFA), which have been implicated in tumor progression59. Accordingly, increased EGR1 and reduced levels of NAB2 are frequently observed in prostate cancer60.
As mentioned above, the MBD3 subunit has been shown to directly interact with JUN19, which has an important role in regulating intestinal homeostasis and tumorigenesis61. A MBD3- containing NuRD complex preferentially interacts with an unphosphorylated form of JUN to repress its transcriptional activity. Upon exposure to extracellular stimuli such as growth factors and cytokines, JUN is phosphorylated by JNK, making its interaction with MBD3 inefficient and relieving the transcriptional repression by the NuRD complex. Inactivation of the Mbd3 gene in mice in intestinal crypts leads to increased expression of JUN target genes, resulting in colonic hyperproliferation and increased susceptibility to tumor development19. These examples indicate that the NuRD complex has a dual role in promoting as well as suppressing tumorigenesis and that which of these prevails is probably dependent on cell type as well as the subunit composition of the complex.
Other examples of the NuRD complex associating with proteins that act as tumor suppressors have also been shown in breast cancer cells. ZIP, a zinc finger and G-patch domain-containing protein, acts as a transcriptional repressor to repress genes involved in cell proliferation, survival and migration62. Loss of ZIP results in aggressive tumor growth in vivo in mouse xenografts. Like NAB2, ZIP also exclusively interacts with the Mi-2α and Mi-2β subunits of the NuRD complex.
The histone demethylase LSD1 was also recently found to be associated with the NuRD complex through the MTA subunit in breast cancer cells and to repress transcription of genes in pathways such as TGFβ, focal adhesion, and MAPK. These pathways are involved in cell migration, invasion and EMT in cancer cells7. At the TGFB1 promoter, only an MTA3 containing form of NuRD complex was found to be associated with LSD17. Depletion of LSD1 led to upregulation of TGFβ1 expression and increased invasiveness in vitro and metastatic potential in vivo7. These results further support the unique role of the MTA3 acting in the context of a tumor suppressor in breast cancer.
It is somewhat surprising that the pattern of association with particular subunits or contact surfaces that has emerged from the many studies reported to date lacks unifying features. Rather, most subunits of the complex have been reported as interaction surfaces in one system or another and within individual subunits, in many cases, different transcription factors are implicated in binding to different regions of the indicated NuRD subunits. This lack of clarity points to a compelling need for additional biochemistry and structural biology to ascertain the available protein interaction surfaces on NuRD complex and how they are utilized by transcription factors in diverse biological contexts to elicit a given functional outcome.
Protein modification of and by the NuRD complex
Tumor hypoxia, an environmental cue known to promote angiogenesis, has also been shown to induce MTA1 expression in breast cancer cells63. MTA1 recruits HDAC1 to deacetylate hypoxia-inducible factor-1α (HIF-1α), the master regulator of the hypoxia transcriptional program63. The deacetylated form of HIF-1α is stabilized and protected from rapid turnover, thus enhancing transcriptional activation of downstream targets, including those involved in angiogenesis and cancer metastasis. Similarly, an MTA1 or MTA2 containing NuRD complex can promote deacetylation of p53 to block p53-dependent transcriptional activation, and inhibit its function in mediating growth arrest and apoptosis64,65. Inactivation of p53 function by the NuRD complex probably represents another mechanism that facilitates tumor growth and progression.
Subunits of the NuRD complex are also subject to posttranslational modification that alters their function. Several recent reports substantiate a role for MTA1 acetylation in gene activation. For example, MTA1 is a transcriptional activator of breast cancer amplified sequence 3 (BCAS3), a gene overexpressed in breast cancer and implicated in enhancing anchorage independent growth66. Only lysine 626 acetylated MTA1 in association with ERα at an intronic enhancer is able to efficiently recruit Pol II to promote BCAS3 transcription66. In breast cancer cells, only an acetylated form of MTA1 was found to repress Gαi2 transcription, leading to activation of the Ras-Raf pathway and was able to transform Rat1 fibroblasts67. An acetylated form of MTA1 is also implicated in DLBCL. In DLBCLs, MTA1 occupies the promoter as well as an enhancer region in the 7th intron of the Pax5 gene68, a B-cell specific transcription factor. Only the acetylated form of MTA1 efficiently recruited Pol II to the Pax5 promoter68. Other NuRD complex subunits have also been shown to have post-translational modifications such as phosphorylation and acetylation69-71. However, functional roles for these modifications have yet to be determined. Regardless, studies of the acetylated form of MTA1 have provided evidence that the NuRD complex can act as a direct transcriptional activator as well as its known role as a transcriptional repressor. Post-translational modifications on NuRD complex subunits likely represent another level through which the biological functions of this complex are regulated.
Recruitment of MBD2 to hypermethylated gene promoters to mediate gene silencing
An aberrant DNA methylation pattern is frequently observed in cancer. Cancer cells often exhibit genome-wide hypomethylation, which is thought to contribute to genome instability72. In contrast, promoter CpG islands are frequently hypermethylated in cancer, and are strongly associated with transcriptional silencing73. Promoter hypermethylation is a widespread mechanism in promoting transcriptional repression of tumor suppressor genes, including INK4A, RB1 and BRCA174. In addition to preventing binding of transcription activators, methylated CpGs can also recruit MBD family proteins and their associated chromatin remodeling enzymes to form repressive chromatin to ensure gene silencing75. MBD2 has been shown to associate with several hypermethylated promoters in cancer cells, including the CDKN2A locus (which encodes INK4A and ARF) in colon cancer76,77. While it remains unclear whether MBD2 specifically recruits other NuRD complex subunits to these gene loci, treatment of colon cancer cells with HDAC inhibitor trichostatin A resulted in greater expression of ARF and INK4A than a DNA methyltransferase inhibitor 5-Aza-C77. These data suggest that cooperative actions between MBD2 and HDAC occur at hypermethylated gene loci, which supports an active role for NuRD complex in gene silencing. Consistent with these observations, Mbd2 deficiency in tumor-prone Apcmin/+ mice have suppressed intestinal tumorigenesis78. It remains to be determined to what extent the MBD2 containing NuRD complex promotes gene silencing at hypermethylated promoters in cancer.
Non-transcriptional roles of the NuRD complex in maintaining genome stability
In addition to transcriptional regulation, emerging data indicate that the NuRD complex also has important roles in other processes that ensure proper DNA replication, cellular proliferation and protection of genome integrity69,79-82. Strict regulation of these processes is critical in protecting cells from malignant transformation. Rapidly proliferating lymphocytes uniquely accumulate a high local concentration of the NuRD complex, or NuRD foci, at pericentromeric heterochromatin on chromosomes 1, 9, 16 during S phase of the cell cycle79. These NuRD foci colocalize with proteins present at active replication forks, such as PCNA and chromatin assembly protein CAF-1, suggesting a role for the NuRD complex in regulating DNA replication and/or subsequent chromatin assembly at these chromosomal regions. Interestingly, the polycomb core complex PRC1, which localizes to pericentromeric heterochromatin in many cell types, is absent in lymphocytes containing NuRD foci, suggesting a unique role of the NuRD complex during lymphocyte proliferation. Coincidentally, cells derived from patients with Immunodeficiency, Centromeric instability and Facial anomalies (ICF) syndrome, due to loss-of-function mutation in DNA methyltransferase 3B, have aberrant hypomethylated pericentromeric heterochromatin83,84. However, B lymphocytes from these patients preferentially exhibit chromosomal instability resulting in defective differentiation85. It is plausible that an MBD2 containing NuRD complex targets the densely methylated regions at pericentromeric heterochromatin in lymphocytes to ensure proper chromatin assembly during cellular proliferation. Whether a similar mechanism is employed by rapidly dividing tumor cells is unknown.
In addition to its involvement in chromatin assembly, the NuRD complex also regulates the G1/S cell-cycle transition69. Manipulation of the NuRD subunits CHD4 69,81,86and MTA281 by RNA interference or of MTA1 by genetic means87 led to a blockade at the G1/S phase transition with accumulation of p21. In U2OS cells, the absence of the NuRD complex prevented deacetylation of p53. Accumulation of stabilized p53 protein led to increased expression of its downstream target p21, leading to cell cycle arrest69. In mouse embryonic fibroblasts, genetic depletion of MTA1 led to destabilization of p53. Nonetheless, p21 levels were also increased. Subsequent investigation revealed that, in the mouse embryonic fibroblast and in mouse tissue, MTA1 and NuRD complex directly regulate p21 levels via a p53 independent mechanism85. As of this writing, it is unclear why these two studies, which describe a similar biological outcome, do so via very different mechanisms. Further experimentation will be required to resolve the mechanistic discrepancies. Irregardless, these analyses collectively indicate that the NuRD complex can have multiple roles at different stages of the cell cycle to regulate cell proliferation, and some functions appear to be cell-type specific events.
In the past year, several groups also reported a novel function of the NuRD complex in regulating DNA damage responses, a role that had previously been ascribed to MTA1 88. A genome-wide RNA interference screen in C. elegans identified egr-1, a homolog of MTA2, as a factor that protects against DNA damage induced by ionizing radiation (IR)81. IR results in chromosomal double-strand breaks (DSBs), and adequate DNA repair mechanisms are necessary to prevent apoptosis or aberrant transformation. The NuRD complex is rapidly recruited to sites of double strand breaks69,80,81,86. This recruitment is dependent on the activity of the poly(ADP ribose) polymerase (PARP), which incorporates poly(ADP ribose) (PAR) chains at sites of DNA damage69,80. The presence of PAR chains recruits several DNA repair proteins, as well as the NuRD complex. CHD4 was found to contain PAR-binding motifs in its amino-terminal region69. Depletion of CHD4 results in hypersensitivity to DNA damage resulting from IR exposure, and accumulation of unrepaired breaks at sites of DNA damage69,80,81. Loss of CHD4 also results in CDC25A degradation and p21 accumulation, leading to cell cycle delay86. CHD4 or MTA2 depleted cells failed to fully activate the G2/M checkpoint, due to the inability of cells to activate the RNF8–RNF168-mediated histone ubiquitylation pathway, which is required for accumulation of checkpoint and repair proteins including BRCA181,86. In addition to promoting DNA repair, there is also evidence that the presence of the NuRD complex at sites of DNA damage acts to suppress transcription80. At sites of DNA damage, there is rapid loss of nascent RNA and elongating RNA polymerase, which was not the case in CHD4 or MTA1 depleted cells. Collectively, these results suggest that the NuRD complex has a critical role in DNA damage response by both recruiting DNA repair proteins and promoting transcriptional repression, in order to facilitate the repair process. Interestingly, NuRD complex has been implicated as an active regulator of both G1/S and G2/M progression checkpoints. These observations highlight the multiple roles played by NuRD in chromosomal biology. In one case, progression through the G1/S boundary, the defect appears to be deregulated transcription. In the second, G2/M progression, the mechanism involves a defect in histone modification that impacts checkpoint control.
Targeting of NuRD subunits for cancer therapy?
Recent progress in understanding the function of NuRD subunits and their specificity in different types of cancer, as discussed above, should set the path for designing effective cancer therapeutic agents that target this complex. However, as the NuRD complex has roles in both promoting and suppressing tumor growth, even within the same tumor type, more knowledge of the fundamental biology downstream of NuRD will be required. Given the current state of the field, MTA1 would seem a prime therapeutic target. It is widely overexpressed in many types of cancer and is downstream of important pathways such as MYC in transformation processes38,39.
The NuRD complex contains histone deacetylase subunits, so HDAC inhibitors may represent one potential therapeutic avenue for targeting NuRD function. A recent study demonstrated that HDAC inhibitors have selective preference for different types of HDAC complexes89 suggesting that targeting specific HDAC complexes may be feasible with enzyme inhibitors. However, it remains unclear whether one could selectively target tumor promoting activities while sparing tumor suppressive functions with this class of drugs.
As the NuRD complex frequently associates with tissue-specific transcription factors to regulate transcription, drugs modulating the activity or interactions of these proteins may represent a more selective approach to inhibit undesirable NuRD functions in cancer cells. Emerging evidence points to the possibility that post-translational modifications of NuRD subunits can modulate their function within the complex, potentially offering additional drug targets.
Conclusions and future directions
When the NuRD complex was first characterized, its subunit composition suggested a role in transcriptional repression. Although many examples of transcriptional repression have been demonstrated, it is now clear that the NuRD complex is multi-functional and participates in many aspects of chromosomal biology, including transcriptional activation, protein modification, DNA repair and DNA replication. In the context of cancer, the NuRD complex has roles in both promoting as well as suppressing tumorigenesis. As the interaction with other proteins represents a major mechanism of its functional specificity, whether and how the NuRD complex contributes to cancer development is dependent on cell type. The microenvironment and transcriptional program of each cell type will dictate subunit composition of the NuRD complex and its interaction partners.
Given the broad role of the NuRD complex in cancer biology, it is surprising that expression of MTA1 subunit only has been shown to be deregulated during tumor progression in various types of cancer. Other chromatin modification complexes, such as the SWI/SNF complex and Polycomb repressive complexes, also have well established roles in cancer47,48. Several subunits of the SWI/SNF complex function as tumor suppressors, and loss-of-function mutations in these subunits have been found in various human cancers. In contrast, the Polycomb proteins have important roles in maintaining cancer stem cell populations, and cancer cells often have increased expression of polycomb proteins. Similarly, MLL1, a histone 3 lysine 4 methyltransferase that functions in a large nuclear complex, is frequently involved in chromosomal translocations in various hematopoietic malignancies90. MLL fusions have also been found to impart leukemic stem cell properties90. As mentioned above, NuRD complex plays important roles in the maintenance and function of hematopoietic stem cells25,26, one can speculate that it may also participate in regulating the transcriptional program in leukemic or other types of cancer stem cells.
As NuRD complex is an integral component of the DNA repair machinery, one might anticipate loss of NuRD complex function, particularly in tumor types characterized by chromosomal instability. Furthermore, aging cells have loss of expression of Mi-2 subunits91, which could contribute to genome instability and cancer susceptibility during cellular aging. Along this line, loss of CHD4 expression has been observed in gastric and colorectal cancer cases with microsatellite instability92, supporting the role of NuRD complex in maintenance of genome integrity in these regions. However, loss-of-function mutations of NuRD complex subunits have only been infrequently observed in cancer in limited studies92-95. For example, a truncating mutation of HDAC2 has also been documented in sporadic carcinomas with microsatellite instability95, although it is not clear whether the loss of HDAC2 function in these cases is in the context of NuRD complex or other HDAC containing nuclear complexes. Ongoing cancer genome sequencing projects may provide insights into the prevalence of NuRD mutations in different types of cancer and reveal patterns of association of loss of function of specific subunits with unique aspects of tumor biology similar to those observed in other chromatin remodeling complexes.
Although mutations have not been observed in NuRD subunits with high frequency in cancer, it is possible that subunit composition of the NuRD complex is perturbed by signaling cascades in cancer cells without disrupting the expression level of individual subunits, leading to loss of function or aberrant genomic targeting of the complex. Recent reports showing the importance of acetylation of MTA1 in facilitating its interaction with oncogenic transcriptional complexes suggest that post-translational modification on NuRD complex subunits maybe critical in determining its function. High throughput screens of compounds with biologic activity in tumor cells may lead to new insights into the contributions of NuRD complex to tumorigenesis as well as provide new therapeutic avenues. Furthermore, while the core composition of the NuRD complex is well characterized, only a handful of tissue-specific transcription factors associating with the complex have been characterized. Identification of binding partners of different variant forms of the NuRD complex and determination of genomic localization in both normal and abnormal tissue, a goal of current genome association studies, will facilitate generation of models relating the biological functions of NuRD complex to cancer.
Summary.
The NuRD complex is a multi-subunit chromatin remodeling complex containing 2 core subunits (Mi2 and HDAC1/2) with enzymatic functions. Mi2 catalyzes ATP-dependent chromatin remodeling, while HDAC1/2 mediates histone/protein deacetylation.
All subunits of the complex are encoded by multiple gene paralogs. Combinatorial assembly of these paralogs contributes to targeting and function of the complex.
The MTA1 subunit is widely overexpressed in many types of cancer and is associated with poor prognosis.
Unlike other chromatin remodeling complexes with well defined roles in cancer, the NuRD complex can promote or suppress tumorigenesis dependent on context.
NuRD complex recruitment to specific loci is mediated by multiple mechanisms, including recruitment by transcription factors and direct interaction with methylated DNA.
Emerging evidence suggests non-transcriptional roles of NuRD complex in maintenance of genome stability, including DNA replication, chromatin assembly, and DNA repair.
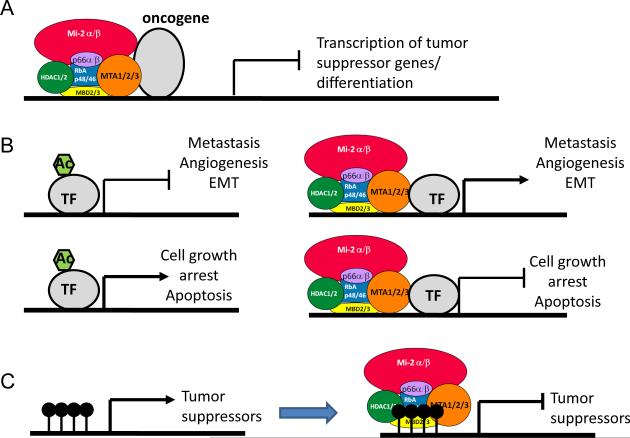
Figure 2. Mechanisms by which the NuRD complex interacts with the different factors to promote cancer development.
A. Recruitment of NuRD complex by tissue-specific transcription factor to gene promoters to mediate transcriptional repression. Several known oncogenes have been shown to recruit the NuRD complex to suppress transcription of tumor suppressor genes. B. Post-translational modification of transcription factor by NuRD complex to modulate downstream transcriptional activities. In hypoxic breast cancer cells, MTA1 recruits HDAC1 to promote deacetylation of HIF1α, leading to stabilization of HIF1α and its transcriptional program63. Conversely, deacetylation of p53 by the NuRD complex results in inactivation of p53, rendering cells resistant to cell growth arrest and apoptosis64,65. C. An MBD2 containing NuRD complex targeting hypermethylated promoters of tumor suppressor genes to mediate transcriptional silencing75-77.
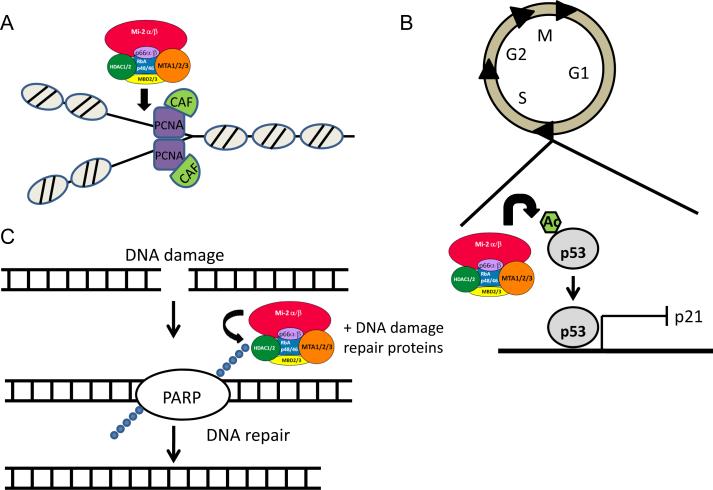
Figure 3. Non-transcriptional mechanisms by which NuRD complex maintain genome stability.
A. Co-localization of the NuRD complex with DNA replication machineries such as CAF1 and PCNA during the S phase of the cell cycle suggests a role of the complex in chromatin assembly during and/or post DNA replication79. B. NuRD complex promoting G1/S phase transition during cell cycle progression by promoting deacetylation of p53. Loss of p53 function results in inactivation of p21 to allow cell cycle progression69. C. Recruitment of NuRD complex to site of DNA damage to facilitate the DNA repair process. At sites of double-stranded breaks, PARP incorporates poly (ADP ribose) chains that can recruit the NuRD complex in addition to other DNA repair proteins to facilitate the repair process69,80.
Acknowledgements
We thank members of the Wade lab for critical comments and suggestions for this manuscript. We apologize to those whose work is not cited due to space limitation. Our research is funded by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (Project number Z01ES101965 to PAW).
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 3.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 4.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–6. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 5.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 8.Wade PA, et al. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–6. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 9.Brackertz M, Boeke J, Zhang R, Renkawitz R. Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J Biol Chem. 2002;277:40958–66. doi: 10.1074/jbc.M207467200. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, et al. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol Cell Biol. 2002;22:536–46. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marhold J, Brehm A, Kramer K. The Drosophila methyl-DNA binding protein MBD2/3 interacts with the NuRD complex via p55 and MI-2. BMC Mol Biol. 2004;5:20. doi: 10.1186/1471-2199-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Brackertz M, Gong Z, Leers J, Renkawitz R. p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006;34:397–406. doi: 10.1093/nar/gkj437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita N, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [This study is the first to demonstrate association of MTA subunit to tissue-specific transcription factor to mediate transcriptional repression.] [DOI] [PubMed] [Google Scholar]
- 16.Le Guezennec X, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–51. doi: 10.1128/MCB.26.3.843-851.2006. [This study demonstrates that MBD2 and MBD3 form mutually exclusive NuRD complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–9. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–77. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 19.Aguilera C, et al. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–5. doi: 10.1038/nature09607. [This study demonstrates the protein-protein interaction property of the MBD domain within Mbd3.] [DOI] [PubMed] [Google Scholar]
- 20.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–18. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–8. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–64. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–6. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida T, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–89. doi: 10.1101/gad.1642808. [This study investigated the function of the NuRD complex in hematopoietic stem cels using Mi-2β conditional knockout mice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Jia S, Wang S, Wang Y, Meng A. Mta3-NuRD complex is a master regulator for initiation of primitive hematopoiesis in vertebrate embryos. Blood. 2009;114:5464–72. doi: 10.1182/blood-2009-06-227777. [DOI] [PubMed] [Google Scholar]
- 27.Gao H, et al. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci U S A. 2009;106:11258–63. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–34. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Hong W, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–78. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Z, et al. FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J. 2010;29:457–68. doi: 10.1038/emboj.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cismasiu VB, et al. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–64. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 33.Grabarczyk P, et al. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- 34.Cobb BS, et al. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14:2146–60. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 36.Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One. 2009;4:e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pencil SD, Toh Y, Nicolson GL. Candidate metastasis-associated genes of the rat 13762NF mammary adenocarcinoma. Breast Cancer Res Treat. 1993;25:165–74. doi: 10.1007/BF00662141. [DOI] [PubMed] [Google Scholar]
- 38.Nicolson GL, et al. Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clin Exp Metastasis. 2003;20:19–24. doi: 10.1023/a:1022534217769. [DOI] [PubMed] [Google Scholar]
- 39.Zhang XY, et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci U S A. 2005;102:13968–73. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis. 2009;26:215–27. doi: 10.1007/s10585-008-9233-8. [DOI] [PubMed] [Google Scholar]
- 41.Fujita N, et al. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–19. doi: 10.1016/s0092-8674(03)00234-4. [This paper demonstrates that MTA family members form distinct NuRD complexes.] [DOI] [PubMed] [Google Scholar]
- 42.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazumdar A, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–7. doi: 10.1038/35050532. [This paper shows that MTA1 expression can be upregulated by the heregulin/HER2 pathway in breast cancer cells and represses ER activity.] [DOI] [PubMed] [Google Scholar]
- 44.Molli PR, Singh RR, Lee SW, Kumar R. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–80. doi: 10.1038/sj.onc.1210839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar R. Another tie that binds the MTA family to breast cancer. Cell. 2003;113:142–3. doi: 10.1016/s0092-8674(03)00274-5. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Stephens LC, Kumar R. Metastasis tumor antigen family proteins during breast cancer progression and metastasis in a reliable mouse model for human breast cancer. Clin Cancer Res. 2006;12:1479–86. doi: 10.1158/1078-0432.CCR-05-1519. [This paper demonstrate the opposing expression pattern of MTA1 and MTA3 during breast cancer progression.] [DOI] [PubMed] [Google Scholar]
- 47.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 48.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 49.Jaye DL, et al. The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol. 2007;213:106–15. doi: 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 50.Kusam S, Dent A. Common mechanisms for the regulation of B cell differentiation and transformation by the transcriptional repressor protein BCL-6. Immunol Res. 2007;37:177–86. doi: 10.1007/BF02697368. [DOI] [PubMed] [Google Scholar]
- 51.Satterwhite E, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–20. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- 52.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–32. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 53.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–9. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 56.Fu J, et al. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–89. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morey L, et al. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol Cell Biol. 2008;28:5912–23. doi: 10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2006;281:15129–37. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 59.Adamson ED, Mercola D. Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol. 2002;23:93–102. doi: 10.1159/000059711. [DOI] [PubMed] [Google Scholar]
- 60.Abdulkadir SA, et al. Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum Pathol. 2001;32:935–9. doi: 10.1053/hupa.2001.27102. [DOI] [PubMed] [Google Scholar]
- 61.Sancho R, et al. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009;28:1843–54. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li R, et al. ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 2009;28:2763–76. doi: 10.1038/emboj.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J. 2006;25:1231–41. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–81. doi: 10.1038/35042612. [This paper was the first to demonstrate that NuRD complex can regulate tumor suppressors via post-translational modifications.] [DOI] [PubMed] [Google Scholar]
- 65.Moon HE, Cheon H, Lee MS. Metastasis-associated protein 1 inhibits p53-induced apoptosis. Oncol Rep. 2007;18:1311–4. [PubMed] [Google Scholar]
- 66.Gururaj AE, et al. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci U S A. 2006;103:6670–5. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohshiro K, et al. Acetylation-dependent oncogenic activity of metastasis-associated protein 1 co-regulator. EMBO Rep. 2010;11:691–7. doi: 10.1038/embor.2010.99. [This paper shows that post-translational modification of MTA1 is important for its concogenic activities.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balasenthil S, et al. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007;67:7132–8. doi: 10.1158/0008-5472.CAN-07-0750. [DOI] [PubMed] [Google Scholar]
- 69.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [This paper demonstrates poly(ADP-ribos)-dependent recruitment of the NuRD complex to sites of DNA damage and shows that NuRD plays a role in G1/S transition during the cell cycle by modulating p53 function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 71.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 72.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–8. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 73.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 74.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–37. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sansom OJ, Maddison K, Clarke AR. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Pract Oncol. 2007;4:305–15. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- 77.Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci U S A. 2001;98:4990–5. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sansom OJ, et al. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34:145–7. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- 79.Helbling Chadwick L, Chadwick BP, Jaye DL, Wade PA. The Mi-2/NuRD complex associates with pericentromeric heterochromatin during S phase in rapidly proliferating lymphoid cells. Chromosoma. 2009;118:445–57. doi: 10.1007/s00412-009-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [This paper also demonstrates poly(ADP-ribose)-dependent recruitment of NuRD complex to sites of DNA damage. It further suggests a role of NuRD complex in repressing transcription at sites of DNA damage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smeenk G, et al. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–9. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brehm A, et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–58. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu GL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–91. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 84.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–7. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blanco-Betancourt CE, et al. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood. 2004;103:2683–90. doi: 10.1182/blood-2003-08-2632. [DOI] [PubMed] [Google Scholar]
- 86.Larsen DH, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li DQ, et al. MTA1 coregulator regulates p53 stability and function. J Biol Chem. 2009;284:34545–52. doi: 10.1074/jbc.M109.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li DQ, et al. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci U S A. 2009;106:17493–8. doi: 10.1073/pnas.0908027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–65. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 90.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 91.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–7. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58:660–8. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- 93.Bader S, et al. MBD1, MBD2 and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers. Oncogene. 2003;22:3506–10. doi: 10.1038/sj.onc.1206574. [DOI] [PubMed] [Google Scholar]
- 94.Zhu Y, Harrison DJ, Bader SA. Genetic and epigenetic analyses of MBD3 in colon and lung cancer. Br J Cancer. 2004;90:1972–5. doi: 10.1038/sj.bjc.6601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ropero S, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–9. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 96.Musselman CA, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–87. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mansfield RE, et al. Plant Homeodomain (PHD) Fingers of CHD4 Are Histone H3-binding Modules with Preference for Unmodified H3K4 and Methylated H3K9. J Biol Chem. 2011;286:11779–91. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouazoune K, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–40. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–7. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–40. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 101.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–73. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 102.Thiagalingam S, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 103.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 104.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 105.Oliver AW, et al. Crystal structure of the proximal BAH domain of the polybromo protein. Biochem J. 2005;389:657–64. doi: 10.1042/BJ20050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Humphrey GW, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–24. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 107.Roche AE, et al. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44:352–60. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murzina NV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–85. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]