Abstract
Ubiquitin C-terminal Hydrolase L1 (UCH-L1) has oncogenic properties and is highly expressed during malignancies. We recently documented that Epstein-Barr virus (EBV) infection induces uch-l1 expression. Here we show that Kaposi's Sarcoma-associated herpesvirus (KSHV) infection induced UCH-L1 expression, via cooperation of KSHV Latency-Associated Nuclear Antigen (LANA) and RBP-Jκ and activation of the uch-l1 promoter. UCH-L1 expression was also increased in Primary Effusion Lymphoma (PEL) cells co-infected with KSHV and EBV compared with PEL cells infected only with KSHV, suggesting EBV augments the effect of LANA on uch-l1. EBV latent membrane protein 1 (LMP1) is one of the few EBV products expressed in PEL cells. Results showed that LMP1 was sufficient to induce uch-l1 expression, and co-expression of LMP1 and LANA had an additive effect on uch-l1 expression. These results indicate that viral latency products of both human γ-herpesviruses contribute to uch-l1 expression, which may contribute to the progression of lymphoid malignancies.
INTRODUCTION
Ubiquitin C-terminal Hydrolase-L1 (UCH-L1) is a cysteine hydrolase that contains the typical active site triad of cysteine, histidine, and aspartic acid and catalyzes hydrolysis of C-terminal esters and amides of ubiquitin (Larsen et al., 1996). In adult humans, UCH-L1 is normally exclusively expressed in the brain and cells of the reproductive system (Kwon et al., 2004; Setsuie and Wada, 2007). Although the physiological function of UCH-L1 in neurons is still unclear, mutations in the uch-l1 gene have been associated with Parkinson's and Alzheimer's diseases (Betarbet et al., 2005). Functional activities, other than acting as an ubiquitin hydrolase, have been proposed for UCH-L1. First, UCH-L1 can dimerize resulting in ubiquitin ligase activity (Liu et al., 2002). Second, in neurons, the stabilization of mono-ubiquitin proteins is not dependent on UCH-L1 deubiquitinating activity (Osaka et al., 2003; Setsuie and Wada, 2007), a finding that points to an ubiquitin-independent function for UCH-L1.
Besides the high levels of expression of UCH-L1 in the brain and reproductive system, de novo expression of UCH-L1 has been detected in numerous cancers, such as lung (Hibi et al., 1999; Kim et al., 2008), colorectal (Loeffler-Ragg et al., 2005), bladder (Yang et al., 2006) and breast cancer (Miyoshi et al., 2006), and points to the involvement of this protein in the oncogenic transformation of cells. High levels of UCH-L1 were also observed in transformed cells of lymphoid origin such as Burkitt lymphoma (Ovaa et al., 2004) and multiple myeloma (Otsuki et al., 2004). Recent studies demonstrate that inhibition of the expression of UCH-L1 reduces the tumorigenic phenotype of transformed cells, including virus-transformed B-lymphocytes (Bheda et al., 2009a; Kim et al., 2008; Rolen et al., 2008). UCH-L1 also associates with cytoskeletal components, including microtubules (Bheda et al., 2010; Kabuta et al., 2008) and actin filaments (Basseres et al., 2010), and it physically associates with mitotic spindles (Bheda et al., 2010), which suggests a potential role in the regulation of mitosis. Furthermore, oncogenic transcription factors, such as B-Myb and β-catenin/TCF, up-regulate the expression of the uch-l1 gene (Bheda et al., 2009b; Long et al., 2003). Together, these findings strongly support the idea of an oncogenic function for UCH-L1, and although the physiological roles of UCH-L1 and the regulation of its expression in normal and transformed cells remain largely unexplored, it has become clear that this multifunctional protein of the ubiquitin system UCH-L1 participates in diverse cellular processes.
Both EBV and KSHV are members of the -herpesvirus subfamily. EBV, the first human tumor virus discovered, causes or is closely associated with both lymphoid and epithelial malignancies, and KSHV is the causative agent of Kaposi's Sarcoma and Primary Effusion Lymphoma (PEL) (Pagano, 2009; Sin et al., 2007). Both viruses produce significant pathology in immunodeficient hosts, most commonly with patients with AIDS (Pagano, 2009; Sin et al., 2007).
During cell transformation by EBV, viral oncoproteins disrupt a variety of host signaling pathways that affect the host ubiquitin system (Pagano, 2009; Shackelford and Pagano, 2005, 2007). The EBV primary oncogene LMP1 inhibits Siah1 ubiquitin ligase and stabilizes the expression of β-catenin (Jang et al., 2005). LMP1 also induces the regulatory ubiquitination of IRF7 (Ning et al., 2008) as well as downregulates the activity of IRF7 via the activation of the ubiquitin-editing enzyme A20 (Ning and Pagano). EBNA1 competes with p53 to interact with HAUSP, the p53 deubiquitinating enzyme, thus indirectly targeting p53 for ubiquitination and degradation (Holowaty and Frappier, 2004; Holowaty et al., 2003). EBNA3C, which possesses intrinsic deubiquitinating activity, inhibits the p53 and Rb pathways by two different mechanisms: deubiquitination of MDM2 and recruitment of SCF4 ligase (Saha et al., 2009; Ying and Xiao, 2006).
The main KSHV protein that directly or indirectly affects the host ubiquitin system is Latency-Associated Nuclear Antigen (LANA), which is expressed in all KSHV latently infected cells and modulates cellular pathways that may contribute to tumorigenesis (Wen et al.). LANA physically associates with p53 and inhibits p53-mediated transcriptional activity and apoptosis (Friborg et al.). LANA also inactivates expression of the tumor suppressor retinoblastoma (Rb) and releases the transactivator E2F, which induces cells to progress through the G1/S cell cycle checkpoint. In addition, LANA interacts with the bromodomain-containing protein RING3/Brd2 and further stimulates cell-cycle progression. Together, p53 regulation and cell-cycle progression are highly regulated by the host ubiquitin system. Finally, LANA rescues β-catenin from phosphorylation-dependent ubiquitination and destruction by interacting with GSK3 (Fujimuro et al., 2007).
LANA functions as a transcriptional modulator of multiple cellular and viral promoters, including its own (Sin et al., 2007). It can both activate as well as repress transcription of multiple viral and cellular genes through a variety of mechanisms. Furthermore, LANA can act as a transcriptional modulator both directly and indirectly. LANA binds DNA, including the KSHV terminal repeat and its own promoter and regulates gene expression. LANA also interacts with p53 and down-regulates its transcriptional activity; however, it can also bind pRb and activate E2F-dependent gene transcription. Additionally, LANA regulates transcription by binding to and inhibiting the histone transferase activity of CREB-binding protein (CBP). Finally, LANA associates with cellular chromatin and remains associated with chromosomes during cell division.
Recently, we have shown that immortalization of peripheral blood mononuclear cells (PBMCs) with EBV activates uch-l1. In type III EBV latency, EBV Nuclear Antigen 2 (EBNA2) forms complexes with the transcription factor PU.1, activating the uch-l1 promoter and inducing UCH-L1 RNA and protein expression (Bheda et al., 2011). In addition, HPV16-mediated transformation induces uch-l1 expression in normal keratinocytes (Rolen et al., 2009). We therefore hypothesize that in the process of cellular transformation, tumor viruses activate the uch-l1 promoter, thus inducing UCH-L1 protein expression and dysregulation of the host ubiquitin system. We now show, for the first time, that infection of normal human endothelial cells with KSHV results in increased endogenous UCH-L1 expression in these cells and that KSHV LANA along with RBP-Jκ activates the uch-l1 promoter. In addition, we demonstrate that EBV LMP1 can also activate the uch-l1 promoter and increase levels of UCH-L1. Finally, we find that in a primary effusion lymphoma (PEL) cell line dually infected with EBV and KSHV endogenous UCH-L1 RNA and protein levels are increased to a greater extent than in PELs infected with KSHV only. These observations demonstrate a mechanism by which KSHV and EBV infections lead to cell transformation and suggest that infection with multiple tumor viruses may have an additive effect on UCH-L1 expression.
MATERIALS AND METHODS
Cells
NIH 3T3 and Cos-7 cells were cultured in Dulbecco Modified Eagle Medium (DMEM) (Sigma) supplemented with 10% FBS (Sigma) and penicillin–streptomycin (Sigma). Primary Effusion Lymphoma cell lines BC-1 (contains both KSHV and EBV genomes) and BC-3 (contains KSHV), were cultured in RPMI 1640 medium (Gibco) plus 10% heat-inactivated FBS, 100 units/ml penicillin–streptomycin, 1% sodium bicarbonate (Sigma) and 0.5% -ME (Sigma). All cell lines were maintained at 37 °C in 5% CO2 in air.
Plasmids
pcDNA LANA-Flag construct was a gift from Dr. Dirk Dittmer. pECE-RBP-Jκ construct was a gift from Dr. Paul Ling. pGL3-UCH-L1 promoter reporter construct was amplified and cloned as described (Bheda et al., 2009a). pcDNA LMP1 has been previously described (Bentz et al.; Bentz et al., 2011; Ning et al., 2008).
Luciferase Reporter Assays
For luciferase assays, cells were seeded in 6-well plates and transiently transfected with the use of Fugene HD (Roche Diagnostics) with UCH-L1p-Luc promoter plasmid, β-gal constructs, and indicated effector plasmids. The total amount of DNA in all transfections was kept constant with empty vector. Luciferase assays were performed 48 h post-transfection as specified by the manufacturer (Promega). All reporter-assay results are from three independent experiments prepared in triplicate and have been normalized for β-gal activity.
Reverse Transcriptase PCR
NIH 3T3 cells were transiently transfected with a total of 2 µg of DNA with the Fugene HD reagent (Roche Diagnostics). Cells were collected 48 h post-transfection for RT-PCR analysis. Total RNA was extracted with the use of Agilent’s Total RNA isolation mini kit per manufacturer’s instructions (Agilent Technologies). 500 ng of total RNA were used for RT-PCR reactions using the one step RT-PCR kit (Qiagen) as per manufacturer’s instructions at an annealing temperature of 55°C. Samples were analyzed on 1 % agarose gel. Primers used:
UCH-L1: 5’-GGATGGCCACCTCTATGAAC-3’, 5’-AGACCTTGGCAGCGTCCT-3’
GAPDH: 5’-AGGTGAAGGTCGGAGTCAACG-3’, 5’-AGGGGTCATTGATGGCAACA-3’.
Chromatin Immunoprecipitation
ChIP assays were performed using Active Motif ChIP-IT enzymatic kit (Active Motif) per manufacturer’s instructions. KR4 cells were fixed with 37% formaldehyde (1% final concentration) for 10 min at 37°C; the reaction was stopped with cold 0.125 M glycine solution for 5 min at RT. The cells were then washed twice with PBS and collected in 0.5 ml digestion buffer with 1× protease inhibitors. Chromatin was sheared with shearing enzyme for 10 min at 37°C to obtain an average of 200–1000 bp fragments. Sheared chromatin was incubated overnight at 4°C with Protein G magnetic beads, and RBP-Jκ antibody (Santa Cruz- H50 X). Immunoprecipitations were performed per the manufacturer’s instructions; cross-linking was reversed by incubating immunoprecipitated complexes with 5 M NaCl and RNase A (final concentration 25 µg /ml) for 2 h at 65°C followed by Proteinase-K (final concentration 50 g/ml) treatment for 2 h at 42°C. PCR reactions were performed with 5 µl precipitated DNA with primer pairs flanking consensus RBP-Jκ sites in UCH-L1 promoter. PCR conditions: one cycle, 95°C for 2 min; 30 cycles at 95°C for 30 s, 55°C for 30 sec, and 72°C for 2 min, and a final extension at 72°C for 10 min. The primers used in the reaction were:
Site 1 (5’ CCTGTTGAATTTGTGCT 3’; 5’ CGCCGGTGAGATAATCTG 3’)
Site 2/3 (5’ GCTCCATACACTCAAGGAAC 3’; 5’ GCCAGACGCACTGTGA 3’)
Western blotting
Total cell lysates were resolved on 12% SDS-PAGE, transferred to PVDF membrane (GE Healthcare), blocked in 5% milk-Tris-buffered saline solution, and incubated at 4°C overnight with UCH-L1 (1:7500, Invitrogen) and GAPDH (1:5000, Sigma) antibodies followed with horse-radish peroxidase-conjugated secondary antibodies. Proteins were detected with Super Signal West Pico Chemiluminescence Detection Kit (Pierce Biotechnology, Rockford, IL, USA) and exposed to Kodak XAR-5 film.
Immunoprecipitation
KR4 cells were lysed with buffer containing 50 mM Tris-HCl, pH 7.6, 1% NP-40, 0.25% Na-deoxycholate, 1 mM EDTA, 1 mM Na3Ov4, 1 mM NaF and complete protease inhibitor mixture (Roche Diagnostics). Cell lysates were incubated with anti-Flag beads (Sigma) incubated withat 4°C overnight, washed four times with protein lysis buffer, and then eluted from protein anti-Flag beads with 2× Laemmli's buffer.
2D-Gel Electrophoresis
Cells were harvested and washed three times with PBS. The cell pellets were lysed with lysis buffer (10 mM Tris, pH 7.4, and 0.3% SDS) and incubated for 30 minutes on ice. Equivalent micrograms of protein were subjected to 2D gel electrophoresis by the UNC Proteomics Center. The 2D gels were stained with Coomasie blue. Unique spots were identified and sequenced by MALDI TOF/TOF mass spectrometry.
LMP1 Knockdown
BC-1 and BC-3 cells were transiently transfected with siRNA specific for LMP1 (siRNA LMP1 5’-GGAAUUUGCACGGACAGGCUU-3’) or with a two-base mutation (siRNA mut 5’-GGAAUGUGCACAGACAGGCUU-3’) using Amaxa® Cell Line Nucleofector® Kit V. Nucleofections were performed at 0 and 24 hours, and cells were harvested at 72 hours. RNA was isolated and RT-PCR was performed for GAPDH, UCH-L1, and LMP1 (as described above). pmaxGFP® Vector was used as a transfection control; the transfection efficiencies were approximately 30%.
RESULTS
KSHV infection of endothelial cells induces the expression of UCH-L1
Human Umbilical Vein Endothelial Cells (HUVECs) were infected with a recombinant KSHV virus expressing green fluorescent protein (GFP), and a stable KSHV-HUVEC cell line was generated as described before (Wang et al., 2006). Equivalent amounts of HUVECs and KSHV-HUVECs were subjected to 2D protein gel electrophoresis analysis, and gels were stained with Coomassie Blue dye. Spots that were differentially expressed in the HUVEC and the KSHV-HUVEC cells were excised from the gel and identified by mass spectrometry (Figure 1A). One protein upregulated in KSHV-HUVECs, but not HUVECs, was the cysteine hydrolase UCH-L1 (Figure 1A), suggesting that KSHV infection induces the expression of UCH-L1.
Figure 1. UCH-L1 expression is induced in endothelial cells after infection with KSHV.
(A) Equivalent amounts of HUVEC and KSHV-HUVEC cell lysates were subjected to 2D gel electrophoresis analysis following which the gels were stained with Coomassie blue. Two of the differentially expressed spots, identified by mass spectrometry, were UCH-L1 and GFP (arrows). (B) RNA from HUVEC and KSHV-HUVEC was isolated and subjected to RT-PCR using uch-l1 and β-actin primers. (C) Equivalent amounts of KSHVHUVEC and HUVEC cell lysates were subjected to SDS-PAGE and immunoblotted with UCH-L1 or actin antibodies. Relative expression was determined by densitometry. All results are shown as the mean ± standard deviation for experiments performed in triplicate.
To determine whether UCH-L1 was upregulated at the transcriptional level, RNA was isolated from these cells, and RT-PCR was performed. KSHV-HUVECs had increased levels of UCH-L1 RNA compared with the uninfected cells (Figure 1B). Similar results were observed when examining UCH-L1 protein levels (Figure 1C); where UCH-L1 levels in infected cells were greater than uninfected cells. Relative UCH-L1 expression was determined for experiments performed in triplicate (Figure 1B and 1C), and results showed a significant (p < 0.05) increase in UCH-L1 gene and protein expression in KSHV-infected HUVECs. These results corroborate the 2D proteomic analysis and demonstrate that KSHV infection induces the expression of UCH-L1.
KSHV LANA interacts with UCH-L1 and induces the endogenous expression of UCHL1
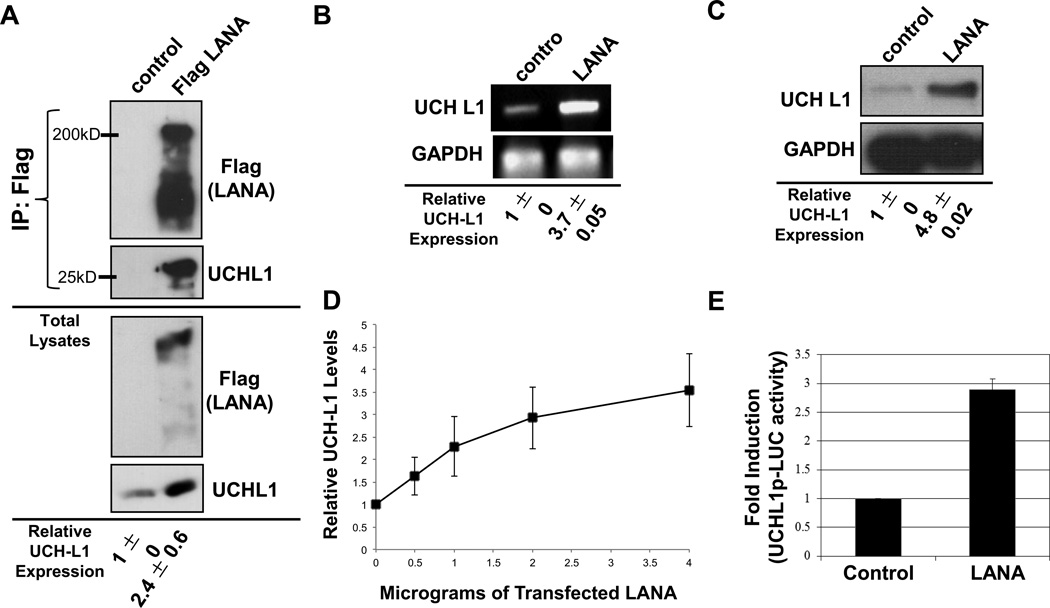
To begin to decipher the KSHV-specific protein(s) required for the observed KSHV-induced upregulation of UCH-L1 expression, we focused on LANA, which is expressed in the majority of KSHV-infected cells. First, the ability of KSHV LANA to interact with UCH-L1 was examined in Cos-7 cells, which express intermediate levels of endogenous UCH-L1 (Bheda et al., 2009a). Immunoprecipitations showed that an interaction between overexpressed LANA and endogenous UCH-L1 could be detected (Figure 2A). Additionally, when total cell lysates were probed with UCH-L1-specific antibodies, the results revealed significantly (p < 0.05) higher levels of UCH-L1 (two-fold increase) in LANA-expressing cells compared with vector-containing cells (Figure 2A). NIH 3T3 cells, which express very low levels of UCH-L1 (Bheda et al., 2009a), were used to confirm these findings. Results showed that NIH 3T3 cells transfected with Flag-LANA exhibited more than a three-fold increase (p < 0.001) in the expression of endogenous UCH-L1 RNA and protein levels (Figure 2B and 2C) when compared with control-expressing cells. In addition, LANA induced UCH-L1 expression in a dose-dependent manner (Figure 2D)
Figure 2. KSHV LANA is associated with endogenous UCH-L1 and induces expression of UCH-L1.
(A) Cos7 cells were transfected with control or LANA-Flag expression vectors and harvested 48h post-transfection for immunoprecipitation analysis. LANA-Flag was immunoprecipitated with anti-Flag-agarose beads. IPs and cell lysates were resolved on 10–12% SDS-PAGE and probed with UCH-L1 and Flag antibodies. (B–C) Total RNA and protein were extracted from cells co-transfected with LANA or control expression constructs. (B) RT-PCR analysis was performed using primers specific for uchl1 and gapdh. (C) Western blot analyses for UCH-L1 protein levels in lysates from cells transfected with or without LANA were performed with UCH-L1 antibodies. GAPDH was used as loading control. (D) Cells were transfected with different amounts of Flag-LANA or vector-control expressing plasmids and Western blot analyses to detect UCH-L1 was performed. Relative expression was determined by densitometry, and results are shown as the mean fold change ± standard deviation for experiments performed in triplicate. (E) NIH3T3 cells were co-transfected with control or LANA-Flag expression vectors (350 ng/well) along with UCH-L1p-LUC wildtype reporter plasmid (500 ng/well) and β-gal expression constructs (250 ng/well). Luciferase assays were performed 48 h post-transfection. The data are shown as the mean ± standard deviation for three independent experiments in triplicate and normalized to β-gal activity.
Because UCH-L1 expression appeared to be induced by LANA at the transcriptional level, the ability of LANA to activate the uch-l1 promoter was tested with the use of a UCH-L1p-LUC reporter construct, which contains a minimal endogenous uch-l1 promoter region (Bheda et al., 2009a). Results showed that LANA expression produced a significant (p < 0.05) three-fold increase in activation of the uch-l1 promoter when compared with control cells (Figure 2D and 2E). Together, these data indicate that KSHV LANA induces UCH-L1 expression at the transcriptional level via its ability to activate the uch-l1 promoter.
EBV increases levels of UCH-L1 in dually infected Primary Effusion Lymphoma cells
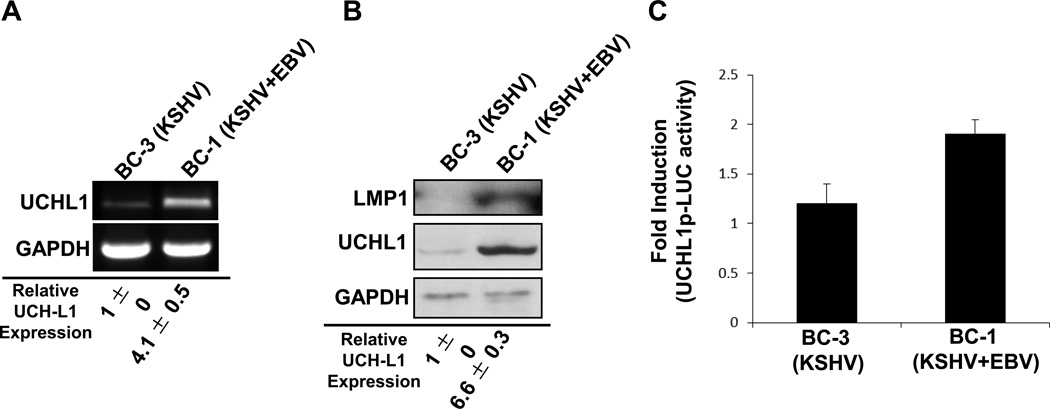
In addition to Kaposi's sarcoma, KSHV is detected in 100% of primary effusion lymphomas (PELs) (Carbone et al., 2000; Carbone and Gloghini, 2005; Fakhari et al., 2006; Sin et al., 2007). PELs, a unique form of non-Hodgkin B-cell lymphomas found only in KSHV-infected patients with AIDS, are an aggressive, rapidly progressing malignancy that is fatal (Carbone and Gloghini, 2005; Petre et al., 2007). Because LANA is one of the KSHV genes that is expressed in PELs (Fakhari et al., 2006), the effect of KSHV on uch-l1 expression in two representative PEL cell lines was examined. We reported previously that naïve B cells contain undetectable levels of UCH-L1 RNA and protein (Bheda et al., 2009a; Bheda et al., 2011). However, analysis of two PEL cell lines (BC-1 and BC-3) revealed detectable yet different levels of UCH-L1 RNA and protein (Figure 3A and 3B). BC-1 cells expressed more than four-fold higher (p < 0.05) levels of both UCH-L1 RNA and protein compared with BC-3 cells. In addition, endogenous uch-l1 promoter activity was significantly (p < 0.05) greater in BC-1 cells than in BC-3 cells. These findings demonstrate that KSHV-mediated cellular transformation can induce uch-l1 expression.
Figure 3. Endogenous UCH-L1 expression is greater in primary effusion lymphoma cells co-infected with EBV.
(A–B) Total RNA and protein were extracted from BC-3 and BC-1 cell lines. (A) RT-PCR was performed with uch-l1- and gapdh-specific primers. (B) Western blot analyses were performed with UCH-L1 and GAPDH antibodies. Relative expression was determined by densitometry. Results are shown as the mean ± standard deviation for experiments performed in triplicate. (C) Endogenous uch-l1 promoter activity was determined in BC-3 (positive for KSHV only) and BC-1 (co-infected with KSHV and EBV) PEL cells. Cells (2 × 105) were nucleofected with UCH-L1p-LUC reporter and β-gal constructs. Luciferase assays were performed 48 h post-transfection. The experiments were done in triplicate and normalized to β-gal activity.
One of the major differences between BC-1 and BC-3 cells is that BC-3 cells are only infected with KSHV while BC-1 cells are infected with both KSHV and EBV (Carbone et al., 2000; Carbone and Gloghini, 2005; Fakhari et al., 2006; Sin et al., 2007). While the role of EBV in PEL co-infection has been little explored and remains obscure, there are reports that suggest that EBV and KSHV can regulate each other’s viral gene expression (Fan et al., 2005b; Groves et al., 2001a; Krithivas et al., 2000; Xu et al., 2007).
EBV LMP1 induces the expression of UCH-L1
We have implicated EBNA2 in the EBV-mediated induction of UCH-L1 expression (Bheda et al.), however, PEL cells exhibit a restricted expression pattern of EBV proteins and lack detectable expression of EBNA2 as well as EBNA3–6 (Callahan et al., 1999). Instead, dually infected PEL cells express low levels of LMP1(Carbone et al., 2000; Carbone and Gloghini, 2005; Fakhari et al., 2006; Sin et al., 2007), a constitutively active transmembrane receptor that indirectly activates host-cell transcription (Hatzivassiliou and Mosialos, 2002; Lam and Sugden, 2003; Li and Chang, 2003; Zheng et al., 2007). Analysis of the uch-l1 promoter revealed putative binding sites for NF-κ B, STATs, AP1, c-Jun, SP1, SP3 and AP2- all of which are the major downstream targets of signaling pathways activated by LMP1.
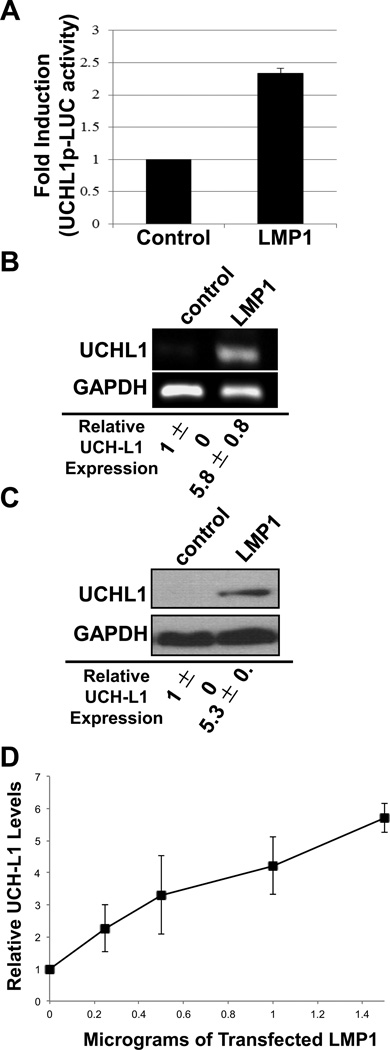
To investigate whether LMP1 can affect endogenous uch-l1 expression, reporter assays were performed to examine the activation of the endogenous uch-l1 promoter. Results showed that LMP1 expression correlated with a significant (p < 0.05) 2.5-fold increase in the activation of the uch-l1 promoter when compared with control-expressing cells (Figure 4A). In addition, over five-fold increases (p < 0.05) in levels of endogenous UCH-L1 RNA and protein were detected in cells expressing LMP1 compared with control cells (Figure 4B and 4C). LMP1 also induced the expression of UCH-L1 in a dosedependent manner (Figure 4D). These data indicate that EBV LMP1 can induce the UCHL1 endogenous expression in cells by activating its promoter.
Figure 4. EBV LMP1 induces expression of UCH-L1.
(A) NIH3T3 cells were co-transfected with control or LMP1-Flag expression vectors (250 ng/well), along with UCH-L1p-LUC wild type reporter plasmid (500 ng/well) and β-gal expression constructs (250 ng/well). Luciferase assays were performed 48 h post-transfection. The data represent three independent experiments performed in triplicate and normalized to β-gal activity. (B–C) Total RNA and protein were extracted from cells co-transfected with LMP1 or control. (B) RT-PCR was performed with specific primers for uch-l1 (gapdh was used as a control). (C) Western blot analyses for UCH-L1 protein levels in lysates from cells transfected with or without LMP1 were performed with UCH-L1 antibodies. GAPDH was the loading control. (D) Cells were transfected with different amounts of Flag-LANA or vector-control expressing plasmids and Western blot analyses to detect UCH-L1 was performed. Relative expression was determined by densitometry. Results are shown as the mean fold change ± standard deviation for experiments performed in triplicate.
KSHV LANA and EBV LMP1 together induce the expression of UCH-L1
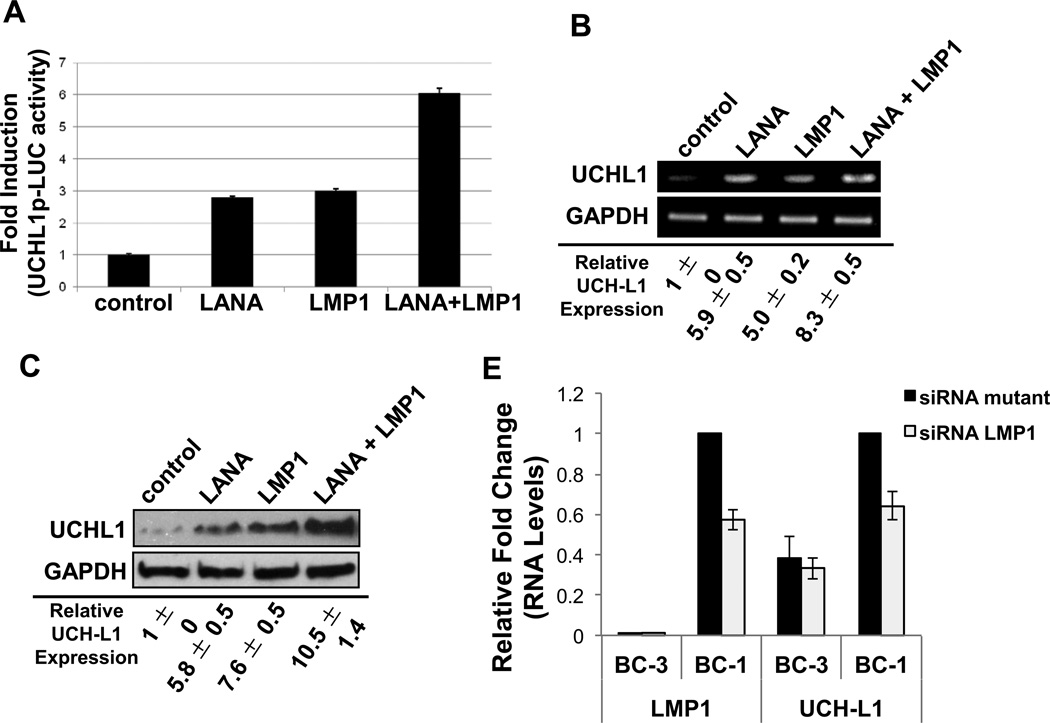
Because data showed enhanced activation of the uch-l1 promoter as well as increased UCH-L1 RNA and protein expression in cells co-infected with KSHV and EBV, we next investigated if LANA and LMP1 could have an additive effect on uch-l1 expression. Reporter assays confirmed the previous data demonstrating that expression of LANA or LMP1 resulted in significant (p < 0.05) increased activation of the uch-l1 promoter. Furthermore, when LANA and LMP1 were co-expressed, there was an additive effect on activation of the endogenous promoter (Figure 5A). These findings were confirmed by examining UCH-L1 RNA and protein levels (Figure 5B and 5C). Levels of UCH-L1 were significantly (p < 0.05) greater when LANA and LMP1 were co-expressed than when these oncoproteins were expressed alone, which was still significantly (p < 0.05) greater than in cells expressing the vector control.
Figure 5. KSHV LANA and EBV LMP1 have additive effects on the expression of UCH-L1.
(A) NIH3T3 cells were co-transfected with control, Flag-LANA, and/or pcDNA3-LMP1 expression vectors (250 ng/well), along with UCH-L1p-LUC wild type reporter plasmid (500 ng/well) and β-gal expression constructs (250 ng/well). Luciferase assays were performed 48 h post-transfection. The data represent three independent experiments performed in triplicate and normalized to β-gal activity. (B–C) Total RNA and protein were extracted from cells co-transfected with the control, Flag-LANA, and/or pcDNA3-LMP1 expression constructs. (B) RT-PCR was performed with specific primers for uch-l1 (gapdh was used as a control). (C) Western blot analyses for UCH-L1 protein levels in lysates were performed. GAPDH was the loading control. (D) BC-1 and BC-3 cells were transfected with siRNA LMP1. Transfection with a siRNA LMP1 mutant, where two nucleotides were mutated, served as a control. RNA was harvested and RT-PCR was performed with specific primers for uch-l1 and LMP1 (gapdh was used as a control). The fold change in relative uch-l1 and LMP1 RNA levels (relative to gapdh) was determined. Results are shown as the mean ± standard deviation for experiments performed in triplicate.
Further confirmation for the additive effects of LMP1 was obtained by knocking down LMP1 expression in PEL cells. BC-3 and BC-1 were transfected with either a LMP1-specific siRNA or a mutant siRNA, in which two bases were changed (Figure 5D). Results showed approximately 40% knockdown in LMP1 RNA levels in BC-1 cells (dually infected with EBV and KSHV), which corresponded with a significant (p < 0.05) 35% decrease in relative uch-l1 levels. No differences in uch-l1 expression were observed in BC-3 cells, which do not express LMP1. These findings confirm the additive effect of LMP1 on uch-l1 expression in dually infected PEL cells.
Activation of the uch-l1 promoter by RBP-Jκ
Finally, the mechanism by which LANA and LMP1 induce the uch-l1 promoter was explored. Analysis of the UCH-L1 promoter sequence with the use of PATCH software (www.gene-regulation.com) revealed 3 partial putative RBP-Jκ̃ binding sites. LANA interacts with RBP-Jκ, so the ability of LANA to interact with RBP-Jκ to activate the uch-l1 promoter was tested. Reporter assays revealed that LANA and RBP-Jκ separately activated the uchl-1 promoter to modest, yet significant (p < 0.05), levels. However, co-expression of LANA and RBP-Jκ resulted in significant (p < 0.05) additive activation of the endogenous promoter (Figure 6A). These results suggest that that KSHV LANA protein activates the uch-l1 promoter via its interaction with RBP-Jκ.
Figure 6. Activation of the uch-l1 promoter by KSHV-LANA and EBV occurs via RBP-Jκ.
(A) NIH3T3 cells were co-transfected with control or RBP-Jκ (100 ng/well) or LANA (350 ng/well) or RBP-Jκ and LANA together, along with UCHL1p-LUC wild type plasmid (500 ng/well) and β-gal expression constructs (250 ng/well). The control DNA was used as filler DNA to maintain the total amount of DNA constant. Luciferase assays were performed 48 h post-transfection. The data represent three independent experiments prepared in triplicate and normalized to β-gal activity. (B) ChIP/PCR analyses were performed to determine binding of RBP-Jκ factor to the putative partial binding sites on the UCH-L1 promoter with the use of specific RBP-Jκ antibody in KR4 LCLs. Normal IgG was used as negative control. PCR reactions were performed with primers targeting the partial RBP-Jκ-binding sites (see Materials and Methods), and amplified DNA products were resolved in 2% agarose gels.
The ability of RBP-Jκ to activate the uch-l1 promoter during EBV infection was confirmed by ChIP assays. Using the EBV-transformed B cell line KR4, which express high levels of LMP1 as well as high levels of endogenous UCH-L1 (Bheda et al., 2009a), RBP-Jκ-specific antibodies were used to pull down RBP-Jκ DNA complexes. Non-immunoprecipitated DNA was used as input DNA, and an isotype-matched IgG antibody served as negative controls. PCR analysis of portions of the uch-l1 promoter revealed that RBP-Jκ bound to the UCH-L1 promoter through at least 2 of the 3 partial RBP-Jκ binding sites (Figure 6B).
Taken together these data suggest that both KSHV and EBV encode latency-associated proteins (LANA and LMP1) that independently induce the expression of UCH-L1 through the activation of RBP-Jκ. With dual infection, which is detected in more than 60% of PELs, KSHV LANA and EBV LMP1 can have an additive effect on the induction of the expression of UCH-L1, thus potentially enhancing the tumorigenic phenotype in these cells.
Discussion
Our studies are the first to identify a role for KSHV LANA and EBV LMP1 in the activation of UCH-L1 expression. During cell transformation by either virus, the induction of the expression of UCH-L1 is a prominent cellular response (Bheda et al., 2010; Bheda et al., 2009a; Bheda et al., 2011; Bheda et al., 2009b). Because UCH-L1 expression is linked to multiple, observed tumorigenic phenotypes in cells, including increases in cell proliferation, adhesion, migration, and invasion as well as changes in cell morphology and inhibition of apoptosis, (Bheda et al., 2009a; Kim et al., 2008; Rolen et al., 2008), the data presented point to a mechanism by which both viruses can induce the expression of UCH-L1 and contribute to the oncogenicity of these viruses (Figure 7). Furthermore, the two representative PEL cell lines, one infected only with KSHV and one dually infected with KSHV and EBV, revealed that the co-infected cells expressed higher levels of UCH-L1. Indeed, co-expression of KSHV LANA and EBV LMP1 was associated with significant increased activation of the uch-l1 promoter as well as UCH-L1 RNA and protein and pointed to an additive response by these two very different viral proteins. Knockdown of LMP1 significantly lessened the additive effect of LMP1 in dually infected PEL cells. Together, these data strongly support the hypothesis that that both of these transforming human γ-herpesviruses activate the uch-l1 promoter, inducing UCH-L1 expression.
Figure 7. Proposed Model of the Induction of uch-l1 following transformation of B cells by KSHV and/or EBV.
Naïve B cells are infected and transformed by KSHV and/or EBV. EBV-mediated transformation results in EBNA2 and LMP1 expression, resulting in the activation of the uch-l1 promoter via PU.1 and RBP-Jκ, respectively, and elevated UCH-L1 expression. KSHV-mediated transformation results in LANA expression, resulting in the activation of the uch-l1 promoter via RBP-Jκ and elevated UCH-L1 expression. Transformation mediated by a dual KSHV/EBV infection results in expression of KSHV LANA and EBV LMP1. Both viral proteins independently activate the uch-l1 promoter, at least in part through RBP-Jκ, resulting in enhanced activation of the uch-l1 promoter and an additive elevation of UCH-L1 expression. Known tumorigenic phenotypes associated with elevated UCH-L1 expression include increased cell proliferation, migration, invasion, and adhesion as well as changes in cell morphology and inhibition of apoptosis.
We recently documented that EBV EBNA2 activates the uch-l1 promoter in type III EBV latency (Bheda et al.), resulting in increased UCH-L1 expression (Bheda et al.; Soni et al., 2007). However, EBNA2 cannot be responsible for up-regulation of UCH-L1 in co-infected PELs because the type III latency promoter Cp is not active in EBV-positive PEL cells, and EBNA2 is not expressed (Carbone et al., 2000; Carbone and Gloghini, 2005; Fakhari et al., 2006; Sin et al., 2007). Rather, dually infected PEL cells exhibit a restricted expression pattern of EBV products and may express low levels of LMP1 (Callahan et al., 1999). LMP1 expression was detected in co-infected PEL cells, LMP1 expression alone was capable of inducing the activation of the uch-l1 promoter and UCH-L1 expression, and knockdown of LMP1 resulted in decreased uch-l1 expression. These findings identify a second EBV latency protein that induces the expression of UCH-L1 (Figure 7). However, because LMP1 levels are low in dually infected PEL cells (Callahan et al., 1999), and knockdown of LMP1 did not completely abrogate the increase in uch-l1 expression observed in BC-1 cells compared to BC-3 cells, these findings cannot eliminate the possibility that EBNA1 (Bornkamm, 2009; Kaul et al., 2007) or EBV-encoded non-polyadenylated RNAs (EBER1 and EBER2) contribute to the up-regulation of uch-l1. Both EBNA1 and the EBERs have been shown to play roles in malignant transformation, and because we propose that during transformation oncogenic viruses activate the uch-l1 promoter; it is possible and probable that EBNA1 and the EBERs may also induce UCH-L1 expression either directly or indirectly. Therefore, the higher levels of endogenous UCH-L1 RNA and protein we detected in EBV-positive PELs (Figure 3) and EBV-transformed cells (Bheda et al.) may be the cumulative result of more than one EBV and/or KSHV products.
The role of EBV in PEL co-infection is still unclear. However, there is evidence for interactions between the two viruses: In vitro EBV infection of KSHV-infected PEL enhances the tumorigenicity of the singly infected PEL in SCID mice (Xu et al., 2007); dually-infected compared with singly KSHV-infected PELs express a unique set of cellular genes (Fan et al., 2005a); KSHV LANA activates the expression of EBV latent membrane protein 1 (LMP1) (Groves et al., 2001b), but reduces the expression of EBV EBNA1 and EBNA2 (Krithivas et al., 2000). Because we show that co-expression of LANA and LMP1 enhanced activation of the uch-l1 promoter and increased expression of UCH-L1, it is possible that their additive effects on UCH-L1 expression also occur during endogenous infection. The effect of UCH-L1 on the tumorigenic phenotypes of cells has been well documented (Bheda et al., 2009a; Kim et al., 2008; Rolen et al., 2008). We have specifically studied the cellular changes resulting from knockdown of uch-l1 in EBV-transformed B cells (Bheda et al., 2009a). Our previous results have documented functions for EBV-induced uch-l1 expression in cell proliferation, adhesion, and migration as well as inhibition of apoptosis (Bheda et al., 2009a). Similar results were observed in different cell lines (independent of viral protein expression) (Bheda et al., 2009a), suggesting a universal role for uch-l1 in these phenotypic changes regardless of cell origin and method of transformation.
The finding that KSHV LANA can itself induce the expression of EBV LMP1 (Groves et al., 2001b) suggests there is a second mechanism through which UCH-L1 levels are augmented in co-infected cells: LANA activates the expression of LMP1, which in turn activates the uch-l1 promoter, resulting in greater levels of UCH-L1. Altogether, because UCH-L1 expression is associated with the tumorigenic phenotype of transformed cells (Bheda et al., 2009a; Kim et al., 2008; Rolen et al., 2008), these data suggest that UCHL1 expression may contribute to enhanced tumorigenesis in PEL.
During EBV infection, EBNA2 interacts with PU.1 to activate the uch-l1 promoter (Bheda et al.). We now document that EBV, as well as KSHV, also induces UCH-L1 expression via RBP-Jκ. Our findings show that RBP-Jκ expression enhanced LANA-induced activation of the uch-l1 promoter and that endogenous RBP-Jκ binds to endogenous uch-l1 promoter sequences in transformed B-cells. While RBP-Jκ itself can activate the uch-l1 promoter, indicating that RBP-Jκ binds to the promoter independent of viral protein expression, the strong combined effect of LANA and RBP-Jκ co-expression on the activity of the promoter suggests that LANA interacts with RBP-Jκ and enhances activation of UCH-L1 expression. However, each factor most likely activates the endogenous promoter through independent mechanisms as well. Due to the overlap of the RBP-Jκ sites with other transcription factor binding sites, including NF-κB binding sites, we did not undertake mutational analysis of the partial RBP-Jκ binding sites, but these data do strongly suggest that RBP-Jκ is important in the activation of the uch-l1 promoter.
In addition to RBP-Jκ, LANA also interacts with SP1, STAT3, c-JUN (Verma et al., 2007), and the uch-l1 promoter has binding sites for each of these transcription factors. LMP1 also activates multiple signaling pathways resulting in the downstream activation of these factors. Therefore, SP1, STAT3, and c-JUN are likely candidates to contribute to LANA- and LMP1-induced activation of the uch-l1 promoter and will be the subject of future studies.
UCH-L1 can also up-regulate its own promoter (Bheda et al., 2009b). Previously, UCH-L1 was one of many proteins identified that co-precipitated with LANA, and here we document that LANA does interact with UCH-L1. Therefore, a third mechanism by which LANA induces UCH-L1 expression is through its interaction with UCH-L1, which would result in a positive feed-back loop, further enhancing UCH-L1 expression following KSHV-mediated transformation.
Together, these findings support our hypothesis that in the process of cellular transformation, tumor viruses activate the uch-l1 promoter, thus inducing UCH-L1 expression. We previously documented that EBV-induced transformation induced UCH-L1 expression via EBNA2 and PU.1 (Bheda et al.) (Figure 7), and now we show KSHV also induces the expression of UCH-L1. Specifically, two viral proteins, LANA and LMP1, which are essential in the viral transformation process, were documented to be sufficient to activate the uch-l1 promoter via RBP-Jκ, resulting in increased protein expression. Dual expression of LANA and LMP1, both endogenously and exogenously, had an additive effect on UCH-L1 expression (Figure 7). Therefore, in the future, it would be interesting to determine if co-infection with other pairs of viruses, such as HPV and EBV, also enhances UCH-L1 expression, which in light of their different mechanisms may combine to enhance viral pathology.
Finally, we observed that KSHV-infected endothelial cells display upregulated UCH-L1 and that KSHV LANA induces the expression of uch-l1 in endothelial cells. Thus, UCH-L1 may also play a role in the pathogenesis of Kaposi sarcoma.
Infection of endothelial cells with KSHV induced UCH-L1 expression.
KSHV LANA is sufficient for the induction of uch-l1.
Co-infection with KSHV and EBV (observed in some PELs) results in the additive induction of uch-l1.
EBV LMP1 also induced UCH-L1 expression.
LANA- and LMP1-mediated activation of the uch-l1 promoter is in part through RBP-Jκ
Acknowledgements
This work was supported by grants from the NCI (CA163217, JSP and BD; CA096500, BD) and supplemental funding from the NCI to UNC Lineberger Comprehensive Cancer Center and CFAR. GB was supported by grant CA160786. JS was supported by grant AI085545. We would like to thank Dirk Dittmer for the KSHV LANA expression plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Basseres E, Coppotelli G, Pfirrmann T, Andersen JB, Masucci M, Frisan T. The ubiquitin C-terminal hydrolase UCH-L1 promotes bacterial invasion by altering the dynamics of the actin cytoskeleton. Cellular microbiology. 2010;12:1622–1633. doi: 10.1111/j.1462-5822.2010.01495.x. [DOI] [PubMed] [Google Scholar]
- Bentz GL, Shackelford J, Pagano JS. Epstein-Barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J Virol. 86:12251–12261. doi: 10.1128/JVI.01407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz GL, Whitehurst CB, Pagano JS. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J Virol. 2011;85:10144–10153. doi: 10.1128/JVI.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, Greenamyre JT. Ubiquitin-proteasome system and Parkinson's diseases. Experimental neurology. 2005;191(Suppl 1):S17–S27. doi: 10.1016/j.expneurol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell cycle (Georgetown, Tex. 2010;9:980–994. doi: 10.4161/cc.9.5.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda A, Shackelford J, Pagano JS. Expression and functional studies of ubiquitin C-terminal hydrolase L1 regulated genes. PloS one. 2009a;4:e6764. doi: 10.1371/journal.pone.0006764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda A, Yue W, Gullapalli A, Shackelford J, Pagano JS. PU.1-dependent regulation of UCH L1 expression in B-lymphoma cells. Leukemia & lymphoma. 2011;52:1336–1347. doi: 10.3109/10428194.2011.562571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda A, Yue W, Gullapalli A, Whitehurst C, Liu R, Pagano JS, Shackelford J. Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and betacatenin/ TCF signaling. PloS one. 2009b;4:e5955. doi: 10.1371/journal.pone.0005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm GW. Epstein-Barr virus and its role in the pathogenesis of Burkitt's lymphoma: an unresolved issue. Semin Cancer Biol. 2009;19:351–365. doi: 10.1016/j.semcancer.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Callahan J, Pai S, Cotter M, Robertson ES. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: Potential switches in latent gene expression due to coinfection. Virology. 1999;262:18–30. doi: 10.1006/viro.1999.9876. [DOI] [PubMed] [Google Scholar]
- Carbone A, Cilia AM, Gloghini A, Capello D, Perin T, Bontempo D, Canzonieri V, Tirelli U, Volpe R, Gaidano G. Primary effusion lymphoma cell lines harbouring human herpesvirus type-8. Leukemia & lymphoma. 2000;36:447–456. doi: 10.3109/10428190009148391. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A. AIDS-related lymphomas: from pathogenesis to pathology. British journal of haematology. 2005;130:662–670. doi: 10.1111/j.1365-2141.2005.05613.x. [DOI] [PubMed] [Google Scholar]
- Fakhari FD, Jeong JH, Kanan Y, Dittmer DP. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J Clin Invest. 2006;116:735–742. doi: 10.1172/JCI26190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Kim SC, Chima CO, Israel BF, Lawless KM, Eagan PA, Elmore S, Moore DT, Schichman SA, Swinnen LJ, Gulley ML. Epstein-Barr viral load as a marker of lymphoma in AIDS patients. J Med Virol. 2005a;75:59–69. doi: 10.1002/jmv.20238. [DOI] [PubMed] [Google Scholar]
- Fan W, Bubman D, Chadburn A, Harrington WJ, Cesarman E, Knowles DM. Distinct subsets of primary effusion lymphoma can be identified based on their cellular gene expression profile and viral association. Journal of Virology. 2005b;79:1244–1251. doi: 10.1128/JVI.79.2.1244-1251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friborg J, Hamilton-Therkildsen M, Homoe P, Kristensen C, Hui A, Liu FF, Weinreb I. A spectrum of basaloid morphology in a subset of EBV-associated "lymphoepithelial carcinomas" of major salivary glands. Head and neck pathology. 6:445–450. doi: 10.1007/s12105-012-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Hayward SD, Yokosawa H. Molecular piracy: manipulation of the ubiquitin system by Kaposi's sarcoma-associated herpesvirus. Reviews in medical virology. 2007;17:405–422. doi: 10.1002/rmv.549. [DOI] [PubMed] [Google Scholar]
- Groves AK, Cotter MA, Subramanian C, Robertson ES. The latency-associated nuclear antigen encoded by Kaposi's Sarcoma-Associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. Journal of Virology. 2001a;75:9446–9457. doi: 10.1128/JVI.75.19.9446-9457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Cotter MA, Subramanian C, Robertson ES. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J Virol. 2001b;75:9446–9457. doi: 10.1128/JVI.75.19.9446-9457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou E, Mosialos G. Cellular signaling pathways engaged by the Epstein-Barr virus transforming protein LMP1. Front Biosci. 2002;7:d319–d329. doi: 10.2741/hatziva. [DOI] [PubMed] [Google Scholar]
- Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am J Pathol. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowaty MN, Frappier L. HAUSP/USP7 as an Epstein-Barr virus target. Biochem Soc Trans. 2004;32:731–732. doi: 10.1042/BST0320731. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J Biol Chem. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- Jang KL, Shackelford J, Seo SY, Pagano JS. Up-regulation of beta-catenin by a viral oncogene correlates with inhibition of the seven in absentia homolog 1 in B lymphoma cells. Proc Natl Acad Sci U S A. 2005;102:18431–18436. doi: 10.1073/pnas.0504054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuta T, Setsuie R, Mitsui T, Kinugawa A, Sakurai M, Aoki S, Uchida K, Wada K. Aberrant molecular properties shared by familial Parkinson's disease-associated mutant UCH-L1 and carbonyl-modified UCH-L1. Human molecular genetics. 2008;15:1482–1496. doi: 10.1093/hmg/ddn037. [DOI] [PubMed] [Google Scholar]
- Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81:10352–10361. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2008 doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- Krithivas A, Young DB, Liao GL, Greene D, Hayward SD. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. Journal of Virology. 2000;74:9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sakurai M, Sato Y, Lee WW, Ishii Y, Kyuwa S, Noda M, Wada K, Yoshikawa Y. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol Reprod. 2004;71:515–521. doi: 10.1095/biolreprod.104.027565. [DOI] [PubMed] [Google Scholar]
- Lam N, Sugden B. CD40 and its viral mimic, LMP1: similar means to different ends. Cell Signal. 2003;15:9–16. doi: 10.1016/s0898-6568(02)00083-9. [DOI] [PubMed] [Google Scholar]
- Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- Li HP, Chang YS. Epstein-Barr virus latent membrane protein 1: structure and functions. J Biomed Sci. 2003;10:490–504. doi: 10.1007/BF02256110. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Loeffler-Ragg J, Skvortsov S, Sarg B, Skvortsova I, Witsch-Baumgartner M, Mueller D, Lindner H, Zwierzina H. Gefitinib-responsive EGFR-positive colorectal cancers have different proteome profiles from non-responsive cell lines. Eur J Cancer. 2005;41:2338–2346. doi: 10.1016/j.ejca.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Long EM, Long MA, Tsirigotis M, Gray DA. Stimulation of the murine Uchl1 gene promoter by the B-Myb transcription factor. Lung Cancer. 2003;42:9–21. doi: 10.1016/s0169-5002(03)00279-4. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Nakayama S, Torikoshi Y, Tanaka S, Ishihara H, Taguchi T, Tamaki Y, Noguchi S. High expression of ubiquitin carboxy-terminal hydrolase-L1 and -L3 mRNA predicts early recurrence in patients with invasive breast cancer. Cancer Sci. 2006;97:523–529. doi: 10.1111/j.1349-7006.2006.00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Campos AD, Darnay BG, Bentz GL, Pagano JS. TRAF6 and the three C-terminal lysine sites on IRF7 are required for its ubiquitination-mediated activation by the tumor necrosis factor receptor family member latent membrane protein 1. Mol Cell Biol. 2008;28:6536–6546. doi: 10.1128/MCB.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Pagano JS. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J Virol. 84:6130–6138. doi: 10.1128/JVI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, Harada T, Hara Y, Kimura I, Chiba S, Namikawa K, Kiyama H, Noda M, Aoki S, Wada K. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Human molecular genetics. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- Otsuki T, Yata K, Takata-Tomokuni A, Hyodoh F, Miura Y, Sakaguchi H, Hatayama T, Hatada S, Tsujioka T, Sato Y, Murakami H, Sadahira Y, Sugihara T. Expression of protein gene product 9.5 (PGP9.5)/ubiquitin-C-terminal hydrolase 1 (UCHL-1) in human myeloma cells. British journal of haematology. 2004;127:292–298. doi: 10.1111/j.1365-2141.2004.05205.x. [DOI] [PubMed] [Google Scholar]
- Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci U S A. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano J. EBV Diseases. Springer; 2009. [Google Scholar]
- Petre CE, Sin SH, Dittmer DP. Functional p53 signaling in Kaposi's sarcoma-associated herpesvirus lymphomas: implications for therapy. J Virol. 2007;81:1912–1922. doi: 10.1128/JVI.01757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolen U, Freda E, Xie J, Pfirmann T, Frisan T, Masucci MG. The Ubiquitin C-terminal Hydrolase UCH-L1 regulates B-cell proliferation and integrin activation. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolen U, Freda E, Xie JJ, Pfirrmann T, Frisan T, Masucci MG. The ubiquitin C-terminal hydrolase UCH-L1 regulates B-cell proliferation and integrin activation. Journal of Cellular and Molecular Medicine. 2009;13:1666–1678. doi: 10.1111/j.1582-4934.2008.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol. 2009;83:4652–4669. doi: 10.1128/JVI.02408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochemistry international. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shackelford J, Pagano JS. Targeting of host-cell ubiquitin pathways by viruses. Essays in biochemistry. 2005;41:139–156. doi: 10.1042/EB0410139. [DOI] [PubMed] [Google Scholar]
- Shackelford J, Pagano JS. Role of the ubiquitin system and tumor viruses in AIDS-related cancer. BMC biochemistry. 2007;8(Suppl 1):S8. doi: 10.1186/1471-2091-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin SH, Roy D, Wang L, Staudt MR, Fakhari FD, Patel DD, Henry D, Harrington WJ, Jr, Damania BA, Dittmer DP. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood. 2007;109:2165–2173. doi: 10.1182/blood-2006-06-028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni V, Cahir-McFarland E, Kieff E. LMP1 TRAFficking activates growth and survival pathways. Advances in experimental medicine and biology. 2007;597:173–187. doi: 10.1007/978-0-387-70630-6_14. [DOI] [PubMed] [Google Scholar]
- Verma SC, Lan K, Robertson E. Structure and function of latency-associated nuclear antigen. Current topics in microbiology and immunology. 2007;312:101–136. doi: 10.1007/978-3-540-34344-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dittmer DP, Tomlinson CC, Fakhari FD, Damania B. Immortalization of primary endothelial cells by the K1 protein of Kaposi's sarcoma-associated herpesvirus. Cancer Research. 2006;66:3658–3666. doi: 10.1158/0008-5472.CAN-05-3680. [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nature cell biology. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Xu DS, Coleman T, Zhang J, Fagot A, Kotalik C, Zhao LJ, Trivedi P, Jones C, Zhang LW. Epstein-Barr virus inhibits Kaposi's sarcoma-associated herpesvirus lytic replication in primary effusion lymphomas. Journal of Virology. 2007;81:6068–6078. doi: 10.1128/JVI.02743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YC, Li X, Chen W. Characterization of genes associated with different phenotypes of human bladder cancer cells. Acta Biochim Biophys Sin (Shanghai) 2006;38:602–610. doi: 10.1111/j.1745-7270.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Ying H, Xiao ZX. Targeting retinoblastoma protein for degradation by proteasomes. Cell cycle (Georgetown, Tex. 2006;5:506–508. doi: 10.4161/cc.5.5.2515. [DOI] [PubMed] [Google Scholar]
- Zheng H, Li LL, Hu DS, Deng XY, Cao Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cellular & molecular immunology. 2007;4:185–196. [PubMed] [Google Scholar]