Abstract
Activators of σ54-RNA polymerase holoenzyme couple ATP hydrolysis to formation of an open complex between the promoter and RNA polymerase. These activators are modular, consisting of an N-terminal regulatory domain, a C-terminal DNA-binding domain, and a central activation domain belonging to the AAA+ superfamily of ATPases. The AAA+ domain of Sinorhizobium meliloti C4-dicarboxylic acid transport protein D (DctD) is sufficient to activate transcription. Deletion analysis of the 3′ end of dctD identified the minimal functional C-terminal boundary of the AAA+ domain of DctD as being located between Gly-381 and Ala-384. Histidine-tagged versions of the DctD AAA+ domain were purified and characterized. The DctD AAA+ domain was significantly more soluble than DctD(Δ1-142), a truncated DctD protein consisting of the AAA+ and DNA-binding domains. In addition, the DctD AAA+ domain was more homogeneous than DctD(Δ1-142) when analyzed by native gel electrophoresis, migrating predominantly as a single high-molecular-weight species, while DctD(Δ1-142) displayed multiple species. The DctD AAA+ domain, but not DctD(Δ1-142), formed a stable complex with σ54 in the presence of the ATP transition state analogue ADP-aluminum fluoride. The DctD AAA+ domain activated transcription in vitro, but many of the transcripts appeared to terminate prematurely, suggesting that the DctD AAA+ domain interfered with transcription elongation. Thus, the DNA-binding domain of DctD appears to have roles in controlling the oligomerization of the AAA+ domain and modulating interactions with σ54 in addition to its role in recognition of upstream activation sequences.
Bacteria often possess a form of RNA polymerase holoenzyme that has σ54 as its sigma factor, referred to as σ54-RNA polymerase holoenzyme or σ54-holoenzyme (1, 20). σ54-Holoenzyme regulates the expression of genes involved in a variety of cellular processes, including nitrogen assimilation and fixation, C4-dicarboxylic acid transport, degradation of aromatic compounds, hydrogen metabolism, flagellar biogenesis, and response to phage infection (13, 20, 39). σ54-Holoenzyme binds to the promoter to form a stable closed complex, but isomerization of this complex to an open complex that can initiate transcription requires a transcriptional activator (21, 30, 37). Activators of σ54-holoenzyme usually bind to sites located 70 bp or more upstream of the promoter, known otherwise as upstream activation sequences (UAS) or enhancers (2, 32). After binding the UAS or enhancer, the activator contacts σ54-holoenzyme bound at the promoter through DNA looping (33, 40). Productive contacts between the activator and σ54-holoenzyme lead to formation of an open complex in a reaction that requires ATP hydrolysis by the activator (46).
Activators of σ54-holoenzyme are modular in structure, generally consisting of an N-terminal regulatory domain, a central domain required for ATP hydrolysis and transcriptional activation, and a C-terminal DNA-binding domain (39). The central domain is well conserved among σ54-dependent activators and for some activators is sufficient to activate transcription if it is expressed at higher than normal levels (7, 8). This domain belongs to the AAA+ superfamily (ATPases associated with various cellular activities) of ATPases (23, 28).
AAA+ proteins are found in all three domains of life and are involved in a variety of cellular processes, including membrane fusion, proteolysis, DNA replication, and transcription (23, 28). In general, the diverse functions of AAA+ proteins are manifested through chaperone-like activities that reorganize protein-protein or protein-DNA interactions (28, 42). AAA+ proteins undergo conformational changes during the ATP hydrolysis cycle which are thought to exert force on the bound substrate, allowing these proteins to carry out their respective biological activities (4, 36).
AAA+ proteins form oligomeric ring structures that often consist of six subunits but may have from five to eight subunits depending on the protein (28, 42, 47). Crystal structures for AAA+ proteins show that the active site for ATP hydrolysis is located between adjacent subunits within the ring structure (28, 42, 47). The ring structures of AAA+ proteins can be created through the regulated assembly of monomers or dimers, the formation a stable ring upon assembly of the subunits, or the formation of a ring from a single polypeptide chain containing multiple AAA+ domains (42). Activators of σ54-holoenzyme appear to form ring structures through the regulated assembly of subunits (18). Crystal structures were reported recently for the ADP-bound form of the ATPase domain alone and the ATPase and adjacent regulatory domains of Aquifex aeolicus NtrC1, a σ54-dependent activator (18). The protein consisting of the ATPase and adjacent regulatory domains crystallized as a dimer, while the ATPase domain by itself crystallized as a heptameric ring.
C4-dicarboxylic acid transport protein D (DctD) activates transcription of dctA, which encodes a permease for C4-dicarboxylic acids in Sinorhizobium meliloti and Rhizobium leguminosarum (25, 34). Removal of the N-terminal regulatory domain of DctD was shown previously to result in a constitutively active form of the protein (6, 17). A large collection of mutant forms of DctD that fail to activate transcription has been generated and characterized (5, 43, 44), and further structural information about these proteins will assist in understanding the function of DctD and other σ54-dependent activators.
We report here on the deletion analysis of S. meliloti DctD, which allowed us to precisely define the minimal functional C terminus of the AAA+ domain of this σ54-dependent activator. The purified DctD AAA+ domain was shown to bind σ54, hydrolyze ATP, and activate transcription in vitro. Interestingly, the DctD AAA+ domain was significantly more soluble than DctD(Δ1-142), a truncated DctD comprised of the AAA+ and C-terminal DNA-binding domains, suggesting that it may be more amenable for structural studies than DctD(Δ1-142).
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli strain DH5α [φ80lacZΔM15 recA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] was used for cloning procedures. E. coli strain JM109 (endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZ ΔM15]) was used to assay the in vivo activities of the products of the mutant dctD genes with 3′ deletions. The histidine-tagged DctD AAA+ domains were expressed in E. coli strain BL21(DE3) [F− ompT(lon) hsdSB(rB− mB−) dcm galλ(DE3)]. Concentrations of antibiotics in the media for selection were 200 μg of ampicillin/ml, 20 μg of kanamycin/ml, and 10 μg of tetracycline/ml.
Construction of 3′ deletions of dctD.
Plasmid pL143 contains codons 143 to 460 of S. meliloti dctD fused to codon 11 of E. coli lacZ from pUC13 and expresses DctD(Δ1-142), which is a truncated, constitutively active form of DctD (15). Plasmids containing various 3′ deletions of dctD were generated by digesting pL143 with EcoRV and then treating the linearized plasmid with nuclease Bal31. EcoRV cuts within dctD between codons 404 and 405. Following treatment with Bal31, the ends of the DNA were filled in with T4 DNA polymerase and the plasmid was digested with HindIII, which cuts upstream of dctD. DNA fragments were isolated from low-melting-point agarose gels and ligated into the HindIII and HincII sites of pUCX, which has translation termination signals in all three reading frames immediately downstream of the HincII site. Plasmid pUCX was constructed by inserting the XbaI linker 5′-CTAGTCTAGACTAG-3′ into the SmaI site of pUC13. Underlined sequences indicate the translation termination signals that were introduced in each of the three reading frames. The fusions in all constructs that were analyzed further were sequenced to identify the 3′ end of dctD and to verify that mutant DctD(Δ1-142) proteins of the expected size were expressed.
Construction of plasmids for expression of DctD AAA+ module.
Plasmid pHX182 carries a truncated S. meliloti dctD allele consisting of codons 141 to 390 which encodes the DctD AAA+ domain and is under control of the T7 promoter in vector pET28a (Novagen). A sequence encoding a hexahistidine tag (amino acid sequence of the tag is MGSSHHHHHHSSGLVPRGSHM) occurs at the 5′ end of the dctD sequence in plasmid pHX182. The dctD allele in plasmid pHX182 was amplified by PCR using the primers 5′-GGCCGAAGCCCATATGGAAGGCCTGC-3′ and 5′-CAATCTTTCCTCGAGGGTCGCTCAGCTTGAGG-3′ with plasmid pYKW3 (44) as the template. Underlined sequences indicate restriction sites for NdeI and XhoI, respectively, and the italics indicate a base change that introduced a translational stop signal. The amplified DNA fragment was inserted into pET28a that had been digested previously with NdeI and XhoI.
Plasmid pHX183 carries a truncated S. meliloti dctD allele consisting of codons 141 to 394 and is also under control of the T7 promoter in plasmid pET28a. A sequence encoding a hexahistidine tag (amino acid sequence of the tag is EHHHHHH) occurs at the 3′ end of the dctD sequence in plasmid pHX183. The dctD allele in plasmid pHX183 was amplified with the primers 5′-CAATCTTTCCTCGAGGGTCGCTC-3′ and 5′-CGAAGCCGCCATGGAAGGCCTGC-3′, with pYKW3 as the template. Underlined sequences indicate restriction sites for XhoI and NcoI, respectively, which were introduced so that the amplified DNA fragment could be cloned into the same sites in plasmid pET28a.
β-Galactosidase assays.
The ability of the various mutant forms of DctD to activate transcription was monitored from dctA′-′lacZ reporter genes carried on pRKRMAZ:+UAS or pRKRMAZ:−UAS (15), which possess or lack the dctA UAS, respectively. E. coli cultures were grown in Luria-Bertani broth, and expression of the DctD proteins was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside to the growth medium as described previously (6). Cells were harvested by centrifugation, cell extracts were prepared by sonication, and spectrophotometric assays of β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside as a substrate were done as described previously (14).
Expression and purification of DctD proteins.
The histidine-tagged DctD AAA+ domain proteins were expressed in E. coli strain BL21 (DE3) by growing the cells in Luria-Bertani broth to an optical density at 600 nm of ∼0.4, at which point 0.5 mM isopropyl-β-d-thiogalactopyranoside was added to the growth medium. After 3 to 4 h, cells were harvested by centrifugation, resuspended in buffer A (20 mM N-[2-hydroxyethyl]piperazine-N′-3-propanesulfonic acid [pH 8.0], 65 mM potassium thiocyanate, 5% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and lysed in a French pressure cell at 12,000 lb/in2. Crude cell lysates were clarified by centrifugation at 10,000 × g for 40 min. The resulting supernatant liquid was applied to a nickel-nitrilotriacetic acid resin (Qiagen), and the histidine-tagged DctD AAA+ domain proteins were eluted from the column with 250 mM imidazole as per the supplier's recommendations. Column fractions containing the histidine-tagged proteins were pooled, dialyzed against buffer B (20 mM Tris-HCl [pH 8.8], 20 mM potassium thiocyanate, 5% glycerol, 1 mM dithiothreitol), and applied to a HiTrap Q anion exchange column (5 ml; Pharmacia). The histidine-tagged DctD AAA+ domain proteins were eluted from the anion exchange column with ∼200 mM KCl in a linear gradient to 1 M KCl in buffer B. DctD(Δ1-142) and an N-terminal histidine-tagged version of DctD(Δ1-142) were purified as described previously (17, 44).
Expression and purification of σ54 proteins.
Plasmid pHX185 is a derivative of plasmid pET28a that carries the S. meliloti rpoN gene (encodes σ54) under control of the T7 promoter in the vector. In plasmid pHX185, S. meliloti rpoN is joined at its 5′ end to a sequence encoding a hexahistidine tag (sequence of the tag is MGSSHHHHHHGLVPRGSHN). S. meliloti rpoN was amplified from plasmid pNtr3.5BE (35) with the primers 5′-GGAGTTTCACACCCATATGGCCTTG-3′ and 5′-CCCGTCATCCAAGCTTTTCAGCGACCGG-3′. Underlined sequences indicate restriction sites for NdeI and HindIII, respectively, which were introduced so that the amplified DNA fragment could be cloned into the same sites in plasmid pET28a.
The histidine-tagged R. meliloti σ54 was expressed in E. coli strain BL21(DE3) by growing the cells in Luria-Bertani broth to an optical density at 600 nm of ∼0.4, at which point 0.5 mM isopropyl-β-d-thiogalactopyranoside was added to the growth medium. After 3 to 4 h, cells were harvested by centrifugation, resuspended in buffer A, and lysed in a French pressure cell at 12,000 lb/in2. Crude cell lysates were clarified by centrifugation at 10,000 × g for 40 min. The resulting supernatant liquid was applied to a nickel-nitrilotriacetic acid resin column, and the histidine-tagged σ54 was eluted from the column with 250 mM imidazole. Fractions containing histidine-tagged σ54 were pooled, dialyzed against buffer C [20 mM N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid (pH 8.0), 20 mM potassium thiocyanate, 5% glycerol, 1 mM dithiothreitol], and applied to a HiTrap heparin column (5 ml; Pharmacia). The histidine-tagged σ54 was eluted with ∼350 mM KCl in a linear gradient to 1 M KCl in buffer C. Native and N-terminal histidine-tagged Salmonella enterica serovar Typhimurium σ54 proteins were expressed and purified as described previously (11, 17).
Native gel analysis and σ54-binding assays for DctD proteins.
The mobilities of proteins on native gels were analyzed after diluting purified DctD(Δ1-142), DctD (141-390), DctD (141-394), or PspFΔHTH to a final concentration of 5 μM monomer in 20 μl of STA buffer (25 mM Tris-acetate [pH 8.0], 8 mM magnesium acetate, 100 mM KCl, 1 mM dithiothreitol, and 3.5% [wt/vol] PEG 6000). PspFΔHTH was kindly provided by Chih Lew (University of California at Los Angeles). Where indicated, ADP-aluminum fluoride (ADP · AlFx, where x is 3 or 4) was prepared in situ in assays by mixing 0.2 mM ADP and 5 mM NaF with the proteins in the STA buffer for 5 min at 30°C and then adding 0.2 mM AlCl3. After 10 min of incubation, samples were loaded onto a 7.5% native polyacrylamide gel and subjected to electrophoresis at 15 V/cm. The electrophoresis and gel buffer contained 25 mM Tris base and 200 mM glycine (pH 8.6). Proteins were visualized by silver staining of the resulting gel using a protein silver staining kit (Amersham Pharmacia) as per the supplier's protocol. Assays for complex formation between σ54 and DctD proteins were carried out essentially as described previously (3). Proteins were diluted into 20 μl of STA buffer, and ADP · AlFx was generated in situ as described above. Final protein concentrations in the reaction mixtures were 1 μM σ54 and 1 μM DctD monomer. Protein complexes were resolved by native gel electrophoresis as described above and visualized by either silver staining or Western blotting, using antiserum directed against S. enterica serovar Typhimurium σ54 or S. meliloti DctD.
ATPase and in vitro transcription assays.
ATP hydrolysis assays were done essentially as described previously (43), using 5 mM (monomer) concentrations of the DctD proteins. Single-round transcription assays were performed as described previously (10). The supercoiled template used was plasmid pJES534, which carried the S. enterica serovar Typhimurium glnA promoter and yields a 155-base transcript that contains no uracil (31). Plasmid pJES534 is reported to have a synthetic module of six directly repeated double-stranded oligonucleotides lacking T residues (21 bp each; AGACACCACAGAGACCACACACA) located at position +28 relative to the glnA promoter and immediately followed by a T-rich sequence that functions in termination of transcription (31). DNA sequencing of plasmid pJES534, however, showed that there are seven directly repeated oligonucleotide elements (B. T. Nixon, unpublished results). Reaction mixtures contained 0.1 to 5 μM DctD AAA+ domain, 60 nM E. coli core RNA polymerase (Epicentre), 100 nM histidine-tagged S. enterica serovar Typhimurium σ54, 20 nM plasmid DNA, 5 U of anti-RNase (Ambion), 4 mM ATP, 400 μM GTP, 5 μM CTP, and 7.5 μCi of [α-32P]CTP (3,000 Ci/mmol). Proteins and DNA template were incubated for 5 min at 37°C, after which ATP was added to stimulate open complex formation. After 10 min of additional incubation, the remaining nucleotides were added to allow synthesis of the transcripts. Heparin was added to the reactions to a concentration of 0.1 mg/ml along with the nucleotides to prevent transcription reinitiation. The resulting transcripts were analyzed on a 6% denaturing polyacrylamide gel, which was exposed to X-ray film to visualize the transcripts.
RESULTS
Identification of the functional C-terminal end of the AAA+ domain of S. meliloti DctD.
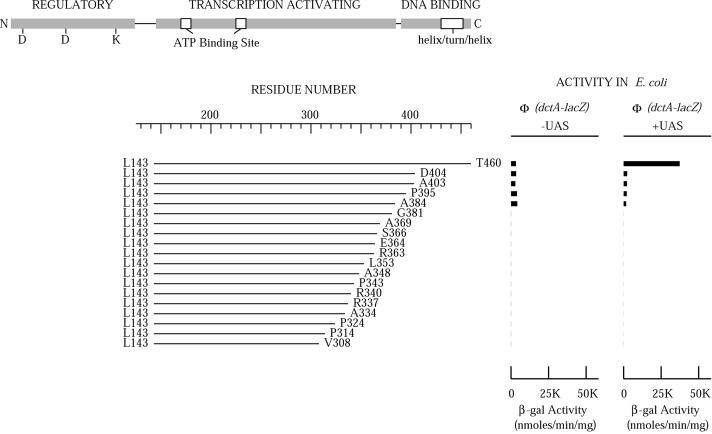
Crystal structures of AAA+ proteins revealed that the AAA+ domain consists of two subdomains, an N-terminal subdomain that has an α/β fold with a nucleotide binding pocket and a C-terminal α-helical subdomain (28). The C-terminal subdomain is variable in size and structurally less conserved than the N-terminal subdomain. To determine the C-terminal boundary of the AAA+ domain of DctD, we generated a series of C-terminal deletion mutants of DctD and examined the activities of these mutant proteins in E. coli. A truncated, constitutively active form of DctD that lacks the N-terminal regulatory domain, referred to as DctD(Δ1-142) (17), was used for this deletion analysis. The abilities of the truncated DctD proteins to activate transcription were assessed in E. coli with dctA′-′lacZ reporter genes that either possessed or lacked the UAS.
The C terminus of wild-type DctD is Thr-460. Eighteen different C-terminal deletion mutants of DctD(Δ1-142) were generated, with the C termini of these mutant proteins ranging from Val-308 to Asp-404 (Fig. 1). Residues Val-416 through Gln-435 are predicted to form a helix-turn-helix motif that recognizes the dctA UAS (8). All of the C-terminal deletions of DctD(Δ1-142) lacked the helix-turn-helix motif, and as expected, those that retained activity were unable to discriminate between the reporter gene that possessed the UAS and the one that lacked the UAS (Fig. 1). Mutants with deletions that extended up to Ala-384 retained the ability to activate transcription, whereas those with deletions that extended up to Gly-381 or beyond were unable to activate transcription. These data indicated that the minimal functional C-terminal boundary of the AAA+ domain of DctD lies between Ala-385 (the residue adjacent to Ala-384) and Gly-381.
FIG. 1.
In vivo activities of C-terminally deleted DctD(Δ1-142) proteins. The diagram at the top shows the three functional domains of DctD. The relative positions of essential aspartate and lysine residues are indicated in the regulatory (receiver) domain. The relative positions of Walker A and B sequences involved in ATP binding are indicated for the central transcription-activating (AAA+) domain, as is the position of the putative helix-turn-helix motif in the DNA-binding domain. The line diagrams indicate the N-terminal and C-terminal ends of the various DctD deletion mutants generated in this study. Thr-460 is the C-terminal residue of the native protein. The activities of the various mutant forms of DctD in E. coli at dctA′-′lacZ reporter genes that either possessed or lacked the UAS are indicated to the right of the line diagrams. None of the DctD protein deletions that extended beyond Ala-384 had activity that was above background levels.
Expression and purification of the DctD AAA+ domain.
A DNA fragment bearing codons 141 to 390 of S. meliloti dctD was cloned into the expression vector pET28a to introduce a sequence encoding a hexahistidine tag at the 5′ end of the dctD sequence [protein referred to as DctD(141-390)]. Ser-390 rather than Ala-384 was chosen for the C-terminal end of the DctD AAA+ domain, since this residue corresponds to the C termini of Helicobacter pylori FlgR and σ54-dependent activators from several chlamydial species, all of which lack the C-terminal DNA-binding domain found in most other σ54-dependent activators (12, 38). In addition, threading of the DctD sequence onto the A. aeolicus NtrC1 structure modeled Ala-384 to Ser-390 as the last helix of the AAA+ α-helical subdomain. A second expression vector for the DctD AAA+ domain was generated by cloning a DNA fragment bearing codons 141 to 394 of S. meliloti dctD into plasmid pET28a. This second plasmid introduced a methionine start codon in front of codon 141 and a sequence encoding a hexahistidine tag located 3′ to codon 394 (protein referred to as DctD(141-394)). Codon 394 was chosen as the 3′ end of the dctD sequence, since it was convenient to introduce a XhoI restriction site at this position for the cloning procedure. Both of these proteins activated transcription from a dctA′-′lacZ reporter gene in vivo, and as expected, failed to discriminate between a dctA′-′lacZ reporter that possessed the UAS and one that lacked the UAS (data not shown).
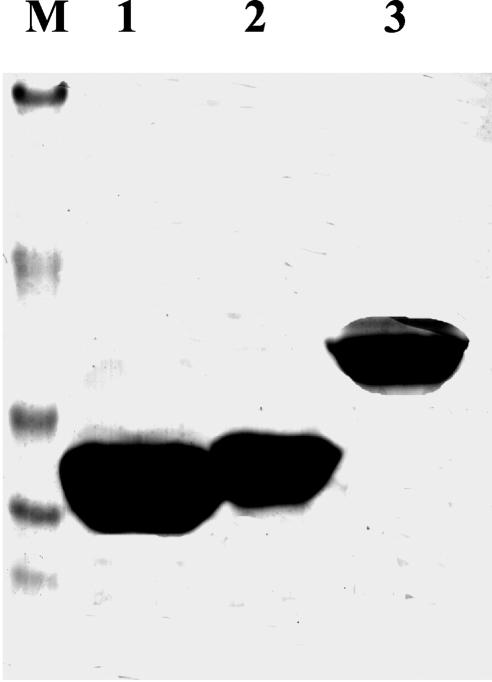
Both DctD(141-390) and DctD(141-394) were purified by affinity chromatography followed by anion exchange chromatography, which resulted in highly purified preparations of the two proteins (Fig. 2). The predicted sizes of DctD(141-390) and DctD(141-394) were 29,444 Da and 28,575 Da, respectively, which was consistent with their migration in the sodium dodecyl sulfate-polyacrylamide gel. Purified DctD(141-390) was soluble to at least 30 mg/ml, while DctD(141-394) was soluble to at least 8 mg/ml. This result was unexpected, since DctD(Δ1-142) is only soluble to about 2mg/ml (17).
FIG. 2.
Purified DctD(Δ1-142) and DctD AAA+ domain proteins. Purified DctD proteins (20 μg each) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were visualized in the gel by staining with Coomassie blue. Lane 1, DctD(141-390); lane 2, DctD(141-394); lane 3, DctD(Δ1-142). Lane M contains the molecular weight markers: bovine serum albumin (98,000), ovalbumin (54,600), carbonic anhydrase (37,400), soybean trypsin inhibitor (29,600), and lysozyme (20,400).
Self-association of the DctD AAA+ domain occurs independently of nucleotides.
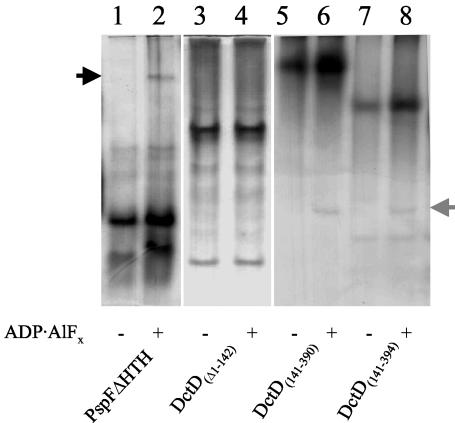
The native gel mobility of E. coli PspFΔHTH, a truncated form of phage shock protein F (PspF) that lacks most of the DNA-binding domain, is altered by the addition of ADP · AlFx (3). The change in native gel mobility of PspFΔHTH caused by ADP · AlFx is attributed to the self-association of the protein, resulting in the formation of a higher-order oligomer (3). To determine if self-association of the DctD AAA+ domain proteins or DctD(Δ1-142) is effected similarly by ADP · AlFx, we examined the behavior of these proteins in a native gel electrophoresis system in the presence or absence of ADP · AlFx. As reported previously (3), ADP · AlFx caused a change in the native gel mobility of PspFΔHTH, which we infer results from an increased association state of the protein (Fig. 3, lanes 1 and 2).
FIG. 3.
Native protein gel mobilities of PspFΔHTH, DctD(Δ1-142), DctD(141-390), and DctD(141-394) in the presence and absence of ADP · AlFx. Approximately 3 μg of purified protein was incubated in the absence (odd-numbered lanes) or presence (even-numbered lanes) of ADP · AlFx. Following electrophoresis, proteins were visualized in the gel by silver staining. Lanes 1 and 2, PspFΔHTH; lanes 3 and 4, DctD(Δ1-142); lanes 5 and 6, DctD(141-390); lanes 7 and 8, DctD(141-394). The arrow to the left of lane 1 indicates the higher-order oligomer of PspFΔHTH that is stabilized by ADP · AlFx. The arrow to the right of lane 8 indicates forms of DctD(141-390) and DctD(141-394) that were observed when the proteins were incubated with ADP · AlFx.
In contrast, the native gel mobility of DctD(Δ1-142) remained unchanged in the presence of ADP · AlFx (Fig. 3, lanes 3 and 4). DctD(Δ1-142) displayed several distinct protein bands on the native gel which represent either different conformations or different oligomeric forms of the protein. In contrast to DctD(Δ1-142), DctD(141-390) and DctD(141-394) migrated predominantly on the native gel as single species in the absence or presence of ADP · AlFx with mobilities that were similar to that of the PspFΔHTH higher-order complex (Fig. 3, lanes 5 to 8). PspFΔHTH was shown previously to self-associate at high concentrations in the absence of nucleotides by native gel electrophoresis (9), but the results shown in Fig. 3 suggested that DctD(141-390) and DctD(141-394) have a greater tendency to form higher-order oligomers in the absence of ADP · AlFx than PspFΔHTH. ADP · AlFx did appear to stabilize the higher-order oligomers of DctD(141-390) and DctD(141-394), however, since the intensities of the bands corresponding to these complexes appeared to increase when ADP · AlFx was present. In addition, a minor, faster-migrating form of DctD(141-390) and one of DctD(141-394) were observed in the presence of ADP · AlFx, which we infer to be different oligomeric forms or conformations of these proteins. Interestingly, the predominant oligomeric form of DctD(141-394) migrated faster than that of DctD(141-390). The DctD(141-390) monomer is slightly larger that that of DctD(141-394) due to its larger histidine tag. Differences in size and amino acid composition between the two DctD AAA+ domain proteins may have accounted for the different mobilities of these proteins in the native gel. Alternatively, these proteins may differ with respect to either their conformations or oligomeric states.
DctD AAA+ domain proteins form a stable ternary complex with σ54 and ADP · AlFx.
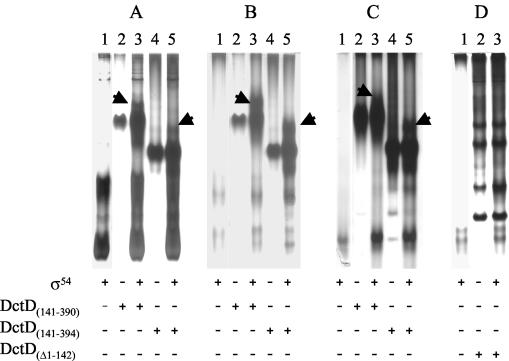
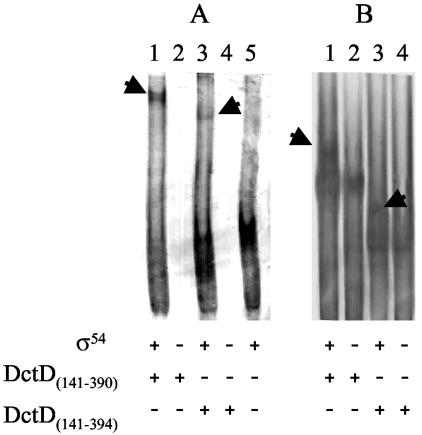
Previous studies with PspF, PspFΔHTH, and the Azotobacter vinelandii NifA AAA+ domain showed that these σ54-dependent activators formed a stable complex with σ54 in the presence of ADP · AlFx (3). Therefore, we examined DctD(Δ1-142) and the DctD AAA+ domain proteins to see if these proteins would similarly form a stable ternary complex with σ54 and ADP · AlFx. Three σ54 proteins were tested in this assay, native S. enterica serovar Typhimurium σ54, an N-terminal histidine-tagged S. enterica serovar Typhimurium σ54, and an N-terminal histidine-tagged S. meliloti σ54.
The S. enterica serovar Typhimurium σ54 proteins migrated on the native gel as two species (Fig. 4A and B, lanes 1 and 3), indicating self-association of the protein. Reaction mixtures in which native or histidine-tagged S. enterica serovar Typhimurium σ54 was incubated with DctD(141-390) or DctD(141-394) and then ADP · AlFx was generated in the mixture revealed bands that migrated immediately above the DctD AAA+ domain and were not present in the control lanes (Fig. 4A and 4B). These new bands were not observed in the absence of ADP · AlFx (data not shown). In some assays a protein band that migrated immediately below the major DctD AAA+ band was also apparent, but this band was not consistently observed. Western blot analysis confirmed that the new complexes observed when σ54 and one of the DctD AAA+ domain proteins were incubated together in the presence of ADP · AlFx contained both σ54 and DctD (Fig. 5). As reported previously with PspF (3), these protein complexes were not observed if the DctD AAA+ domain was incubated with ADP · AlFx before the protein was mixed with σ54 (data not shown). Unique complexes were also observed when DctD(141-390) or DctD(141-394) was incubated with S. meliloti σ54 in the presence of ADP · AlFx (Fig. 4C). We assume that these complexes contain σ54, but we could not verify this by Western blotting since S. meliloti σ54 did not cross-react with the antiserum directed against S. enterica serovar Typhimurium σ54. In contrast to the results with the DctD AAA+ domain, we did not observe complexes between DctD(Δ1-142) and any of the σ54 proteins that we tested (Fig. 4D; data shown only for S. meliloti σ54).
FIG. 4.
ADP · AlFx-dependent complex formation between the DctD AAA+ domain and σ54. Complex formation between DctD proteins and native S. enterica serovar Typhimurium σ54(A), histidine-tagged S. enterica serovar Typhimurium σ54 (B), or N-terminal histidine-tagged S. meliloti σ54 (C and D) was assessed. DctD(141-390), DctD(141-394), and a histidine-tagged version of DctD(Δ1-142) were included in binding reaction mixtures as indicated at the bottom of each lane. Protein complexes were resolved on a native gel and visualized by silver staining. Arrows in panels A to C indicate the complexes formed between σ54 proteins and the DctD AAA+ domain proteins. No complexes were detected between σ54 and DctD(Δ1-142) under the same reaction conditions. Panel A shows binding reactions done with S. enterica serovar Typhimurium σ54, panel B show reactions with the histidine-tagged S. enterica serovar Typhimurium σ54, and panels C and D show reactions with the histidine-tagged S. meliloti σ54.
FIG. 5.
Western blot analysis of ternary complexes. Complexes formed between S. enterica serovar Typhimurium σ54 and DctD(141-390) or DctD(141-394) in the presence of ADP · AlFx were analyzed by Western blotting using antiserum directed against S. enterica serovar Typhimurium σ54 (A) or S. meliloti DctD (B). The proteins included in the binding reaction mixtures are indicated below each lane. The arrows in panel A indicate the presence of σ54 in the ternary complexes, while the arrows in panel B indicate the presence of the DctD AAA+ domain proteins in the ternary complexes. The gel used for panel B was subjected to electrophoresis for a longer period of time to achieve better separation of the ternary complex from the DctD AAA+ domain complex.
In vitro activity of the DctD AAA+ domain.
The purified DctD AAA+ domains were examined for their ability to hydrolyze ATP. The measured ATPase activities of DctD(141-390) and DctD(141-394), which were determined at a single concentration (2.5 μM monomer), were 228 and 175 pmol of ATP hydrolyzed/min/pmol of DctD monomer, respectively. These values were comparable to that of DctD(Δ1-142), which was 251 pmol of ATP hydrolyzed/min/pmol of protein, and somewhat higher than that of an N-terminal histidine-tagged DctD(Δ1-142) protein, which was 107 pmol of ATP hydrolyzed/min/pmol of protein.
DctD(141-390) and DctD(141-394) were also tested for their ability to activate transcription in vitro. Titration of DctD(141-390) and DctD(141-394) in a single-round transcription assay showed that increasing concentrations of these proteins resulted in increased transcript levels (Fig. 6A). A supercoiled plasmid DNA template that carried the S. enterica serovar Typhimurium glnA promoter region upstream of a uracil-free tract was used for the transcription assay. This DNA template was used because σ54-holoenzyme has a higher affinity for the glnA promoter than it does for the S. meliloti dctA promoter.
FIG. 6.
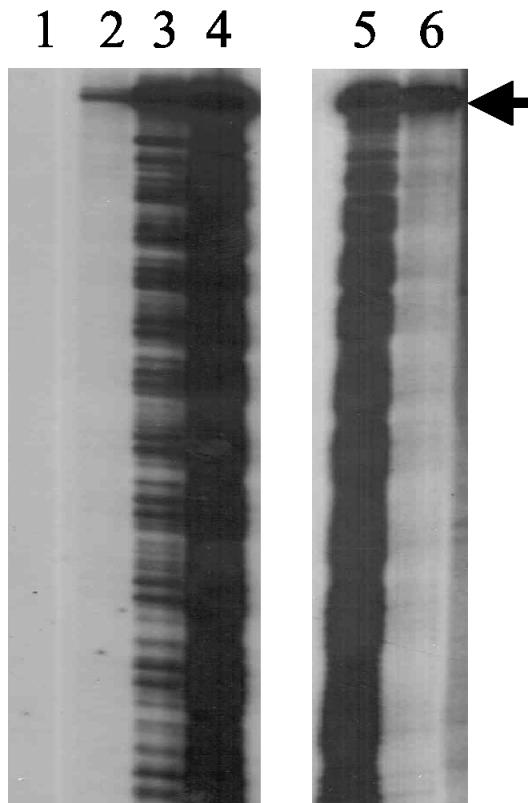
In vitro transcriptional activation by DctD AAA+ domain proteins. Transcriptional activation by DctD(141-390) and DctD(141-394) from the S. enterica serovar Typhimurium glnA promoter was assessed in a single-round in vitro transcription assay. Lanes 1 to 4 contained 100 nM, 500 nM, 1 μM, and 5 μM DctD(141-390) monomer, respectively, and show increased transcript levels with increasing concentrations of DctD(141-390). Lane 5 contained 5 μM DctD(141-390) monomer, and lane 6 contained 5 μM DctD(141-394) monomer. The arrow indicates the full-length transcript, which is ∼155 bases in length.
In transcription assays that contained the higher concentrations of DctD(141-390), a ladder of transcripts that were shorter than the full-length transcript was observed in addition to the full-length transcript (Fig. 6A). The pattern of the shorter transcripts was somewhat regular, with a repeat phase of about 11 nucleotides. This pattern of transcripts was not seen for DctD(141-394) (Fig. 6A), nor was it observed previously for DctD(Δ1-142) (17, 43, 44). Several lines of evidence argue against these shorter transcripts resulting from contaminating RNase activity. First, DctD(141-390) and DctD(141-394) were purified in the same manner and both protein preparations were very homogeneous (Fig. 2), yet the shorter transcripts were evident only in assays in which transcripts were initiated with DctD(141-390). Second, the phasing pattern of the shorter transcripts is not typical of nuclease digestion. Third, both the full-length and shorter transcripts were stable in the transcription assay when the incubation time following transcription initiation was extended from 10 to 60 min (Fig. 7A). Analysis of these data with a PhosphorImager showed that there were no appreciable differences in the levels of the full-length transcripts at the various incubation times, which strongly argued against RNase accounting for the shorter transcripts. Finally, adding DctD(141-390) to assay mixtures in which open complexes were formed with DctD(141-394) did not result in the shorter transcripts (Fig. 7B), which argues further against the DctD(141-390) preparations being contaminated with RNase. We infer from these observations that the shorter transcripts result from DctD(141-390) interfering with the transcription elongation complex when it is present at high levels.
FIG. 7.
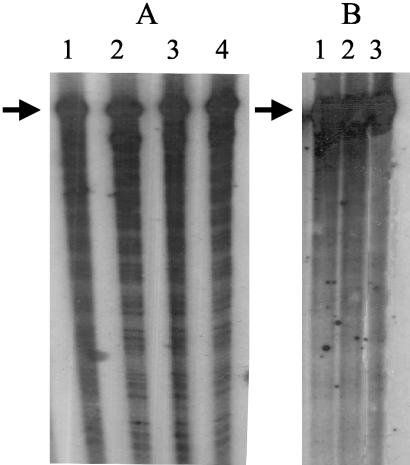
Stability of the transcripts formed in vitro. (A) In vitro transcription assays with 5 μM DctD(141-390) were incubated for various periods of time after the initiation of transcription by the addition of nucleotides to the reaction mixtures. Heparin was included to prevent transcription reinitiation. Assay mixtures were incubated for 10 min (lane 1), 20 min (lane 2), 30 min (lane 3), or 60 min (lane 4) after the addition of the nucleotides. Levels of the full-length transcript in lanes 1 to 4 as analyzed with a PhosphorImager SI (Molecular Dynamics) were virtually identical (volumes were 1.51 × 106, 1.49 × 106, 1.51 × 106, and 1.49 × 106, respectively). (B) Formation of open complexes at the glnA promoter were initiated with 5 μM DctD(141-394), after which DctD(141-390) was added to the reaction mixture at various times. In lane 1, no DctD(141-390) was added to the assay mixture, and the reaction was stopped 10 min after the addition of nucleotides and heparin. In lane 2, 5 μM DctD(141-390) was added immediately after adding the nucleotides and heparin, and the reaction was stopped 10 min later. In lane 3, 5 μM DctD(141-390) was added 10 min after the addition of nucleotides and heparin, and the reaction was stopped after an additional 10 min of incubation. Transcript levels in the three lanes were essentially the same as determined by PhosphorImager analysis (volumes were 3.26 × 104, 3.01 × 104, and 3.10 × 104, respectively).
DISCUSSION
The α-helical subdomain of the DctD AAA+ domain plays an important role in transcriptional activation, since certain substitutions in this region interfere with DctD(Δ1-142) function (43; H. Xu, M. T. Kelly, B. T. Nixon, and T. R. Hoover, submitted for publication). Similarly, mutations in the α-helical subdomains of other σ54-dependent activators, including S. enterica serovar Typhimurium NtrC, Pseudomonas putida XylR, and E. coli PspFΔHTH, have been reported to result in a loss of activity (19, 27, 29, 43). Because the α-helical subdomains of AAA+ proteins are variable in length, we undertook a deletion analysis of the 3′ end of dctD to define the minimal functional C-terminal end of this subdomain in DctD. The results of the deletion analysis indicated that the minimal functional C-terminal end lies somewhere between Gly-381 and Ala-385. This result was somewhat surprising, since threading the DctD sequence onto the A. aeolicus NtrC1 structure (18) modeled the DctD segment Ala-384 to Ser-390 as the last helix of the AAA+ α-helical subdomain, suggesting that DctD does not require the last helix of the α-helical subdomain to function in vivo.
With the exception of the loss of DNA-binding activity, the DctD AAA+ domain was functional in vivo and in vitro. Analysis of the DctD AAA+ domain on the native gel suggested that it migrated predominantly as a single oligomeric species, while DctD(Δ1-142) migrated as several different species. These data suggest a role for the DNA-binding domain in preventing assembly of DctD into a functional oligomeric complex in the absence of binding to the UAS, implying that the DNA-binding domain interacts with the AAA+ domain. DNase I footprinting of DctD(Δ1-142) at the dctA UAS revealed changes in the footprint in response to ADP · AlFx, consistent with the idea that the DNA-binding domain can communicate with the AAA+ domain, at least in the transition state during ATP hydrolysis (45). Alternatively, intermolecular interactions between the DNA-binding domains of DctD monomers may interfere with assembly of a functional oligomeric complex, which may be supported by the observation that introduction of three alanine substitutions in the enhancer recognition helix of S. enterica serovar Typhimurium NtrC eliminates DNA-binding activity and stabilizes the dimeric state of the protein (26).
AAA+ proteins often form hexamers (28), although the crystal structure of the AAA+ domain of A. aeolicus NtrC1 with ADP bound indicated that this protein forms a heptamer (18). We could not ascertain the oligomeric state of the DctD AAA+ domain from the native gel, but future studies will address this issue as well as determining if the predominant oligomeric forms of DctD(141-390) and DctD(141-394) differ from each other. An unexpected result was that the DctD AAA+ domain is significantly more soluble than DctD(Δ1-142). The increased solubility of the DctD AAA+ domain and its more uniform oligomeric nature appear beneficial for structural studies of DctD, since we have recently obtained crystals of DctD(141-390) that diffract X rays.
Unlike DctD(Δ1-142), the DctD AAA+ domain formed a stable complex with σ54 in the presence of ADP · AlFx, suggesting that the DNA-binding domain interferes with formation of the ternary complex. Inhibition may have resulted from steric hindrance by the DNA-binding domain of interactions between the AAA+ domain and σ54. Alternatively, since not all of the DctD(Δ1-142) species observed on the native gel are likely to be capable of forming a complex with σ54 in the presence of ADP · AlFx, the inhibition of ternary complex formation by the DNA-binding domain may have been an indirect effect.
In the in vitro transcription assay with DctD(141-390), a large number of transcripts which were shorter than the full-length transcript were produced, which we infer to result from DctD(141-390) interfering with transcription elongation. The pattern of termination appeared remarkably regular, suggesting that the transcription elongation complex was more susceptible to termination at regular intervals along the DNA template. This phasing may have been influenced by periodicity of the DNA template in the transcription vector, which was constructed using a synthetic module of directly repeated double-stranded oligonucleotides of 21 bp in length (31). DctD(141-394) did not generate the shorter transcripts in the in vitro transcription assay. This may have been because either the DctD segment from Ser-390 to Leu-394 or the histidine tag at the C terminus of DctD(141-394) prevented the AAA+ domain from interfering with transcription elongation.
Inhibition of transcription elongation by DctD(141-390) suggests that it contacts the transcription elongation complex. These interactions could lead to either stalling of the transcription elongation complex on the DNA or the premature release of transcripts from the complex. The failure to observe the regular pattern of shorter transcripts when DctD(141-390) was added to assay mixtures after open complexes were formed with DctD(141-394) suggests that DctD(141-390) must be present before transcription initiation to inhibit elongation. One possible explanation for this is that DctD(141-390) does not always disengage σ54-holoenzyme following open complex formation and remains associated with RNA polymerase upon promoter clearance. If DctD(141-390) can associate with the transcription elongation complex, to which subunit(s) of RNA polymerase might it bind? As is the case with other σ factors, σ54 is thought to dissociate from RNA polymerase following promoter clearance (41), but it is unclear if σ54 could remain associated with RNA polymerase in the transcription elongation complex if DctD(141-390) was also part of the complex. Recent studies have provided evidence that σ70 can remain associated with transcription elongation complexes (22, 24), which at least raises the possibility that σ54 could likewise. Alternatively, DctD(141-390) could engage the β subunit of RNA polymerase, since DctD can be chemically cross-linked to this subunit even in the absence of σ54 (16).
Acknowledgments
This work was funded by award MCB-9974558 to T.R.H. from the National Science Foundation and by Public Health Service grant GM40404 to B.T.N.
We thank Chih Lew for providing purified PspFΔHTH.
REFERENCES
- 1.Buck, M., M.-T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, M., S. Miller, M. Drummond, and R. Dixon. 1986. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature (London) 320:374-378. [Google Scholar]
- 3.Chaney, M., R. Grande, S. R. Wigneshweraraj, W. Cannon, P. Casaz, M.-T. Gallegos, J. Schumacher, S. Jones, S. Elderkin, A. E. Dago, E. Morett, and M. Buck. 2001. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 15:2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLaBarre, B., and A. Brunger. 2003. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 10:856-863. [DOI] [PubMed] [Google Scholar]
- 5.Gao, Y., Y.-K. Wang, and T. R. Hoover. 1998. Mutational analysis of the phosphate-binding loop of Rhizobium meliloti DctD, a σ54-dependent activator. J. Bacteriol. 180:2792-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu, B., J. H. Lee, T. R. Hoover, D. Scholl, and B. T. Nixon. 1994. Rhizobium meliloti DctD, a σ54-dependent transcriptional activator, may be negatively controlled by a subdomain in the C-terminal end of its two-component receiver domain. Mol. Microbiol. 13:51-66. [DOI] [PubMed] [Google Scholar]
- 7.Huala, E., and E. M. Ausubel. 1989. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J. Bacteriol. 171:3354-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huala, E., J. Stigter, and F. M. Ausubel. 1992. The central domain of Rhizobium leguminosarum DCTD functions independently to activate transcription. J. Bacteriol. 174:1428-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jovanovic, G., J. Rakonjac, and P. Model. 1999. In vivo and in vitro activities of the Escherichia coli σ54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J. Mol. Biol. 285:469-483.. [DOI] [PubMed] [Google Scholar]
- 10.Kelly, M. T., I. Ferguson, J. A., and T. R. Hoover. 2000. Transcription initiation-defective forms of σ54 that differ in ability to function with a heteroduplex DNA template. J. Bacteriol. 182:6503-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly, M. T., and T. R. Hoover. 1999. Mutant forms of Salmonella typhimurium σ54 defective in initiation but not promoter binding activity. J. Bacteriol. 181:3351-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo, I. S., and R. S. Stephens. 2003. A developmentally regulated two-component signal transduction system in Chlamydia. J. Biol. Chem. 278:17314-17319. [DOI] [PubMed] [Google Scholar]
- 13.Kustu, S., E. Santero, D. Popham, D. Weiss, and J. Keener. 1989. Expression of σ54(ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 54:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledebur, H., B. Gu, I. J. Sojda, and B. T. Nixon. 1990. Rhizobium meliloti and Rhizobium leguminosarum dctD gene products bind to tandem sites in an activation sequence located upstream of σ54-dependent dctA promoters. J. Bacteriol. 172:3888-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledebur, H., and B. T. Nixon. 1992. Tandem DctD-binding sites of the Rhizobium meliloti dctA upstream activating sequence are essential for optimal function despite a 50- to 100-fold difference in affinity for DctD. Mol. Microbiol. 6:3479-3492. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. H., and T. R. Hoover. 1995. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a σ54-dependent transcriptional activator, interacts with σ54 and the β subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 92:9702-9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. H., D. Scholl, B. T. Nixon, and T. R. Hoover. 1994. Constitutive ATP hydrolysis and transcriptional activation by a stable, truncated form of Rhizobium meliloti DCTD, a σ54-dependent transcriptional activator. J. Biol. Chem. 269:20401-20409. [PubMed] [Google Scholar]
- 18.Lee, S.-K., A. De La Torre, D. Yan, S. Kustu, T. Nixon, and D. E. Wemmer. 2003. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 17:2552-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lew, C. M., and J. D. Gralla. 2002. New roles for conserved regions within a σ54-dependent enhancer-binding protein. J. Biol. Chem. 277:41517-41524. [DOI] [PubMed] [Google Scholar]
- 20.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 21.Morett, E., and M. Buck. 1989. In vivo studies on the interaction of RNA polymerase-σ54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters: the role of NIFA in the formation of an open promoter complex. J. Mol. Biol. 210:65-77. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay, J., A. N. Kapanidis, V. Mekler, E. Kortkhonjia, Y. W. Ebright, and R. H. Ebright. 2001. Translocation of sigma(70) with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106:453-463. [DOI] [PubMed] [Google Scholar]
- 23.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 24.Nickels, B. E., C. W. Roberts, H. Sun, J. W. Roberts, and A. Hochschild. 2002. The σ70 subunit of RNA polymerase is contacted by the λQ antiterminator during early elongation. Mol. Cell 10:611-622. [DOI] [PubMed] [Google Scholar]
- 25.Nixon, B. T., C. W. Ronson, and F. M. Ausubel. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.North, A. K., and S. Kustu. 1997. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J. Mol. Biol. 267:17-36. [DOI] [PubMed] [Google Scholar]
- 27.North, A. K., D. S. Weiss, H. Suzuki, Y. Flashner, and S. Kustu. 1996. Repressor forms of the enhancer-binding protein NtrC: some fail in coupling ATP hydrolysis to open complex formation by σ54-holoenzyme. J. Mol. Biol. 260:317-331. [DOI] [PubMed] [Google Scholar]
- 28.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Martin, J., and V. de Lorenzo. 1996. ATP binding to the σ54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell 86:331-339. [DOI] [PubMed] [Google Scholar]
- 30.Popham, D., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629-635. [DOI] [PubMed] [Google Scholar]
- 31.Porter, S. C., A. K. North, A. B. Wedel, and S. Kustu. 1993. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 7:2258-2272. [DOI] [PubMed] [Google Scholar]
- 32.Reitzer, L. J., and B. Magasanik. 1986. Transcription at glnA of E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 33.Rippe, K., M. Guthold, P. H. von Hippel, and C. Bustamante. 1997. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase σ54 holoenzyme by scanning force microscopy. J. Mol. Biol. 270:125-138. [DOI] [PubMed] [Google Scholar]
- 34.Ronson, C. W., P. M. Astwood, B. T. Nixon, and F. M. Ausubel. 1987. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 15:7921-7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronson, C. W., B. T. Nixon, L. M. Albright, and F. M. Ausubel. 1987. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J. Bacteriol. 169:2424-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouiller, I., B. DeLaBarre, A. P. May, W. I. Weis, A. T. Brunger, R. A. Mulligan, and E. M. Wilson-Kubalek. 2002. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 9:950-957. [DOI] [PubMed] [Google Scholar]
- 37.Sasse-Dwight, S., and J. D. Gralla. 1988. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl. Acad. Sci. USA 85:8934-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su, W., S. Porter, S. Kustu, and H. Echols. 1990. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA 87:5504-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tintut, Y., J. T. Wang, and J. D. Gralla. 1995. A novel bacterial transcription cycle involving σ54. Genes Dev. 9:2305-2313. [DOI] [PubMed] [Google Scholar]
- 42.Vale, R. D. 2000. AAA proteins: lords of the ring. J. Cell Biol. 150:F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y.-K., and T. R. Hoover. 1997. Alterations within the activation domain of the σ54-dependent activator DctD that prevent transcriptional activation. J. Bacteriol. 179:5812-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y.-K., J. H. Lee, J. M. Brewer, and T. R. Hoover. 1997. A conserved region in the σ54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol. Microbiol. 26:373-386. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y.-K., S. Park, B. T. Nixon, and T. R. Hoover. 2003. Nucleotide-dependent conformational changes in the σ54-dependent activator DctD. J. Bacteriol. 185:6215-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, X., M. Chaney, S. R. Wigneshweraraj, J. Schumacher, P. Bordes, W. Cannon, and M. Buck. 2002. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 45:895-903. [DOI] [PubMed] [Google Scholar]