Abstract
Campylobacter jejuni, a gram-negative motile bacterium, secretes a set of proteins termed the Campylobacter invasion antigens (Cia proteins). The purpose of this study was to determine whether the flagellar apparatus serves as the export apparatus for the Cia proteins. Mutations were generated in five genes encoding three structural components of the flagella, the flagellar basal body (flgB and flgC), hook (flgE2), and filament (flaA and flaB) genes, as well as in genes whose products are essential for flagellar protein export (flhB and fliI). While mutations that affected filament assembly were found to be nonmotile (Mot−) and did not secrete Cia proteins (S−), a flaA (flaB+) filament mutant was found to be nonmotile but Cia protein secretion competent (Mot−, S+). Complementation of a flaA flaB double mutant with a shuttle plasmid harboring either the flaA or flaB gene restored Cia protein secretion, suggesting that Cia export requires at least one of the two filament proteins. Infection of INT 407 human intestinal cells with the C. jejuni mutants revealed that maximal invasion of the epithelial cells required motile bacteria that are secretion competent. Collectively, these data suggest that the C. jejuni Cia proteins are secreted from the flagellar export apparatus.
Campylobacter jejuni, a gram-negative motile bacterium, is a frequent cause of human gastrointestinal infections (39). The spectrum of disease observed in C. jejuni-infected individuals ranges from asymptomatic to severe enteritis characterized by fever, severe abdominal cramping, and diarrhea with blood and mucus (2, 4). By analogy with other more extensively characterized bacterial pathogens, the mechanism of C. jejuni-mediated enteritis is proposed to be multifactorial. Previous work has indicated that motility as well as the presence of the flagellum contributes to the ability of C. jejuni to colonize the intestinal tract of animals (33, 36, 42).
The flagellum of C. jejuni is composed of a basal body, hook, and filament. The flagellar filament is comprised of two proteins, FlaA and FlaB, although it appears that FlaA is the preferred subunit (3). While the C. jejuni FlaA and FlaB flagellin proteins are transcribed concomitantly (16), the flaA gene is regulated by σ28 and the flaB gene is regulated by σ54 (3, 16). Hendrixson et al. (16) noted that a C. jejuni isolate deficient in σ28, which is encoded by the fliA gene, is able to assemble a truncated filament composed exclusively of the flagellin protein FlaB. This result indicates that the regulation of flagellar gene expression within C. jejuni differs from the regulation in more intensely studied systems such as that of Salmonella enterica. Unlike flagellar gene expression in C. jejuni, flagellar gene expression in S. enterica is initiated by a master regulator, while late gene expression and motility require σ28 (1).
Previous work in our laboratory has demonstrated that C. jejuni synthesizes a set of proteins during coculture with epithelial cells, a subset of which are secreted. The secreted proteins have been collectively referred to as Campylobacter invasion antigens (Cia proteins) (22). The functions of the secreted proteins are not known; however, insertional mutagenesis of the gene (ciaB) encoding a 73-kDa secreted protein (CiaB) results in a significant reduction in the number of C. jejuni cells internalized compared to a C. jejuni wild-type isolate. The absence of Cia protein secretion in the C. jejuni ciaB mutant is specific, as the invasive phenotype of this organism is restored by complementation in trans with the ciaB gene (37). CiaB lacks an identifiable signal sequence (22). In addition, an environmental stimulus is required to induce Cia protein secretion (37). While these characteristics are reminiscent of virulence-associated type III secretion systems, translation of the complete genome of C. jejuni NCTC 11168 failed to reveal proteins with identity to virulence-associated type III secretory systems (http://www.sanger.ac.uk/Projects/C.jejuni).
Some proteins that have the classical type III secretory apparatus show amino acid sequence similarity with flagellar structural proteins (26, 29). Moreover, evidence is beginning to accumulate that components of the flagellar apparatus participate in the export of virulence determinants in several pathogens. For example, experiments by Young et al. (45) demonstrated that Yersinia secretes flagellar outer proteins (Fops) via the flagellar apparatus. More recently, secretion of virulence-associated proteins from Bacillus thuringiensis has been found to be dependent on flhA, an essential component of the flagellar export apparatus (13). Consistent with the notion that components of the flagellar export apparatus can play a role in the export of virulence-associated proteins in some organisms, we noted that a C. jejuni flhB export mutant was deficient in secretion of the Cia proteins. The purpose of this study was to determine whether the flagellar protein export apparatus is required for secretion of the Cia proteins.
(A portion of this work was presented at the William R. Wiley Award Research Exposition held at Washington State University in Pullman, Wash., 21 February 2001; at the 11th International Workshop on Campylobacter, Helicobacter, and Related Organisms, in Freiburg, Germany, 1 to 5 September 2001; and at the 82nd annual meeting of the Conference of Research Workers in Animal Diseases, St. Louis, Mo., 11 to 13 November 2001.)
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
Human clinical C. jejuni strains F38011 and 81116 were cultured as described previously (38). Plates were supplemented with kanamycin sulfate (200 μg/ml) as appropriate. Escherichia coli InvαF′, MRF XL-1 Blue (Tetr), and DH5α were cultured in Luria-Bertani broth or solid medium, supplemented with kanamycin (50 μg/ml) or tetracycline (15 μg/ml) as appropriate, in a 37°C incubator.
Isolation of C. jejuni flagellar mutants.
All of the suicide vectors described below were introduced into C. jejuni F38011 by electroporation. C. jejuni F38011 mutants were identified by acquisition of kanamycin or chloramphenicol resistance or kanamycin sensitivity, and specific gene disruption was confirmed by PCR.
C. jejuni strain NCTC 11168 flaA, flaB, flhB, flgB, flgC, flgE2, fliD, and fliI gene sequences were obtained from the Sanger Center website (http://www.sanger.ac.uk/Projects/C.jejuni). Two flgE genes have been identified in NCTC 11168 (Cj0043 and Cj1729c); however, based on previously published work showing sequence similarity of the flgE2 product to other hook proteins (18, 25), flgE2 (Cj1729c) was targeted. The flhB (Cj0335), flgB (Cj0528c), flgC (Cj0527c), flgE2 and fliD (Cj0548), and fliI (Cj0195) genes in C. jejuni F38011 were disrupted by recombination via a single crossover event between the chromosomal gene and an internal fragment of the homologous gene in a suicide vector harboring the aphA3′ gene encoding kanamycin resistance (21). The C. jejuni flhB (forward primer, 5′-AAA AAA CAG AAG AAC CCA CG; reverse primer, 5′-GTG CAA CCA TAG TAT AAA GCT C), flgB (forward primer, 5′-CAT TTA AAT CAA AAG AAC TGG; reverse primer, 5′-TCC ATC AAG AGC TGT TAT C), flgC (forward primer, 5′-AAG TGA TTT TGA TAT TAG TGG; reverse primer, 5′-GTA GCT TCA ATC AAA TCT GC), flgE2 (forward primer, 5′-AGT GGT TTA AAT ATA GGA ACT TCA AG; reverse primer, 5′-AAA GAA TCA TAA ATT TCA AGC C), fliD (forward primer, 5′-TGC GAA AAG AAA AGT TGT AGG; reverse primer, 5′-ATT TGC ATC TTC AGT AGT TCC), and fliI (forward primer, 5′-CTC GAG GCT ACA AGT ATA GAA ATT CGT GG; reverse primer, 5′-GAG CTC GTT GCA CTT GCA CTT CCT AC) internal gene fragments were amplified with Taq polymerase (Invitrogen) and gene-specific primers as part of a PCR.
In addition to the single-crossover insertion mutants generated above, a deletion-insertion mutation was constructed in the fliA (Cj0061c) gene by allelic replacement via a double crossover event. With a similar strategy, a filament-negative C. jejuni F38011 mutant was generated by deletion of the 3′ end of the flaA (Cj1338c) gene and the 5′ end of the flaB (Cj1339c) gene. Target genes in C. jejuni F38011 were disrupted by allelic replacement via a double-crossover event between the target chromosomal gene and a plasmid-borne copy of the target gene containing an internal deletion. To aid in the selection of the double-crossover mutations, the deletions were marked by the cat gene, encoding chloramphenicol resistance. The final vectors were constructed with standard molecular biology techniques. Briefly, the 5′ end of the fliA gene with flanking DNA was amplified with the F1 and R1 primer set (F1, 5′-TTG GAT CCT TGG AAG ACA TTT TAA TAG AAG; R1, 5′-AAC CGC GGA AAG CTA GCC ACA AGC TCA TCT TGC TCT TTC). The italics represent restriction sites that were used to facilitate the cloning and construction of the final vector.
The fliA F1 primer contained a BamHI restriction site, and the fliA R1 primer contained NheI and SacII restriction sites. The 3′ end of the fliA gene with a flanking region was amplified with a second set of primers (F2, 5′-TTG CTA GCC ACG AAG TGC TAG ATG ATC TTA AAG; R2, 5′-AAC CGC GGA TTT CTT TGA TTT CAT CTT TAT C). The fliA F2 primer contained an NheI restriction site and the fliA R2 primer contained a SacII restriction site. Following ligation of DNA fragments harboring the 5′ and 3′ regions of the fliA gene, the cat gene was ligated into the NheI restriction site. Portions of the flaA and flaB genes were amplified with the primer sets FlaAF1/FlaAR1 and FlaBF1/FlaBR1 (FlaAF1, 5′-GGA TTC CCG TTA AAG CAA CTA TAG GTG CAA; FlaAR1, 5′-GCT AGC CCT GAT ACT GAA CTA ATA GAA TTG; FlaBF2, 5′-CT A GCT AGC GGT CAA GCT ATA AAC AAT GGT AAT GAT C; and FlaBR2, 5′-TTG GAT CCT TAT TGA AGA AGT TTT AAA ACA TTT TGC TGC). The italics represent restriction sites that were used to facilitate the cloning and construction of the final vector. The flaAF1 and flaBR2 primers contained a BamHI restriction site, and the flaAR1and flaBF2 primers contained an NheI restriction site. Amplification with FlaAF1/FlaAR1 resulted in deletion of the 3′ end of flaA, whereas amplification with FlaBF2/FlaBR2 resulted in deletions at both the 5′ and 3′ ends of flaB. Following ligation of DNA fragments harboring the truncated flaA and flaB genes, the cat gene was ligated into the NheI restriction site, and the deletion-insertion mutation was crossed onto the F38011 chromosome.
Phenotypic analysis of the C. jejuni flagellar mutants.
Motility assays were performed with Mueller-Hinton (MH) medium supplemented with 0.4% Select Agar (Gibco-BRL). A 10-μl suspension of each bacterial isolate was spotted on the surface of semisolid medium. Motility plates were incubated at 37°C under microaerophilic conditions for 48 h. C. jejuni strains were also analyzed by transmission electron microscopy (TEM). Bacterial suspensions prepared from MH agar plates with phosphate-buffered saline were added dropwise to Formvar-coated copper grids. Bacteria were stained with 1% phosphotungstic acid. Samples were analyzed with a 1200 EX transmission electron microscope (JEOL).
Complementation analysis.
A 2,032-bp fragment containing the entire flaA gene and flanking DNA sequences was amplified from C. jejuni NCTC 11168 by PCR with the primers 5′-GGA TCC TAA AAC GCA TTT CAT CAC AGC (forward primer; BamHI linker is italic in each case) and 5′-GGA TCC GAT TAA AGC AAA AAG TGT TC (reverse primer). The forward primer is 199 bp upstream of the AUG methionine initiation codon, and the reverse primer extends 114 bp beyond the UAG stop codon. Following an intermediate cloning step into pCR2.1 (Invitrogen), the gel-purified insert was ligated into the BamHI site of pMEK80 (37). The resultant shuttle plasmid, designated pMEK3502, was introduced into a C. jejuni strain 81116 flaA flaB mutant (GRK7) by electroporation (15). Transformants were identified as described above. The C. jejuni pMEK3503 shuttle vector (flaB+) was generated in a similar fashion with the primers 5′-GGA TCC CAA AAT GTT TTA AGA TTA CTA CAG (forward primer) and 5′-GGA TCC TTT TTG CTT GGG TTT ATG CAC (reverse primer). The forward primer is 172 bp upstream of the AUG methionine initiation codon, and reverse primer extends 161 bp beyond the UAA stop codon. The amplified PCR product containing the flaB gene and flanking DNA sequences was 2,052 bp.
Analysis of C. jejuni secreted proteins.
C. jejuni cells were metabolically labeled with [35S]methionine as described elsewhere (19). For each sample, the supernatant fluid was treated in an identical manner. Briefly, the supernatant fluids were concentrated fourfold by the addition of 5 volumes of ice-cold 1 mM HCl-acetone. The pellets were air dried and resuspended in an equal volume of water. Equal volumes of the concentrated samples were then loaded in the wells of a sodium dodecyl sulfate-12.5% polyacrylamide gel. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with the discontinuous buffer system described by Laemmli (24). In most instances, gels were treated with Amplify (Amersham Life Sciences) according to the manufacturer's instructions, and autoradiography was performed with Kodak BioMax MR film at −70°C. Alternatively, the proteins were electrophoretically transferred to polyvinylidene fluoride membranes (Immobilon P; Millipore Corp., Bedford, Mass.) for immunoblot analyses.
RNA isolation and reverse transcription-PCR.
Bacteria were grown in Mueller-Hinton broth with 0.05% deoxycholate under microaerobic conditions overnight, pelleted, and resuspended in 200 μl of TE (10 mM Tris Cl, 1 mM EDTA, pH 7.4) buffer. The bacteria were then treated with 100 μl of 50 mM glucose-25 mM Tris-Cl-10 mM EDTA-200 μl of 0.1 M NaOH-1% SDS-150 μl of 5 M potassium acetate and centrifuged for 5 min at 13,000 × g. All solutions were pretreated with 0.1% diethyl pyrocarbonate (Sigma). The supernatant was treated with an equal volume of phenol-chloroform (1:1). The resulting supernatant was added to an equal volume of 90% isopropanol and centrifuged for 5 min at 13,000 × g. The pellet was then washed in 70% isopropanol, pelleted by centrifugation as above, dried by vacuum aspiration, and incubated with 50 U of RNase-free DNase (Roche, Indianapolis, Ind.) for 15 min at 37°C. After DNase treatment, an equal volume of 7.5 M sodium acetate was added as well as 2 volumes of 90% isopropanol, centrifuged for 20 min at 4°C, washed in 70% isopropanol, and recentrifuged. The pellet was resuspended in 50 μl of RNase-free water (27).
Ten micrograms of RNA was used for cDNA preparation with 3 μl of the random primers provided with the ProSTAR First Strand reverse transcription-PCR kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Then, 2.5 μl of the cDNA was subjected to PCR amplification with primers within the coding region of the ciaB primers (forward primer, 5′-CTA TGC TAG CCA TAC TTA GGC; reverse primer, 5′-GCC CGC CTT AGA ACT TAC) as well as primers within the asp gene (asp-S3 forward primer, 5′-CCA ACT GCA AGA TGC TGT ACC; asp-S6 reverse primer, 5′-TTA ATT TGC GGT AAT ACC ATC) (12). The cycling conditions were as follows: one cycle of 5 min at 94°C; five cycles of 30 s at 94°C, 30 s at 45°C, and 1 min at 72°C; 20 cycles of 30 s at 94°C, 30 s at 49°C, and 1 min at 72°C; and a final extension of 5 min at 72°C. The resulting products were resolved by electrophoresis through 1% agarose in Tris-borate-EDTA buffer, and bands were visualized by UV light after ethidium bromide staining.
Examination of the interactions of C. jejuni with INT 407 cells.
Adherence and internalization assays were performed with INT 407 cells (human embryonic intestine cells; ATCC CCL 6) as previously described (20). Briefly, each well of a 24-well tissue culture tray was seeded with 105 cells/well and incubated for 18 h at 37°C in a humidified, 5% CO2 incubator. The cells were rinsed with Eagle's minimal essential medium-1% fetal bovine serum and inoculated with approximately 5 × 107 CFU of a bacterial suspension. Tissue culture trays were centrifuged at 600 × g for 5 min and incubated at 37°C in a humidified, 5% CO2 incubator. For binding, the infected monolayers were incubated for 30 min and rinsed three times with phosphate-buffered saline, and the epithelial cells were lysed with a solution of 0.25% (wt/vol) sodium deoxycholate. The suspensions were serially diluted, and the number of viable, adherent bacteria was determined by counting the resultant colonies on MH/blood plates.
To measure bacterial internalization, the infected monolayers were incubated for 3 h, rinsed three times with Eagle's minimal essential medium-1% fetal bovine serum, and incubated for an additional 3 h in Eagle's minimal essential medium-1% fetal bovine serum containing a bactericidal concentration (250 μg ml−1) of gentamicin (31). The number of internalized bacteria was determined as outlined above. The number of adherent bacteria for C. jejuni F38011 was typically 106 CFU and the number of internalized bacteria was 5.0 × 104 CFU; for actual values, refer to Table 1. Because adherence is a prerequisite for C. jejuni invasion, the values reported were transformed with the following equation: [(number of internalized bacteria)/(number of adherent bacteria)] × 100. Significance between samples was determined with Student's t test following log10 transformation of the data. Two-tailed P values were determined for each sample, and a P value of <0.01 was considered significant. All assays were repeated a minimum of three times.
TABLE 1.
Phenotypes displayed by wild-type isolates and isogenic flagellar protein export and filament mutants
| Background strain | Relevant characteristic | FlaA synthesis | Filament assembly | Motility | Secretion | Adherencea | Invasionb | I/Ac |
|---|---|---|---|---|---|---|---|---|
| F38011 | Wild type | + | + | + | + | 100 ± 5 | 100 ± 20 | 5.3 |
| flhB | − | − | − | − | 47 ± 9 | 0.24 ± 0.04 | 0.03 | |
| flgB | − | − | − | − | 40 ± 6 | 0.46 ± 0.1 | 0.06 | |
| flgC | − | − | − | − | 73 ± 2 | 2.1 ± 0.5 | 0.13 | |
| flgE2 | − | − | − | − | 13 ± 2 | 0.26 ± 0.1 | 0.11 | |
| fliD | + | + | + | + | 59 ± 8 | 44 ± 4 | 4.0 | |
| ciaB | + | + | + | − | 223 ± 15 | 1.9 ± 0.7 | 0.02 | |
| 81116 | Wild type | + | + | + | + | 100 ± 9 | 100 ± 27 | 2.7 |
| flaA flaB | − | − | − | − | 191 ± 27 | 10 ± 3 | 0.14 | |
| flaA | − | Truncated | − | + | 145 ± 19 | 70 ± 7 | 1.3 | |
| flaA flaB/pMEK3502 (flaA+) | + | + | + | + | 127 ± 18 | 29 ± 1 | 0.63 | |
| flaA flaB pMEK3503 (flaB+) | − | Truncated | − | + | 255 ± 27 | 43 ± 7 | 0.46 |
As a percentage of the wild-type value, which was 9.4 × 105 ± 0.5 × 105.
As a percentage of the wild-type value, which was 5.0 × 104 ± 1 × 104.
I/A, [(number of internalized bacteria)/(number of adherent bacteria)] × 100.
Other analytical procedures.
Freshly prepared serum against the C. jejuni CiaB protein was generated in a New Zealand White rabbit as described elsewhere (22). Immunoblot analyses were performed with a 1:50 dilution of a rabbit anti-C. jejuni flagellin serum or a rabbit anti-CiaB protein serum as described elsewhere (15). Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G and 4-chloro-1-naphthol (Sigma) as the chromogenic substrate.
RESULTS
Generation of C. jejuni flagellar mutants.
Experiments were performed to determine if the Cia proteins are secreted from the flagellar apparatus. A C. jejuni F38011 flhB flagellar export apparatus mutant was found to be nonmotile (Mot−) and also failed to secrete the Cia proteins (S−) (Table 1), which is consistent with the notion that the flagellar export apparatus is required for Cia protein secretion. Additional experiments were undertaken to test this preliminary finding. Insertion mutations were generated in the C. jejuni F38011 genes encoding the flagellar basal body (flgB and flgC) and hook (flgE2) and in the putative filament cap protein (fliD). In addition, we used previously published strains of C. jejuni 81116 with disruptions in either flaA (GRK17) or flaA and flaB (GRK7) (15). For clarity, we will refer to the C. jejuni 81116 flaA mutant throughout the remainder of the text as a flaA (flaB+) mutant. After confirming each mutation by gene-specific PCR, we assessed whether each mutant was motile, synthesized the FlaA filament protein, and assembled a filament.
Phenotypically, the C. jejuni flgB, flgC, flgE2 and flaA flaB flagellar mutants exhibited a Mot− phenotype on motility agar, failed to synthesize FlaA, as determined by immunoblot analysis, and did not assemble a filament, as judged by TEM (Table 1 and data not shown). However, a 62-kDa immunoreactive band of decreased intensity, likely corresponding to the FlaB protein, was detected in whole-cell lysate preparations of C. jejuni F38011 flgB, flgC, and flgE2 mutants as well as in the C. jejuni 81116 flaA (flaB+) mutant with a flagellin polyclonal serum. The 62-kDa immunoreactive band was not detected in the C. jejuni 81116 flaA flaB mutant. Collectively, these data indicate that synthesis of the FlaA protein was greatly reduced in the C. jejuni F38011 flgB, flgC, and flgE2 flagellar export mutants, which is consistent with the findings of Matz et al. (28), who determined that there was a significant decrease in the level of flaA gene expression in a C. jejuni strain containing a defective flagellar export apparatus. A truncated filament was observed in the C. jejuni 81116 flaA (flaB+) mutant. These results are consistent with the findings of Wassenaar et al. (41), who showed that motility correlated with the synthesis and assembly of a FlaA but not a FlaB filament. Wassenaar et al. (41) also observed truncated filaments in a flaA (flaB+) mutant.
A mutation in the C. jejuni fliD gene resulted in bacteria with a motility-impaired phenotype. Although TEM examination revealed that the C. jejuni fliD mutant displayed full-length filaments when harvested directly from motility agar plates, the same strain displayed truncated filaments when harvested from broth cultures (not shown). In fact, the C. jejuni fliD mutant grown in broth resembled the C. jejuni 81116 flaA (flaB+) mutant with respect to filament structure. Yokoseki et al. (44) found that an S. enterica serovar Typhimurium fliD mutant also formed minute swarms on motility agar plates but did not produce filaments in liquid medium. The investigators proposed that the motility of the S. enterica serovar Typhimurium fliD mutant was due to flagellin monomers which could not freely diffuse in the motility agar being assembled into filaments (44).
Secretion of the Cia proteins requires the intact flagellar apparatus.
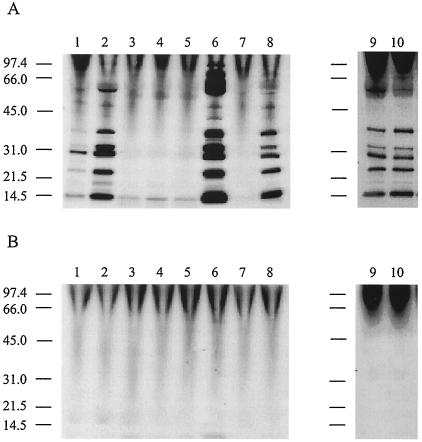
Secretion assays were performed in the presence and absence of fetal bovine serum (FBS) to determine whether the C. jejuni mutants were capable of Cia protein export. FBS serves as an artificial signal to stimulate the synthesis and secretion of the Cia proteins (37). The Cia proteins were readily identifiable in the supernatant fluids of C. jejuni F38011 and 81116 wild-type strains as well as the C. jejuni F38011 fliD mutant and the C. jejuni 81116 flaA (flaB+) mutant (Fig. 1A). However, the Cia proteins were not detected in the supernatant fluids from the flgB, flgC, and flgE2 mutants or the flaA flaB mutant. In agreement with previous work indicating that a stimulus is required to induce Cia secretion, the Cia proteins were not detected in the supernatant fluids when FBS was omitted from the labeling medium (Fig. 1B).
FIG. 1.
C. jejuni Cia protein secretion requires a functional flagellar apparatus. C. jejuni cells were precultured on Mueller-Hinton plates (lane 1) and Mueller-Hinton plates supplemented with deoxycholate (lanes 2 to 10), and labeled in minimal essential medium in the presence (A) and absence (B) of FBS. Lanes: 1, C. jejuni F38011 (Mueller-Hinton plate); 2, C. jejuni F38011 (Mueller-Hinton deoxycholate plate); 3, C. jejuni F38011 flgB mutant; 4, C. jejuni F38011 flgC mutant; 5, C. jejuni F38011 flgE2 mutant; 6, C. jejuni 81116; 7, C. jejuni 81116 flaA flaB (GRK7) mutant; 8, C. jejuni 81116 flaA (flaB+) (GRK17) mutant; 9, C. jejuni F38011; and 10, C. jejuni F38011 fliD mutant. C. jejuni strains were deemed positive for protein secretion based on the presence of multiple bands in a lane (i.e., panel A, lanes 1, 2, 6, 8, 9, and 10). The positions of molecular mass standards are indicated on the left (in kilodaltons).
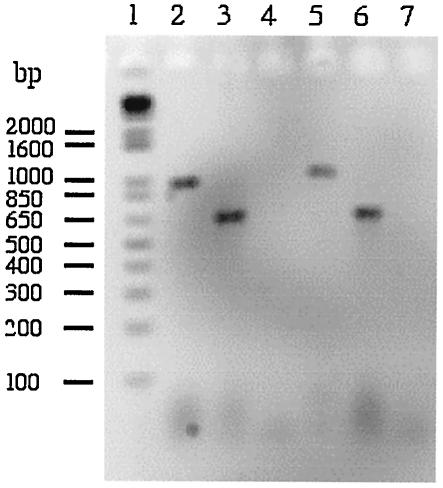
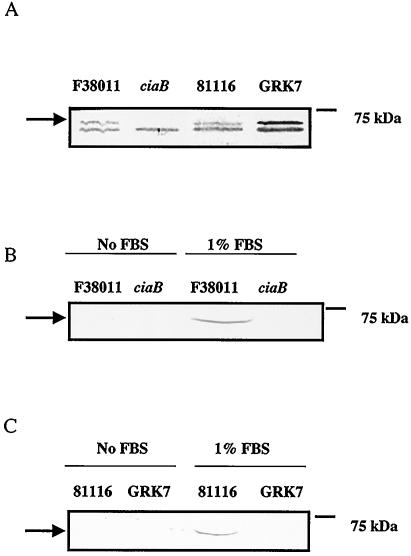
To determine if the ciaB gene was expressed in the flaA flaB mutant, reverse transcription-PCR analysis was performed (Fig. 2). A band representing the ciaB transcript was clearly evident in this mutant, demonstrating that ciaB is expressed. To demonstrate that the Cia protein is indeed one of the proteins secreted from the C. jejuni wild-type isolate, whole-cell lysates and supernatant fluids of the C. jejuni wild-type isolate and the C. jejuni flaA flaB mutant were analyzed by immunoblotting with a CiaB antibody (Fig. 3). A band corresponding in size to the CiaB protein was detected in whole-cell lysates of both the C. jejuni F38011 and 81116 wild-type strains and the C. jejuni 81116 flaA flaB mutant, but not in the whole-cell lysate of the C. jejuni F38011 ciaB mutant (Fig. 3A). In addition, CiaB was detected in the supernatant fluids of the C. jejuni wild-type F38011 and 81116 isolates, but only when FBS was added to the medium (Fig. 3, Panels B and C). However, CiaB was not detected in the supernatant fluids of the C. jejuni ciaB or flaA flaB mutants even when FBS was added to the media. These data show that the CiaB protein is synthesized, but is not secreted, in the C. jejuni flaA flaB mutant.
FIG. 2.
Detection of a ciaB transcript in the C. jejuni 81116 wild-type isolate and C. jejuni 81116 flaA flaB mutant by reverse transcription-PCR. C. jejuni were cultured on Mueller-Hinton plates supplemented with deoxycholate. PCR was performed with gene-specific primers and a cDNA that was generated with RNA extracted from the C. jejuni 81116 wild-type isolate (lanes 2 to 4) and the C. jejuni 81116 flaA flaB (GRK7) mutant (lanes 5 to 7). Lanes: 1, DNA ladder (Invitrogen, Carlsbad, Calif.); 2, PCR performed with the ciaB gene-specific primers; 3, PCR performed with the asp gene-specific primers; 4, PCR performed with the ciaB gene-specific primers where the reverse transcriptase enzyme was omitted from the cDNA synthesis step; 5, PCR performed with the ciaB gene-specific primers; 6, PCR performed with the asp gene-specific primers, and 7, PCR performed with the ciaB gene-specific primers where the reverse transcriptase enzyme was omitted from the cDNA synthesis step.
FIG. 3.
Detection of the CiaB protein in C. jejuni wild-type isolates. Secretion assays were performed as outlined in Materials and Methods. Bacterial whole-cell lysates (A) and supernatant fluids (B and C) were analyzed by SDS-PAGE coupled with immunoblot analysis with the CiaB antibody. (A) Lanes: 1, C. jejuni F38011 wild-type strain; 2, C. jejuni F38011 ciaB mutant; 3, C. jejuni 81116 wild-type strain; 4, C. jejuni 81116 flaA flaB (GRK7) mutant. (B) Lanes: 1, C. jejuni F38011 wild-type strain minus FBS; 2, C. jejuni F38011 ciaB mutant minus FBS; 3, C. jejuni F38011 wild-type strain with FBS added to the medium to induce Cia protein export; 4, C. jejuni F38011 ciaB mutant with FBS added to the medium. (C) Lanes: 1, C. jejuni 81116 wild-type strain minus FBS; 2, C. jejuni 81116 flaA flaB (GRK7) mutant minus FBS; 3, C. jejuni 81116 wild-type strain with FBS added to the medium to induce Cia protein export; 4, C. jejuni 81116 flaA flaB (GRK7) mutant with FBS added to the medium to induce Cia protein export. The arrows indicate the position of the CiaB protein.
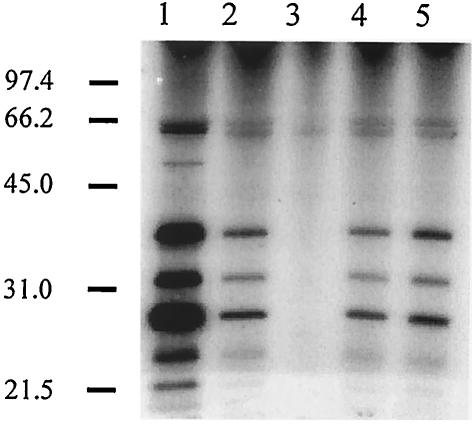
To address whether the Cia proteins are secreted from a C. jejuni 81116 flaA+ flaB strain, the C. jejuni flaA flaB mutant was transformed with a Campylobacter shuttle plasmid harboring an intact flaA gene. As a control, the C. jejuni flaA flaB mutant was also transformed with the same C. jejuni shuttle plasmid harboring an intact flaB gene. Immunoblot analysis with antiflagellin serum revealed that both the C. jejuni flaA flaB/flaA+ and C. jejuni flaA flaB/flaB+ transformants synthesized a filament protein, presumably FlaA and FlaB, respectively (Table 1). In addition, assays with metabolic labeling coupled with autoradiography revealed that the Cia proteins were secreted from the C. jejuni flaA flaB/flaA+ transformant, although not to the same extent as from the wild-type strain (Fig. 4). Similarly, a reduced amount of Cia proteins was detected in the supernatant fluid from the flaB+ transformant versus the C. jejuni flaA (flaB+) mutant (Fig. 4). Based on these data, it is clear that the lack of CiaB export from the C. jejuni flaA flaB mutant is not due to the absence of ciaB gene expression or CiaB protein synthesis. These results further suggest that an intact flagellar structure (containing the basal body, hook, and at least a partial filament) is required for Cia protein secretion.
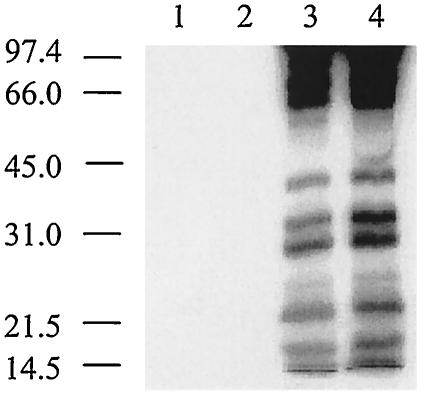
FIG. 4.
Transformation of a C. jejuni flaA flaB strain with a recombinant plasmid harboring either flaA or flaB restores Cia secretion. C. jejuni cells were precultured on Mueller-Hinton plates supplemented with deoxycholate and labeled in minimal essential medium in the presence of FBS. Lanes: 1, C. jejuni 81116 wild-type strain; 2, C. jejuni 81116 flaA flaB (GRK7) mutant harboring pMEK3502 (flaA+); 3, C. jejuni 81116 flaA flaB (GRK7) mutant; 4, C. jejuni 81116 flaA flaB (GRK7) mutant harboring pMEK3503 (flaB+); 5, C. jejuni 81116 flaA (flaB+) (GRK17) mutant. The positions of molecular mass standards are indicated on the left (in kilodaltons).
Cia protein export is independent of σ28.
The C. jejuni flaA gene is transcribed from a σ28-specific promoter (35), and the C. jejuni flaB gene is transcribed from a σ54-specific promoter (16). A mutation was generated in the fliA gene of C. jejuni F38011, which encodes σ28, to determine whether this sigma factor is responsible for cia gene transcription. As predicted, the C. jejuni fliA mutant was nonmotile, most likely due to the lack of flaA transcription. Indeed, an immunoreactive band of decreased intensity, corresponding in mass to the FlaB protein, was detected in the mutant, as judged by immunoblot analysis of whole-cell lysates with antiflagellin polyclonal serum (not shown). The C. jejuni fliA mutant also resembled the C. jejuni 81116 flaA (flaB+) mutant with respect to filament structure, as judged by TEM analysis. Finally, secretion assays revealed that the C. jejuni fliA mutant was secretion competent (Fig. 5). This result demonstrates that a sigma factor other than σ28 is required for transcription of the cia genes.
FIG. 5.
C. jejuni protein secretion is independent of σ28. C. jejuni cells were harvested from Mueller-Hinton agar plates labeled in minimal essential medium in the absence (lanes 1 and 2) and presence (lanes 3 and 4) of FBS as outlined in Materials and Methods. Lanes: 1, C. jejuni F38011 (without FBS); 2, C. jejuni fliA mutant (without FBS); 3, C. jejuni F38011 (with FBS); 4, C. jejuni fliA mutant (with FBS). The positions of molecular mass standards are indicated on the left (in kilodaltons).
Maximal C. jejuni invasion requires Cia protein secretion and motility.
In vitro assays were performed to assess the contribution of motility and Cia protein secretion in C. jejuni uptake (Table 1). Because C. jejuni binding is a prerequisite for invasion (19), C. jejuni-INT 407 cell contact was promoted by centrifugation. Despite this effort to minimize motility-dependent effects on C. jejuni-host cell association, a reduction was noted in binding of all C. jejuni flagellar mutants (flhB, flgB, flgC, flgE2, and fliD) to the INT 407 cells compared to the C. jejuni wild-type strain. Therefore, the data are presented as the percentage of adherent bacteria that were internalized to more readily identify those organisms with deficiencies in invasive potential. With the exception of the C. jejuni fliD mutant, a significant (P < 0.01) decrease was noted in internalization efficiency of the C. jejuni flhB, flgB, flgC, and flgE2 flagellar mutants compared to the C. jejuni wild-type strain. Noteworthy is that the C. jejuni ciaB mutant displayed a Mot+ S− phenotype but exhibited an invasion phenotype that was similar to that of the flhB, flgB, flgC, and flgE2 flagellar export and structure mutants. Based on these results, it appears that motility, in the absence of Cia protein secretion, is not sufficient for maximal C. jejuni invasion of epithelial cells.
Binding and internalization assays were also performed with the C. jejuni 81116 wild-type strain, isogenic flaA flaB (Mot−, S−), and C. jejuni 81116 flaA (flaB+) (Mot−, S+) mutants (Table 1). Significant (P < 0.01) differences were noted in the invasive potentials of the C. jejuni 81116 flaA flaB and flaA (flaB+) mutants compared to the wild-type strain. A significant difference was also noted in the invasiveness of the C. jejuni flaA flaB and flaA flaB+ mutants compared to one another, with a greater number of the flaA (flaB+) mutant organisms internalized. Wassenaar et al. (41) also reported that a flaA (flaB+) strain was capable of invading INT 407 epithelial cells as long as cell-to-cell contact was promoted by centrifugation. Complementation of the flaA flaB mutant with a shuttle vector, in trans, harboring either the flaA or flaB gene resulted in transformants that displayed characteristics similar to that of the C. jejuni 81116 wild-type strain or the C. jejuni 81116 flaA (flaB+) mutant, respectively. However, transformation of the C. jejuni flaA flaB mutant with a shuttle plasmid harboring a functional flaA or flaB gene did not fully restore Cia secretion or the percentage of bacteria internalized to the expected levels. A possible explanation for the diminished secretion rates and corresponding reduction in internalization efficiency is that an increase in the amount of FlaA or FlaB within a cell may interfere with Cia secretion. In agreement with the results presented in Table 1, these data suggest that the secretion of the Cia proteins from the flagellar export apparatus contributes significantly to C. jejuni host cell invasion.
The C. jejuni F38011 fliD mutant displayed a filament structure similar to that of the C. jejuni 81116 flaA (flaB+) mutant. Given this finding, we hypothesized that the C. jejuni F38011 fliD mutant grown in liquid medium should be internalized at a level comparable to that of the C. jejuni 81116 flaA (flaB+) mutant, given that both mutants displayed truncated flagellar filaments. Invasion assays were performed with the C. jejuni F38011 fliD mutant grown in liquid medium and the C. jejuni F38011 fliD mutant grown on solid medium as an appropriate control (Table 2). Consistent with the results shown in Table 1 with the C. jejuni 81116 flaA (flaB+) mutant, a twofold difference in the ratio of internalized to adherent cells was obtained with the F38011 fliD mutant cultured in broth compared to the wild-type strain. Based on this finding as well as that obtained with the C. jejuni fla mutants, it appears that organisms that have a filament and are Mot+ are 1.5- to 2.5-fold more invasive than organisms that have a filament and are Mot−. However, the C. jejuni F38011 wild-type strain (S+, Mot+) was found to be approximately 50-fold more invasive than the C. jejuni ciaB isogenic mutant (S−, Mot+), which highlights the importance of the secretion-competent phenotype.
TABLE 2.
Phenotypes displayed by F38011 wild-type isolates and isogenic filament cap mutants
| Relevant characteristic | FlaA synthesis | Filament assembly | Motility | Secretion | Adherencea | Invasiona | I/A |
|---|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | 100 ± 14 | 100 ± 1 | 5.5 |
| fliD mutant (plate cultured) | + | + | + | + | 85 ± 6 | 64 ± 5 | 4.1 |
| fliD mutant (broth cultured) | + | Truncated | NDb | ND | 77 ± 6 | 29 ± 3 | 2.1 |
| flgC mutant | − | − | − | − | 52 ± 14 | 0.58 ± 0.2 | 0.06 |
Percentage of the value for the wild type.
ND, not determined.
DISCUSSION
Gram-negative bacteria possess at least six different mechanisms to actively transport proteins across the bacterial membranes (reviewed in reference 5). Of these six pathways, protein secretion induced upon contact of the bacteria with host cells has been referred to as the type III secretion pathway (10). Requirements of type III secretion pathways include the absence of a cleavable, hydrophobic amino-terminal signal sequence in the secreted protein, export of the protein across the bacterial inner and outer membranes without a periplasmic intermediate, and a signal to induce secretion (23). Most but apparently not all type III secreted proteins require chaperones (7, 8). We have demonstrated elsewhere that C. jejuni synthesize a novel set of proteins upon coculture with epithelial cells, some of which are secreted (22, 37). The secreted proteins were termed the Campylobacter invasion antigens (Cia proteins) because they were found to be required for maximal invasion of intestinal epithelial cells by C. jejuni (22, 30, 37). Because the Cia proteins are synthesized and secreted in response to an environmental stimulus and the secreted CiaB protein is not processed, the Cia proteins appear to conform to the criteria for type III proteins. However, a BLAST search of the C. jejuni genome revealed that the only apparent type III export system in C. jejuni is the flagellar apparatus.
A considerable amount of evidence exists that motility is essential for the maximal colonization of animals by C. jejuni (32, 33, 34, 36, 42). In parallel with these studies, additional work has been done to dissect the importance of motility versus the actual flagellum in the interaction of C. jejuni with cultured epithelial cells (15, 41, 43). Investigators have targeted genes encoding various flagellar structural components, and while discrepancies have been reported with respect to the phenotypes of particular mutants (14, 43), there appears to be a consensus among investigators that motility plays a role in C. jejuni pathogenesis. Moreover, motility and the expression of the flaA gene are clearly necessary for maximal invasion of eukaryotic cells and for the translocation of C. jejuni across polarized cells (15, 41). Perhaps more relevant to this study, differences in the invasive potential of C. jejuni flaA (flaB+) and C. jejuni flaA flaB strains were noted in earlier studies; C. jejuni flaA (flaB+) strains have been reported to be more invasive than a C. jejuni flaA flaB strain (15, 41). Also noteworthy is that the invasiveness of a C. jejuni flaA (flaB+) strain is enhanced 10-fold by promoting bacterium-host cell contact via centrifugation; in contrast, the centrifugation step did not change the invasive potential of the C. jejuni wild-type strain (40). Based on the difference observed in the invasive potential of the C. jejuni flaA (flaB+) strain versus the C. jejuni flaA flaB strain, Grant et al. (15) concluded that the flagellar structure played a role in internalization that was independent of motility. Prior to this study, it was unclear how the flagellum could have any effect on C. jejuni host cell invasion other than by conferring motility or acting directly as an adhesin.
Given our previous work suggesting that the Cia proteins are secreted in a type III-dependent manner and the absence of a type III secretion system dedicated to the export of virulence proteins in the C. jejuni genome, experiments were performed to determine if the flagellum serves as the Cia export apparatus. Mutations that abolished flagellin export (flhB, flgB, flgC, and flgE2), filament structure (fliD, flaA, flaB), and filament synthesis (fliA) were generated. With these mutants, we have shown that C. jejuni motility and virulence are linked. Specifically, we demonstrate that the C. jejuni Cia proteins are secreted via the flagellar export apparatus. The secretion system utilized by C. jejuni appears to be unique in that either one of the filament proteins is required for Cia protein secretion.
To test whether components of the flagellar apparatus serve as the C. jejuni Cia export apparatus, two separate experiments were performed. Mutations were generated in several flagellar structural genes in C. jejuni strain F38011 to determine if the loss of an operational flagellar apparatus resulted in the loss of Cia export. In addition, with two mutants of C. jejuni strain 81116 that were defective in expression of either one or both flagellin filaments as well as Cia protein export, we tested whether restoration of a flagellar filament also restored Cia protein synthesis. Mutations that affected either the export of flagellar components (flhB) or the nonfilament structural components (flgB, flgC, and flgE2) resulted in an S− phenotype. Comparable results were obtained with a second C. jejuni strain, 81116, in which the genes encoding the flagellin filament (flaA and flaB) were mutated. Complementation of the flagellar filament defect in 81116 with either flaA or flaB restored the organism's ability to secrete the Cia proteins. To ensure that the S− phenotype exhibited by the C. jejuni 81116 flaA flaB mutant was not unique to a particular strain, a C. jejuni F38011 flaA flaB mutant was generated. The C. jejuni F38011 flaA flaB mutant also exhibited an S− phenotype (not shown). Therefore, the genetic evidence presented is consistent with Cia protein secretion through the flagellar export system.
Insertion mutagenesis of fliD (Cj0548) and flgC (Cj0527c) was expected to have a polar effect on downstream gene expression. As the downstream genes in these putative operons are also expected to be associated with flagellar biosynthesis, we predicted that the phenotype associated with polarity on the downstream genes would be similar to that of the targeted gene. With regard to flgB and flgC, fliE (Cj0525c) and pbpB (Cj0524) would also be affected. In the case of fliD, fliS (Cj0549) and a hypothetical open reading frame of no known function (Cj0550) would have been affected.
The fact that the amount of FlaA protein was reduced in the whole-cell lysates of the C. jejuni flgB, flgC, and flgE2 mutants, as judged by immunoblot analysis with a flagellin antiserum, raised the possibility that C. jejuni may possess the FlgM anti-sigma factor. In bacteria such as Salmonella, Yersinia, and Helicobacter pylori, the negative regulator FlgM inhibits flagellin transcription in response to a defective hook-basal body complex (6, 9, 11). A protein corresponding to a putative FlgM homolog that shows similarity with the recently identified FlgM protein from H. pylori (9) has been identified in the genome of C. jejuni NCTC 11168 (Cj1464). In S. enterica serovar Typhi, σ28 is involved in regulating gene expression of type III proteins (13). Given this fact, it is clear that the expression of virulence genes in S. enterica serovar Typhi is affected, albeit indirectly, by FlgM and the assembly of the flagellar export apparatus. Regardless, the Cia proteins are secreted in a C. jejuni fliA (σ28) mutant. Therefore, the cia genes cannot be subject to transcriptional regulation via a mechanism involving the anti-sigma factor FlgM. Our results are in agreement with those of Jagannathan et al. (17), who observed that a C. jejuni fliA mutant displayed truncated flagella; this finding indicates that σ28 is not responsible for the transcription of the genes encoding the flagellar export apparatus in C. jejuni.
The ATPase FliI plays an essential role in flagellar apparatus assembly and in flagellar protein export. To address whether the S− phenotype of some of the C. jejuni mutants was due to regulatory effects, the expression of ciaB was analyzed in a C. jejuni F38011 fliI mutant. After the mutant was generated, RNA was extracted from bacteria that were cultured in Mueller-Hinton broth with 0.05% deoxycholate under microaerobic conditions. Importantly, ciaB was transcribed in the C. jejuni F38011 fliI mutant, as judged by reverse transcription-PCR. In addition, the CiaB protein was synthesized in the C. jejuni F38011 fliI mutant, as judged by immunoblot analysis with a CiaB-specific antibody.
The results of this study are consistent with the hypothesis that the flagellar type III secretion pathway is required for Cia protein export. Secretion of the Cia proteins requires a functional basal body and hook and at least one of the filament proteins. Coupled with the metabolic labeling experiments in which the C. jejuni strains were examined for protein secretion, the adherence and internalization data indicate that the difference in the invasiveness of the C. jejuni flaA flaB+ and C. jejuni flaA flaB strains is a result of Cia secretion. Based on the phenotypes of the C. jejuni ciaB mutant (Mot+, S−), it is also evident that motility, in the absence of Cia protein secretion, is not sufficient for C. jejuni invasion of epithelial cells. We believe that the data presented here reveal what had formerly been unclear about the Cia protein export apparatus and the relationship between C. jejuni motility and host cell invasion.
Acknowledgments
We thank Gary A. Flom for assistance in generation of the fliA suicide vector, which was used to generate the C. jejuni F38011 fliA mutant. We also thank Randal Eckert, Gary A. Flom, Nicole Lindstrom, and Chris Davitt for assistance in the preparation and examination of samples for TEM and Amy M. Keech for performing reverse transcription-PCR. We are grateful to Scott Minnich (Department of Microbiology, Molecular Biology, and Biochemistry, University of Idaho, Moscow, Idaho) and Anthony Garza (School of Molecular Biosciences, Washington State University) for helpful discussions and reviewing the manuscript.
This work was supported by a grant from the National Institutes of Health (grant DK50567) awarded to M.E.K.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M., and M. J. Blaser. 1995. Campylobacter jejuni and the expanding spectrum of related infections. Clin. Infect. Dis. 20:1092-1099. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., P. Guerry, and T. J. Trust. 1993. The Campylobacter σ54 flaB flagellin promoter is subject to environmental regulation. J. Bacteriol. 175:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J., J. G. Wells, R. A. Feldman, R. A. Pollard, and J. R. Allen. 1983. Campylobacter enteritis in the United States. A multicenter study. Ann. Intern. Med. 98:360-365. [DOI] [PubMed] [Google Scholar]
- 5.Büttner, D., and U. Bonas. 2002. Port of entry-the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 6.Chadsey, M. S., and K. T. Hughes. 2001. A multipartite interaction between Salmonella transcription factor σ28 and its anti-sigma factor FlgM: implications for σ28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915-929. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., and O. Schneewind. 2000. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 8:214-220. [DOI] [PubMed] [Google Scholar]
- 9.Colland, F., J.-C. Rain, P. Gounon, A. Labigne, P. Legrain, and H. De Reuse. 2001. Identification of the Helicobacter pylori anti-σ28 factor. Mol. Microbiol. 41:477-487. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daughdrill, G. W., M. S. Chadsey, J. E. Karlinsey, K. T. Hughes, and F. W. Dahlquist. 1997. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, σ28. Nat. Struct. Biol. 4:285-291. [DOI] [PubMed] [Google Scholar]
- 12.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. L. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golden, N. J., and D. W. K. Acheson. 2002. Identification of motility and auto-agglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70:1761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, C. C. R., M. E. Konkel, W. Cieplak, Jr., and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 17.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinsella, N., P. Guerry, J. Cooney, and T. J. Trust. 1997. The flgE gene of Campylobacter coli is under the control of the alternative sigma factor σ54. J. Bacteriol. 179:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konkel, M. E., and W. Cieplak, Jr. 1992. Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect. Immun. 60:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konkel, M. E., M. D. Corwin, L. A. Joens, and W. Cieplak, Jr. 1992. Factors that influence the interaction of Campylobacter jejuni with cultured mammalian cells. J. Med. Microbiol. 37:30-37. [DOI] [PubMed] [Google Scholar]
- 21.Konkel, M. E., S. G. Garvis, S. L. Tipton, D. E. Anderson, Jr., and W. Cieplak, Jr. 1997. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24:953-963. [DOI] [PubMed] [Google Scholar]
- 22.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32:691-701. [DOI] [PubMed] [Google Scholar]
- 23.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galán, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lüneberg, E., E. Glenn-Calvo, M. Hartmann, W. Bär, and M. Frosch. 1998. The central, surface-exposed region of the flagellar hook protein FlgE of Campylobacter jejuni shows hypervariability among strains. J. Bacteriol. 180:3711-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propeller and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, p. 89-90. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Matz, C., A. H. M. van Vliet, J. M. Ketley, and C. W. Penn. 2002. Mutational and transcriptional analysis of the Campylobacter jejuni flagellar biosynthesis gene. flhB. Microbiology 148:1679-1685. [DOI] [PubMed] [Google Scholar]
- 29.Mecsas, J. J., and E. J. Strauss. 1996. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg. Infect. Dis. 2:271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteville, M. R., and M. E. Konkel. 2002. Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 70:6665-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2003. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer membrane protein and microfilament reorganization. Microbiology 149:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131:1973-1980. [DOI] [PubMed] [Google Scholar]
- 33.Nachamkin, I., X. H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newell, D. G. 1986. Monoclonal antibodies directed against the flagella of Campylobacter jejuni: production, characterization and lack of effect on the colonization of infant mice. J. Hyg. 96:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuijten, P. J. M., B. A. M. van der Zeijst, and D. G. Newell. 1991. Localization of immunogenic regions on the flagellin proteins of Campylobacter jejuni 81116. Infect. Immun. 59:1100-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavlovskis, O. R., D. M. Rollins, R. L. Haberberger, Jr., A. E. Green, L. Habash, S. Strocko, and R. I. Walker. 1991. Significance of flagella in colonization resistance of rabbits immunized with Campylobacter spp. Infect. Immun. 59:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera-Amill, V., B. J. Kim, J. Seshu, and M. E. Konkel. 2001. Secretion of the virulence associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J. Infect. Dis. 183:1607-1616. [DOI] [PubMed] [Google Scholar]
- 38.Rivera-Amill, V., and M. E. Konkel. 1999. Secretion of Campylobacter jejuni Cia proteins is contact dependent, p. 225-229. In P. S. Paul and D. H. Francis (ed.), Mechanisms in the pathogenesis of enteric diseases, 2nd ed. Plenum Publishing Corporation, New York, N.Y. [DOI] [PubMed]
- 39.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassenaar, T. M., N. M. Bleumink-Pluym, D. G. Newell, P. J. M. Nuijten, and B. A. M. van der Zeijst. 1994. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect. Immun. 62:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. M. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 43.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 44.Yokoseki, T., K. Kutsukake, K. Ohnishi, and T. Iino. 1995. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 141:1715-1722. [DOI] [PubMed] [Google Scholar]
- 45.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]