Abstract
In Escherichia coli, interactions between the replication initiation protein DnaA, the β subunit of DNA polymerase III (the sliding clamp protein), and Hda, the recently identified DnaA-related protein, are required to convert the active ATP-bound form of DnaA to an inactive ADP-bound form through the accelerated hydrolysis of ATP. This rapid hydrolysis of ATP is proposed to be the main mechanism that blocks multiple initiations during cell cycle and acts as a molecular switch from initiation to replication. However, the biochemical mechanism for this crucial step in DNA synthesis has not been resolved. Using purified Hda and β proteins in a plate binding assay and Ni-nitrilotriacetic acid pulldown analysis, we show for the first time that Hda directly interacts with β in vitro. A new β-binding motif, a hexapeptide with the consensus sequence QL[SP]LPL, related to the previously identified β-binding pentapeptide motif (QL[SD]LF) was found in the amino terminus of the Hda protein. Mutants of Hda with amino acid changes in the hexapeptide motif are severely defective in their ability to bind β. A 10-amino-acid peptide containing the E. coli Hda β-binding motif was shown to compete with Hda for binding to β in an Hda-β interaction assay. These results establish that the interaction of Hda with β is mediated through the hexapeptide sequence. We propose that this interaction may be crucial to the events that lead to the inactivation of DnaA and the prevention of excess initiation of rounds of replication.
DNA replication consists of three sequential steps: initiation, elongation, and termination. In Escherichia coli the initiation of a new round of chromosome replication occurs when the initiation protein, DnaA, binds to a 9-mer DnaA box within the chromosomal origin, oriC (reviewed in references 34 and 41). In vitro studies have shown that the binding of DnaA to oriC in the presence of the DNA structural protein HU or IHF (14, 16, 40) stimulates opening of the DNA duplex by melting the AT-rich 13-mer region in oriC (4, 9, 26). The unwound region then provides an entry site for the DnaB-DnaC helicase, which expands the region of single-stranded DNA. DnaG primase, single-stranded DNA-binding protein, DNA polymerase III holoenzyme (Pol III), and other proteins required for the replication fork formation are then recruited, and bidirectional DNA synthesis is initiated (13).
In normal growth, cells replicate their DNA once before cell division and in E. coli there are at least three different mechanisms that block the occurrence of multiple initiations. The first is the sequestration of oriC by SeqA (33). The newly replicated DNA at the oriC is hemimethylated until acted upon by DNA-adenine methyltransferase (8). SeqA binds to the hemimethylated oriC locus and inhibits the initiation of replication until full methylation is reestablished, with the eclipse time being up to one-third of the cell cycle (8, 33). The second mechanism is the titration of free DnaA protein. The datA locus in E. coli, which contains a cluster of high-affinity DnaA binding sites, can bind 300 to 400 DnaA molecules in vivo (24), thus decreasing the available level of DnaA accessible to oriC (21). Overinitiation is observed when datA is removed from the chromosome, resulting in an increase in the amount of DnaA per cell. Introduction of additional intracellular copies of the datA site in trans is limiting for initiation and cell division (25).
The third mechanism is the accelerated conversion of ATP-DnaA to ADP-DnaA. While the DnaA protein binds to both adenosine nucleotides ATP and ADP (KDATP-DnaA, 0.03 μM; KDADP-DnaA, 1 μM) (18), only ATP-DnaA is active for initiation (reviewed in reference 20). Regulatory inactivation of DnaA, which is the switching of the active ATP-DnaA to the inactive ADP-DnaA, was initially observed in a soluble cell extract that specifically inhibited in vitro replication from an oriC-containing plasmid (18). This activity appears to be mediated by the β-clamp subunit of DNA Pol III (β2) loaded as a sliding clamp on template DNA, interacting with the newly identified Hda (for “homologous to DnaA”) protein (19, 22). Hda protein is one-half the size of DnaA and has high-level sequence homology to the domain III region of DnaA, which contains the ATP binding site (22). Hda also has a role in plasmid initiation, as it is found to interact with initiation protein TrfA of the broad-host-range plasmid RK2 (23).
The mechanisms that operate to allow the loaded β subunit and the Hda proteins to stimulate ATPase activity of DnaA are not known, but it appears to be stimulated by DNA synthesis (28). The results of recent studies of a cold-sensitive DnaA mutant with an A184→V change in the ATP binding motif of DnaA suggested that the inability of the mutant DnaA protein to bind and, hence, also to cleave ATP allows the mutant DnaA to stay bound at oriC. This would prevent the entry of the Pol complex and eventually lead to the arrest of DNA replication (38). This scenario has also been suggested for an ATP cleavage-defective mutant DnaA R335→A (36). Recent studies of the hda86 mutant (22) suggested that overinitiations in this mutant also led to the stalling of DNA replication. These results suggest that DnaA-ATP hydrolysis and concomitant DNA synthesis are coupled and that this coupling is perhaps mediated by the physical interaction of DnaA-Hda-β (19, 29). Whether DnaA-Hda-β interaction directly affects replication fork progression is not known, and it is possible that other unknown factors that interact with DnaA and other components of the replisome are also involved.
Here, we demonstrate that the E. coli Hda protein interacts with the β clamp in vitro. We identify a conserved hexapeptide as a likely β-binding motif located close to the amino terminus of members of Hda family of proteins. This motif, QL[SP]LPL, is related to the previously identified pentapeptide, β-binding motif QL[SD]LF (10). Peptides with this motif are known to inhibit interactions between Pol α and β and between the δ clamp loader and β and also to inhibit in vitro DNA replication by Pol III* (45). We demonstrate that the hexapeptide motif is required for Hda-β interaction, as mutations in the motif severely decrease Hda-β binding. We show that synthetic peptides, containing β-binding consensus pentameric sequence and Hda-derived hexamer motifs, can inhibit binding of Hda to β. Our results suggest that Hda binds directly to β and that it probably signals this interaction to DnaA, possibly via a conformational change, leading to stimulation of the ATPase activity of DnaA and hence inactivation of initiation of further rounds of DNA replication.
MATERIALS AND METHODS
Strains and materials.
E. coli strains XL1-Blue (Stratagene) and TOP10 (Invitrogen) were used as hosts in site-directed mutagenesis and subcloning of PCR inserts. BL21(DE3)pLyS (Novagen) and TB1 (New England Biolabs) were used as hosts for protein expression. Expression vector pET16b was from Novagen, and expression vector pMAL was from New England Biolabs. Antibodies to the maltose-binding protein (MBP) and amylose resin were purchased from New England Biolabs. Monoclonal anti-His tag antibody and Ni-nitrilotriacetic acid (NTA) agarose were purchased from Qiagen. Polyclonal antibodies to β2 were raised in rabbits as previously described (10). Restriction enzymes were from Promega, and chemicals were from Sigma. Proteinase inhibitors were obtained from Roche Molecular Biochemicals. Wild-type β2 was expressed and purified as described by Oakley et al. (39). Peptides (see Table 2) were synthesized and purified as described previously (10).
TABLE 2.
Sequences of synthetic peptides used in the Hda-β2 binding assays
| Peptide | Sequence | Source | Inhibition (μM) of Hda-β interaction (IC50)a |
|---|---|---|---|
| consensus_1 | IG QLDLF GV | 38-40 | |
| consensus_2 | IG QLSLF GV | 31-32 | |
| pepDnaE_4 | IG QADMA GV | DnaE | >500 |
| pepHda_n | PA QLSLPL YL | Hda | 7-8 |
| pepHda_e | IG QLSLPL GV | Hda | 25-28 |
IC50, 50% inhibitory concentration.
The PSI-BLAST searches of the Microbial Genome database at NCBI and MEME analysis.
The growing number of eubacterial genome sequences provides access to large data sets of orthologous proteins. The data sets used in this work were compiled from searches of the Microbial Genome database at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/MICROBES/InProgress.html). The BLAST searches were initiated with the founding member of the Hda family, E. coli Hda (GenBank accession NP_416991; also known as yfgE and b2496). The amino acid sequences of the most distantly related members were then used in searches until no further new members could be identified. Version 3 of MEME software was run on the San Diego Super Computer website (http://meme.sdsc.edu/meme/website/). MEME was run with the following parameters: zero or one motif per sequence, five motifs per alignment, and motifs 2 to 25 amino acids wide for the first analysis. The maximum size of the motif was then reduced until no further members of the protein family were included in the lists of significant matches.
Buffers.
The ingredients for the buffers used were as follows: for lysis buffer A, 50 mM NaH2PO4 (pH 9.0), 300 mM NaCl, 5 mM EDTA, 0.1% (vol/vol) Triton X-100, and 1 mg of lysozyme/ml; for lysis buffer B, 50 mM Tris (pH 7.5), 50 mM NaCl, 12 mM 2-mercaptoethanol, 1 mM benzamidine, 10 μM 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), 0.1% Triton X-100, and 1 mg of lysozyme/ml; for solubilization buffer, 8 M urea, 0.1 M NaH2PO4, 10 mM Tris (pH 8.0), 5 mM 2-mercaptoethanol, 1 mM benzamidine, and 10 μM AEBSF, pH 8.0; for column buffer, 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol; for refolding buffer, 20 mM Tris (pH 7.5), 3 M urea, 500 mM NaCl, 0.01% (vol/vol) Tween 20, 1 mM EDTA, 1 mM benzamidine, 10 mM glutathione (reduced form), and 1 mM glutathione (oxidized form); for protein storage buffer, 20 mM Tris (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 1 mM EDTA, 1 mM benzamidine, and 0.2 mM AEBSF; for wash buffer (WB), 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, and 0.05% (vol/vol) Tween 20; for coating buffer, 100 mM Na2CO3 (pH 9.5); for blocking buffer, 5% skim milk powder in WB; and for buffer C, 10 mM HEPES (pH 7.4), 150 mM NaCl, and 0.05% Tween 20.
Expression and purification of recombinant proteins. (i) Histidine-tagged proteins.
Full-length E. coli β gene (amino acids 1 to 366) was amplified from XL1-Blue genomic DNA with primers 5′-AAAGGCATATGAAATTTACCGTAGAACG-3′ and 5′-AACCAGGATCCTTACAGTCTCATTGGCAGAC-3′ (restriction sites are underlined) and cloned into NdeI and BamHI sites of pET16b to yield pET16b-β expression plasmid. Similarly, full-length Hda (amino acids 1 to 248) was amplified using primers 5′-AACCGCATATGTAAACTTCTCGCGATTTTGTG-3′ and 5′-AACCCTCGAGCTACAACTTCAGAATTTCTTTC-3′ (restriction sites are underlined). The PCR product was digested with NdeI and XhoI and cloned into pET16b. A soluble histidine decamer [a (MGH10SSGHIEGRH)-tagged form of the β2 protein (H10-β2)] was expressed and purified by Ni-NTA affinity resin (Qiagen) according to the manufacturer's protocol. The recombinant H10-Hda was present in the insoluble fraction and was purified from inclusion bodies. Briefly, the inclusion bodies were washed in lysis buffer B, sonicated, collected after centrifugation, and dissolved in solubilization buffer. The solubilized recombinant proteins were purified by affinity chromatography using the Ni-NTA resin. Bound proteins were eluted with a gradient of imidazole. After dialysis, the H10-Hda proteins were refolded in refolding buffer, dialyzed against urea, and stored in protein storage buffer at −80°C.
(ii) MBP-Hda fusion protein.
A full-length Hda open reading frame was amplified from XL1-Blue genomic DNA with primers 5′-ATGGTAAACTTCTCGCGATTTTG-3′ and 5′-AGGGAAGCTTCTACAACTTCAGAATTTCTTTC-3′ (the restriction site is underlined), digested with HindIII, and ligated with pMAL-C2/XmnI HindIII-linearized vector to create an MBP-Hda wild-type fusion protein. The MBP-Hda mutants were obtained by site-directed mutagenesis (see below). The expression and purification of soluble wild-type and mutant MBP-Hda proteins were performed according to the instructions of the manufacturer (New England Biolabs). After affinity purification using amylose resin, protein-containing fractions were pooled and dialyzed at 4°C against protein storage buffer.
Site-directed mutagenesis.
The pCR-Blunt-Hda plasmid (Invitrogen vector pCR-Blunt containing an Hda gene insert) was used as a template for the generation of alanine substitution mutants with a QuikChange site-directed mutagenesis kit (Stratagene). The putative β-binding hexapeptide motif on E. coli Hda (QLSLPL) was replaced by QASAPA in mutant A1 and by QLSAAL in mutant A2. The mutant clones were sequenced using a Big Dye Terminator ready-reaction sequencing kit (Perkin-Elmer Life Sciences) to confirm the presence of the mutation and the authenticity of the remaining Hda sequence.
Microtiter plate binding assay and peptide inhibition assay.
Purified H10-Hda or MBP-Hda was diluted to 0.5 μg/ml in coating buffer and adsorbed onto 96-well microtiter plates (Falcon flexible plates; Becton Dickinson) by overnight incubation (50 μl/well) at 4°C. Subsequent steps were performed at room temperature. Plates were washed with WB, blocked with Blotto (100 μl/well for at least 1 h), and washed again with WB. All washing steps were done by manually submerging the plates in the WB bath (at least 500 ml of WB was used). For binding assays, plates were incubated for 1 h with 0 to 5 μg of wild-type β2/ml in buffer C (100 μl). Purified MBP-paramyosin (New England Biolabs) was used as a control. After binding to wild-type β2 was performed, plates were washed and incubated with rabbit anti-β serum (1: 1,000; 50 μl/well; 1 h). This was followed by a washing step and incubation with sheep anti-rabbit immunoglobulin horseradish peroxidase (1:1,000; 50 μl/well; 1 h). For detection, 2′,2′-azino-bis 3-ethylbenzthiazoline-6-sulfonic acid (ABTS) was used as the substrate. The absorbance of microtiter plates was read at A405 using a Multiskan reader with Ascent software (Labsystems). Triplicate assays were performed for wild-type MBP-Hda and were repeated at least three times for each mutant.
For inhibition assays, the listed peptides (see Table 2) were allowed to associate with wild-type β2 (5 μg/ml in buffer C) in preblocked 96-well microtiter plate (Sarstedt) for 90 min. Samples (50 μl) were then transferred from each well to a corresponding well of the Hda-coated plates. After 15 to 20 min of incubation, the plates were thoroughly washed in WB and treated with rabbit anti-β serum (1:1,000 dilution in WB; 50 μl/well) for 30 min. The assay was developed as described above.
Pulldown assay.
Purified H10-β2 (5 μg) coupled to 50 μl of Ni-NTA resin (2 h at 4°C with rotation) was incubated with 10 μg of wild-type or mutant MBP-Hda protein in buffer C (200 μl) at 4°C for 30 min with constant agitation. After excessive washing with the same buffer (four times with 500 μl), the resin was resuspended in 25 μl of sodium dodecyl sulfate (SDS) sample buffer and the bound proteins were separated in SDS-polyacrylamide gel electrophoresis (SDS-10% PAGE) (Bis-Tris gel; Invitrogen) and visualized by Coomassie staining. As a control, wild-type and mutant MBP-Hda proteins were incubated with the resin lacking H10-β2 under the same conditions to check for nonspecific binding of the protein to the Ni-NTA resin.
Western blotting.
Proteins were separated by SDS-10% PAGE and electroblotted onto a nitrocellulose membrane (Bio-Rad). After incubation for 1 h at room temperature with blocking buffer, Tris-buffered saline (TBS) containing 5% nonfat dry milk, the membrane was incubated with primary antibody for 1 h at room temperature in the same buffer, washed extensively with TBS with 0.05% (vol/vol) Tween 20, and incubated with peroxidase-conjugated secondary antibody for 1 h. After extensive washing with TBS with 0.05% (vol/vol) Tween 20, the blot was developed by precipitation of 4-chloro-1-naphthol as the substrate.
RESULTS
Hda and β2 interaction.
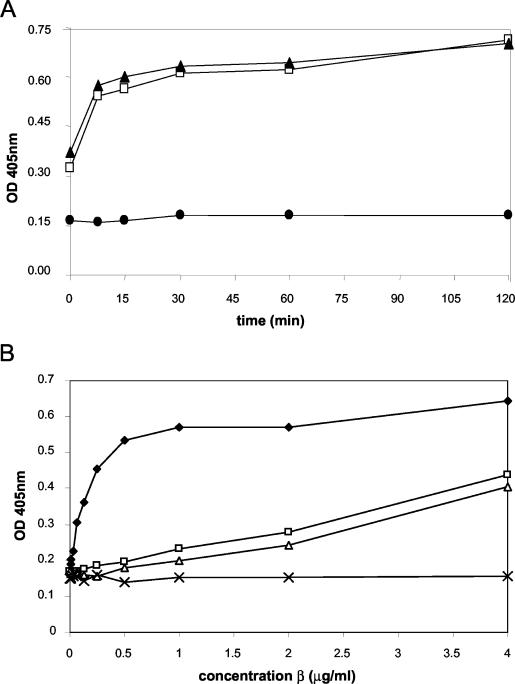
To determine whether Hda interacts directly with β2, we used purified recombinant E. coli MBP-Hda (Hda fused at the amino terminus with the 40-kDa MBP), H10-β2 protein (β protein tagged with a hexahistidine decamer at the amino terminus), and a polyclonal antibody to the β polypeptide in microtiter plate binding assays. In the plate binding assays MBP-Hda was coated onto 96-well microtiter plates and allowed to associate with β2. The MBP-Hda fusion protein was used in the assays, because similarly purified MBP-Hda has previously been shown to be active in in vitro complementation of the hda86 mutant (22). Time courses of 2 and 5 μg of β2/ml binding to MBP-Hda showed that the association was time dependent, reaching saturation in 20 to 25 min (Fig. 1A). Τhe binding curves of increasing concentrations of β2 in wells coated with MBP-Hda or MBP-paramyosin are shown (Fig. 1B). Low levels of background binding to the negative-control protein MBP-paramyosin were observed, while an increasing signal was achieved with increasing concentrations of β2 binding to MBP-Hda. Similar results were obtained when MBP-Hda was replaced in the binding assay with H10-Hda (data not shown).
FIG. 1.
Interactions of Hda with β2. (A) Time courses of two different concentrations of β2 bound to immobilized wild-type MBP-Hda. The binding is concentration dependent and reaches a plateau after 20 to 25 min of incubation. •, no β2; □, 2 μg of β2/ml; ▴, 5 μg of β2/ml. (B) Comparison of the abilities of MBP-Hda wild-type and mutant proteins to bind to β2. MBP-Hda wild-type and mutant proteins were immobilized onto the plate at 0.5 μg/ml. β2 was added followed by the detection of bound β2 with anti-b2 antibody. ♦, MBP-Hda wild type; □, MBP-Hda mutant A1; ▵, MBP-Hda mutant A2; ×, the negative-control MBP-paramyosin.
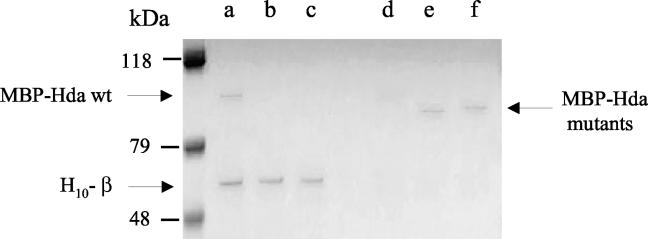
To confirm the results of the plate assay we employed a conventional pulldown technique. In the pulldown experiment, 10 μg of purified MBP-Hda was allowed to bind to 5 μg of β2 immobilized on Ni-NTA agarose resin. After binding, the proteins were resolved by SDS-PAGE and the gel was stained with Coomassie blue. The results (Fig. 2, lane a) confirmed that MBP-Hda binds β2. Purified MBP alone or MBP-paramyosin did not bind β2 (data not shown). This experiment together with the plate assays provides strong evidence for a direct interaction of Hda and β2.
FIG. 2.
Association between Hda and β2 in Ni-NTA resin pulldown experiments. Proteins were analyzed by SDS-10% PAGE and visualized by Coomassie brilliant blue staining. H10-β2 coupled to Ni-NTA resin samples interacted with wild-type (wt) MBP-Hda (lane a) but not with MBP-Hda mutant A1 (lane b) or MBP-Hda mutant A2 (lane c). The supernatant (unbound protein) of each reaction was also analyzed (lanes d, e, and f for MBP-Hda wild type, mutant 1, and mutant 2, respectively).
Identification and distribution of putative orthologues of Hda in the eubacteria.
Through the use of the amino acid sequence of E. coli Hda (gi:6176581), a series of BLAST searches of the completed and unfinished microbial genomes database at NCBI were undertaken. Genes encoding probable orthologues of Hda were identified in most, but not all, proteobacteria. Of the completed genome sequences of proteobacteria deposited in GenBank on 2 September 2002, Hda homologues were not identified in Brucella sp., Campylobacter jejuni, Helicobacter pylori, Buchnera sp., and Vibrio cholerae. Probable Hda orthologues were not identified in any nonproteobacteria.
Identification of a candidate β-binding motif in the amino-terminal domain of Hda proteins.
The binding of the E. coli replication-repair proteins DnaE, PolB, UmuC, DinB, and MutS to β has been shown to involve short amino acid sequences identified as belonging to a pentapeptide motif with the consensus sequence QL[SD]LF (2, 10, 11, 30). A related, but distinct, tripeptide motif (SLF) has been identified in members of the HolA (δ) family of clamp-loading subunits (10, 17). However, members of the Hda family of proteins do not appear to contain either of these motifs.
The previously identified β-binding motifs have frequently been located at the carboxy termini of the proteins or in regions that do not contain domains conserved between different families of proteins (10). To identify regions containing cross-protein family conserved domains, the amino acid sequences of representative Hda family proteins were used to search the NCBI Conserved Domain database. The members of the Hda family are related to the DnaA family, with most sequences examined exhibiting a region with a significant match to the bacterial DnaA domain (bac-dnaA; pfam00308). At the carboxy-terminal end of the Hda proteins the matches with the bac-dnaA domain extended to or very close to the end of the proteins. At the amino terminus two clusters of start points for the regions of significant matches between Hda proteins and the bac-dnaA domain from position 1 of the bac-dnaA domain consensus sequence and from position 37 were observed. The regions of all available Hda proteins from the amino-terminal amino acid to the residues equivalent to position 1 of the bac-dnaA domain (position 28 in E. coli Hda) and to position 37 of the bac-dnaA domain (position 56 in E. coli Hda) cutoffs were then analyzed using MEME (1) motif identification software. The first line of the consensus sequence from the MEME search was taken as the core motif for each of the motifs identified (Table 1).
TABLE 1.
Identification of a conserved hexapeptide motif in the amino-terminal domain of Hda proteinsa
| Dataset and no. of hits | P | Motif |
|---|---|---|
| Hda 12-56b | ||
| 41 | 1.1e-67 | TFDNFY |
| 51 | 1.1e-60 | QLPLPLe |
| 21 | 1.4e-26 | YLPDDE |
| 9 | 3.6e-18 | QEHSGY |
| 4 | 5.5e-7 | YVERLC |
| Hda 12-28c | ||
| 50 | 1.4e-99 | PAQLPLPY |
| Hda motifd | ||
| 41 | 1.4e-45 | QLPLPL |
The amino-terminal regions of the Hda family proteins (equivalent to amino acids 12 to 56 of the E. coli Hda sequence) were analysed with MEME motif identification software.
Redundant data set. Size, 51 sequences.
Redundant data set excluding the Colwellia sp. sequence, which was too short for inclusion. Size, 50 sequences.
Nonredundant data set. Size, 41 sequences.
Conserved sequence is underlined.
The MEME analysis identified a number of motifs close to the amino terminus of the proteins, but only one (QLPLPL) was conserved in almost all putative orthologues of Hda (Table 1). The same motif was identified using a nonredundant data set of the 6-amino-acid motif and up to 10 flanking residues from the Hda proteins. This motif is related to the previously identified QL[SD]LF β-binding motif (10), making it a good candidate for being the β-binding motif. This was further supported by the observation that the Wolbachia sp. homologue of E. coli Hda contained a pentapeptide motif (QLNLF) at the equivalent position. More recently, additional DNA sequences encoding putative Hda orthologues with similar pentapeptide motifs from Anaplasma phagocytophila (QLKLF), Erlichia chaffeensis (QLTLF), and Neorickettsia sennetsu (QLILI) have also been deposited in GenBank. These species all lie in the Rickettsiales group of the α-Proteobacteria. The Rickettsia spp. also lie in this group; they have the difficult-to-classify sequence QQYIF[RH]F, which has elements of both the hexapeptide and pentapeptide motifs.
The Hda QL(SP)LPL motif as part of the β-binding site.
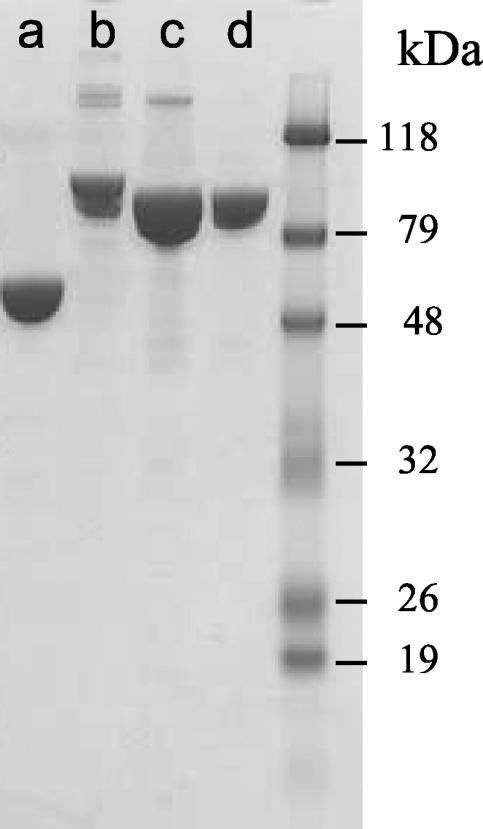
To determine whether the peptide motif constitutes part of the β-binding site, two mutant MBP-Hda proteins were constructed by replacing either the leucine residues or the proline residue with alanine as follows: QLSLPL → QASAPA in mutant A1 and QLSLPL → QLSAAL in mutant A2. The wild-type and mutant proteins were purified using the MBP tag, and the Coomassie-stained protein results are shown in Fig. 3. The alanine substitution mutants of Hda protein were tested in both the plate assay (Fig. 1B) and in the pulldown reactions (Fig. 2, lanes b and c). The results show that the ability of the mutants to bind to β2 was severely limited. These experiments suggest that the interaction of Hda with β2 is mediated through the hexapeptide sequence. Another possible explanation is the incorrect folding of the mutant proteins, although there was no indication that this was the case, as both mutants were soluble and bound amylose resin with high-level affinity. However, the three MBP-Hda fusion proteins were observed to have different migration profiles on SDS-PAGE (Fig. 3). These differences may be due to different amounts of the proteins loaded. In addition, in mutant A2 a proline residue was changed to alanine. Proline is known to make protein less globular and hence tends to retard the migration of the protein. The identity of the higher-molecular-mass bands present in the wild-type and mutant A1 samples is not known. It is possible that they were purification impurities (i.e., indigenous E. coli proteins which have affinity for the amylose resin).
FIG. 3.
Expression of β and MBP-Hda proteins. Coomassie brilliant blue-stained gel of purified proteins resolved in SDS-10% PAGE. Lane a, purified β wild type; lane b, MBP-Had; lane c, MBP-Hda mutant A1; lane d, MBP-Hda mutant A2.
Interaction of Hda with β2 is inhibited by peptides containing the putative β-binding motifs.
The crystal structure of the δ:β complex shows that δ subunit binds to the hydrophobic pocket located between the middle and the carboxy-terminal domain of β2 (17) and that the conserved hydrophobic residues L73F74 of the β-binding motif of δ (SLF) (5) bind in the hydrophobic pocket of β. Previous work by Dalrymple et al. has shown that peptides containing QL[SD]LF, the consensus pentapeptide motif, can inhibit α:β and δ:β interactions in vitro (10). These pentapeptides and a related 16-residue C-terminal peptide of Pol IV with the RQLVLGL β-binding motif can competitively inhibit Pol IV and α subunit DNA Pol activities in vitro (7, 45). If the interaction of β2 with Hda were indeed mediated through the hexapeptide sequence, one would expect that the binding of Hda to β2 would be disrupted by the addition of synthetic peptides containing the putative Hda β-binding motif. That this is the case is summarized in Table 2. Experiments using PepHda_n, which has the natural occurring dipeptide flanking the putative β-binding sequence at 7 to 8 μM, resulted in 50% inhibition of binding of Hda to β2. A 50% inhibition of binding was achieved with 25 to 28 μM pepHda_e that has the dipeptide flanking sequence derived from “DnaE”-flanked counterparts. The addition of the consensus_1 and consensus_2 peptides containing the related pentameric motif also inhibited the binding of Hda to β2 but was not as efficient as that of the Hda peptides (Table 2). Within the discrimination of the plate assays, we found that the native 9-mer Hda peptide (pepHda_n) was consistently better at inhibiting Hda-β2 interaction than pepHda_e and the consensus peptides.
It has been demonstrated that a number of DNA replication-repair proteins such as DnaE (α), the mismatch-repair protein, MutS, and the translesion DNA Pols (PolB, DinB, and UmuDC) share a pentapeptide, QL[SD]LF, and that the δ subunit contains a related SLF motif for binding to the same site on β2 (2, 7, 10). The observation that peptides containing the pentameric consensus sequence were also able to inhibit Hda binding to β2 (Table 2) suggests that the β-binding motif of Hda targets the same site on β2 as other replication-repair proteins. This is further supported by our data that pepHda_n and pepHda_e inhibit α:β and δ:β interactions in vitro (45).
DISCUSSION
In addition to functioning as a sliding clamp for DNA replicative Pols DnaE (α subunit) and PolC, β2 also interacts with the δ subunit of the clamp loader and with other Pols participating in both DNA damage checkpoint control and translesion DNA synthesis such as PolA (31), Pol II (PolB) (3, 15), Pol V (UmuDC) (42, 43), and Pol IV (DinB) (43, 44) and with other mismatch repair proteins, including MutS and LigA (31) and also MutL (37). In this work we have shown that β2 also interacts with Hda, a protein that is involved in the negative regulation of chromosome replication initiation in E. coli. These results suggest that β2 is the central molecule that coordinates DNA replication with DNA repair as well as with the switching from the initiation to elongation modes of replication by regulating inactivation of the initiator protein DnaA. They also support the proposal that the role of Hda in the negative regulation of initiation is in binding to both DnaA and β2, in which the proteins bind to different sites of Hda. It has also been demonstrated that β2 must be bound to DNA for this negative regulation to occur (19), and our experiments show that Hda can bind to β2 in solution. It may be that the requirement for β2 to be bound to DNA in vivo (19) is related to the proximity or orientation of β2 such that Hda can bind both β2 and DnaA.
The results of the protein-protein interaction assays and protein-peptide binding assays of the mutant Hda proteins have identified the QL[SP]LPL motif in the amino-terminal region of Hda as the key site in the interaction of Hda with β2. The relatively high level of conservation of this motif across all but one of the known orthologues of Ηda suggests that this site contributes to the binding of all Hda proteins to their β2 partners. Experiments examining the crystal structures of the clamp loader complex (17) and the 16-residue C-terminal peptide of DNA Pol IV (6, 7) bound to β2 have identified the L73F74 residues in E. coli δ and the residues L14L16 of P16 as being central to the interactions (7, 17). It is likely that the conserved LF in the pentapeptide β-binding motif is equivalent to these residues. Our results confirm the importance of the conserved LPL motif as a key component in the Hda and β2 interaction. Comparison of the hexapeptide motif with the δ and pentapeptide β-binding motifs suggests that the two conserved leucine residues may be equivalent to the LF motifs. It is not clear whether there is a functional biological difference between the pentapeptide and hexapeptide motifs, as a number of the putative Hda orthologues contain pentapeptide motifs. In addition, the hexapeptide motif is found sporadically distributed in a number of families of β-binding proteins containing predominantly pentapeptide motifs, in particular, the DinB and MutS families (11). These observations suggest that there appear to be two functionally equivalent β-binding peptide motifs: the pentapeptide motif is the most common in all known families of β-binding proteins except the Hda family (which has a few members with a pentapeptide motif), while the hexapeptide motif is most frequent in the Hda family and is sporadically distributed across some of the other families and includes the QLVLGL motif from E. coli DNA Pol IV (DinB) (6, 7). Conversion between the two motifs could potentially occur without the loss of β-binding activity.
Recently López de Saro et al. (32) identified the C-terminal seven residues (QVELEFD) of the α subunit of DNA Pol III in E. coli as a β-binding site. However, this site does not appear to be present in all DnaE proteins; in fact, it is only readily identifiable in species closely related to E. coli. This region of the protein is not present at all in Mycoplasma and Ureaplasma species. The QADMF site identified by Dalrymple et al. (10) is readily identifiable in most known members of the DnaE family (including those from Mycoplasma and Ureaplasma spp.) and is in an equivalent location with respect to the Pol domain of the related PolC family of DNA Pols.
The limited distribution of the orthologues of Hda suggests that Hda may have arisen from DnaA by a gene duplication event in the progenitor of the Proteobacteria family. If this was the case, the hexapeptide β-binding motif may have been acquired as a new event independently of the other families of β-binding proteins. The sequence of the β-binding motif and the peptide competition assays suggest that Hda binds to the same site on β2 as DnaE, δ, etc. Experimental data with a number of β-binding proteins suggest that although β2 is a dimer and therefore has two potential binding sites, only one site is occupied (35). In this case, binding of DnaE and synthesis of DNA could not have initiated at the origin until Hda had vacated the β-binding site. This suggests that the stalled DNA replication in DnaA and Hda mutants would not be directly due to the abnormal DnaA/Hda/β interaction, as has been proposed previously (36, 38).
Genes encoding orthologues of Hda are widely, but not universally, distributed in members of the Proteobacteria family. Hda homologues were not identified in the following Proteobacteria species: Brucella sp., Silicibacter pomeryi, C. jejuni, Heliobacter pylori, Buchnera sp., Dichelobacter nodosus, and V. cholerae. Hda orthologues were not identified in any nonproteobacteria. In Bacillus subtilis, YabA, which is also a negative regulator of initiation of rounds of replication, binds to the respective β2 protein (37). Though genes encoding orthologues of Hda are widely distributed in the Proteobacteria family, genes encoding orthologues of YabA are only found in a group of species related to B. subtilis. Both Hda and YabA also bind to the respective members of the DnaA family but despite their apparent functional similarities exhibit no significant amino acid similarity. In addition, no peptide motifs similar to either the penta- or hexa-peptide β-binding motifs have been identified in YabA proteins (11).
As determined by modeling Hda on the structure of the Aquifex aeolicus DnaA (12), the β-binding peptide is likely to be relatively exposed and unstructured in Hda (result not shown). The mechanism by which the stabilization of this interaction leads to a stalling of initiation of DNA replication is not clear, but presumably the establishment of the leading-strand DNA Pol complex at initiation is different in at least some respects from the reinitiation of lagging-strand synthesis at many points around the chromosome. A major difference is that at the initiation of synthesis at the origin of replication, no Pol subunits are closely associated or bound to the DNA whereas at the reinitiation events the Pol is at least in part already associated with the DNA. At some point in the initiation process there must be a handover of the loaded β to the Pol. It is not known whether Hda and the α subunit of DNA Pol III can both bind to a single β2 at the same time. Further work is required to address the precise order of events in the handover of β from a stationary complex of DnaA and Hda at the origin of replication to the mobile complex of DNA Pol III and, thus, the successful initiation of a new round of DNA synthesis.
The prevention of multiple chromosome replication initiation events appears to involve a number of degenerate mechanisms. In E. coli, three systems have been characterized: the regulatory inactivation of DnaA by Hda and β2, inactivation of oriC by the SeqA protein, and the titration of DnaA on the datA locus. B. subtilis does not encode either SeqA (27) or a member of the Hda family. In fact, homologues of SeqA have only been identified in members of the Enterobacteriaceae family, Salmonella spp., members of the Pasteurellaceae family, Colwellia spp., Shewanella putrefaciens, and Vibrio spp. In contrast, orthologues of DnaA are likely to be present in all eubacteria, suggesting that titration of DnaA on datA or an equivalent is likely to be a primary regulatory mechanism. The more limited distribution of Hda, YabA, and SeqA proteins perhaps reflects the more recent evolution of additional layers of control.
Acknowledgments
We thank Roger Pearson for help with the protein purification and Ross Tellam and Rob Seymour for reviewing the manuscript.
REFERENCES
- 1.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Xth Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 2.Becherel, O. J., R. P. P. Fuchs, and J. Wagner. 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1:703-708. [DOI] [PubMed] [Google Scholar]
- 3.Bonner, C. A., P. T. Stukenberg, M. Rajagopalan, R. Eritja, M. O'Donnell, K. McEntee, H. Echols, and M. F. Goodman. 1992. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol. Chem. 267:11431-11438. [PubMed] [Google Scholar]
- 4.Bramhill, D., and A. Kornberg. 1988. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743-755. [DOI] [PubMed] [Google Scholar]
- 5.Bullard, J. M., A. E. Pritchard, M. S. Song, B. P. Glover, A. Wieczorek, J. Chen, N. Janjic, and C. S. McHenry. 2002. A three-domain structure for the delta subunit of the DNA polymerase III holoenzyme delta domain III binds delta′ and assembles into the DnaX complex. J. Biol. Chem. 277:13246-13256. [DOI] [PubMed] [Google Scholar]
- 6.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnouf, D. Y., V. Olieric, J. Wagner, S. Fujii, J. Reinbolt, R. P. P. Fuchs, and P. Dumas. 2004. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J. Mol. Biol. 335:1187-1197. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 9.Crooke, E., R. Thresher, D. S. Hwang, J. Griffith, and A. Kornberg. 1993. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J. Mol. Biol. 233:16-24. [DOI] [PubMed] [Google Scholar]
- 10.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalrymple, B. P., G. Wijffels, K. Kongsuwan, and P. A. Jennings. 2003. Towards an understanding of protein-protein interaction network hierarchies—bioinformatics analysis of DnaN (β)-binding peptide motifs in members of protein families interacting with the eubacterial processivity clamp, the β subunit of DNA polymerase III. Conf. Res. Pract. Inf. Technol. 19:153-159. [Google Scholar]
- 12.Erzberger, J., M. M. Pirruccello, and J. M. Berger. 2002. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 18:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, L., M. J. Davey, and M. O'Donnell. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541-553. [DOI] [PubMed] [Google Scholar]
- 14.Grimwade, J. E., V. T. Ryan, and A. C. Leonard. 2000. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35:835-844. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, A. J., Jr., S. K. Bryan, H. Chen, R. E. Moses, and C. S. McHenry. 1991. Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits. J. Biol. Chem. 266:4568-4573. [PubMed] [Google Scholar]
- 16.Hwang, D. S., and A. Kornberg. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with HU or IHF. J. Biol. Chem. 267:23083-23086. [PubMed] [Google Scholar]
- 17.Jeruzalmi, D., O. Yurieva, Y. Zhao, M. Young, J. Stewart, M. Hingorani, M. O'Donnell, and J. Kuriyan. 2001. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417-428. [PubMed] [Google Scholar]
- 18.Katayama, T., and E. Crooke. 1995. DnaA protein is sensitive to a soluble factor and is specifically inactivated for initiation of in vitro replication of the Escherichia coli minichromosome. J. Biol. Chem. 270:9265-9271. [DOI] [PubMed] [Google Scholar]
- 19.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosome replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 20.Katayama, T., and K. Sekimizu. 1999. Inactivation of Escherichia coli DnaA protein by DNA polymerase III and negative regulations for initiation of chromosomal replication. Biochimie 81:835-840. [DOI] [PubMed] [Google Scholar]
- 21.Katayama, T., K. Fujimitsu, and T. Ogawa. 2001. Multiple pathways regulating DnaA function in Escherichia coli: distinct roles for DnaA titration by the datA locus and the regulatory inactivation of DnaA. Biochimie 83:13-17. [DOI] [PubMed] [Google Scholar]
- 22.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, P. D., T. Banack, D. M. Lerman, J. C. Tracy, J. E. Camara, E. Crooke, D. Oliver, and W. Firshein. 2003. Identification of a novel membrane-associated gene product that suppresses toxicity of a TrfA peptide from plasmid RK2 and its relationship to the DnaA host initiation protein. J. Bacteriol. 185:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa, R., H. Mitsuki, T. Okazaki, and T. Ogawa. 1996. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19:1137-1147. [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication inhibition by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Co., New York, N.Y.
- 27.Kunst, F. N., L. Ogasawara, A. M. Moszer, G. Albertini, V. Alloni, M. G. Bertero, et al. 1997. The complete genome sequence of the gram positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa, K., T. Mizushima, T. Kubota, T. Tsuchiya, T. Katayama, and K. Sekimizu. 1998. A stimulation factor for hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. Biochem. Biophys. Res. Commun. 243:90-95. [DOI] [PubMed] [Google Scholar]
- 29.Kurokawa, K., S. Nishida, A. Emoto, K. Sekimizu, and T. Katayama. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18:6642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenne-Samuel, N., J. Wagner, H. Etienne, and R. P. Fuchs. 2002. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 3:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase and DNA polymerase I. Proc. Natl. Acad. Sci. USA 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López de Saro, F. J., R. E. Georgescu, M. F. Goodman, and M. O'Donnell. 2003. Competitive processivity—clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 22:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 34.Messer, W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26:355-374. [DOI] [PubMed] [Google Scholar]
- 35.Naktinis, V., J. Turner, and M. O'Donnell. 1996. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell 84:137-145. [DOI] [PubMed] [Google Scholar]
- 36.Nishida, S., K. Fujimitsu, K. Sekimizu, T. Ohmura, T. Ueda, and T. Katayama. 2002. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication. J. Biol. Chem. 277:14986-14995. [DOI] [PubMed] [Google Scholar]
- 37.Noirot-Gros, M.-F., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, S. Dusko Ehrlich, and P. Noirot. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci USA 99:8352-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyborg, M., T. Atlung, O. Skovgaard, and F. G. Hansen. 2000. Two types of cold sensitivity associated with the A184→V change in the DnaA protein. Mol. Microbiol. 35:1202-1210. [DOI] [PubMed] [Google Scholar]
- 39.Oakley, A. J., P. Prosselkov, G. Wijffels, J. L. Beck, M. C. Wilce, and N. E. Dixon. 2003. Flexibility revealed by the 1.85 °A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. Sect. D Biol. Crystallogr. 59:1192-1199. [DOI] [PubMed] [Google Scholar]
- 40.Skarstad, K., T. A. Baker, and A. Kornberg. 1990. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 9:2351-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skarstad, K., and E. Boye. 1994. The initiator protein DnaA: evolution, properties and function. Biochim. Biophys. Acta 1217:111-130. [DOI] [PubMed] [Google Scholar]
- 42.Sutton, M. D., T. Opperman, and G. C. Walker. 1999. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc. Natl. Acad. Sci. USA 96:12373-12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. Fuchs. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wijffels, G., B. Dalrymple, P. Prosselkov, K. Kongsuwan, V. C. Epa, P. E. Lilley, S. Jergic, J. Buchardt, S. E. Brown, P. F. Alewood, P. A. Jennings, and N. E. Dixon. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry, in press. [DOI] [PubMed]