Abstract
Hereditary spastic paraplegias (HSPs) are neurodegenerative motor neuron diseases characterized by progressive age-dependent loss of corticospinal motor tract function. Although the genetic basis is partly understood, only a fraction of cases can receive a genetic diagnosis, and a global view of HSP is lacking. By using whole-exome sequencing in combination with network analysis, we identified 18 previously unknown putative HSP genes and validated nearly all of these genes functionally or genetically. The pathways highlighted by these mutations link HSP to cellular transport, nucleotide metabolism, and synapse and axon development. Network analysis revealed a host of further candidate genes, of which three were mutated in our cohort. Our analysis links HSP to other neurodegenerative disorders and can facilitate gene discovery and mechanistic understanding of disease.
Hereditary spastic paraplegias (HSPs) are a group of genetically heterogeneous neurodegenerative disorders with prevalence between 3 and 10 per 100,000 individuals (1). Hallmark features are axonal degeneration and progressive lower limb spasticity resulting from a loss of corticospinal tract (CST) function. HSP is classified into two broad categories, uncomplicated and complicated, on the basis of the presence of additional clinical features such as intellectual disability, seizures, ataxia, peripheral neuropathy, skin abnormalities, and visual defects. The condition displays several distinct modes of inheritance, including autosomal dominant, autosomal recessive, and X-linked. Several loci have been linked to autosomal recessive HSP (AR-HSP), from which 22 genes with mutations have been cloned. However, most of the underlying causes of HSP remain unidentified.
We analyzed 55 families displaying AR-HSP by whole-exome sequencing (WES). We identified the genetic basis in about 75% of the cases, greatly increasing the number of mutated genes in HSP; functionally validated many of these genes in zebrafish; defined new biological processes underlying HSP; and created an “HSPome” interaction map to help guide future studies.
Multiple Genes Are Implicated in HSP
We used WES to identify the genetic causes of AR-HSP in families with documented consanguinity. Selecting from these families without congenital malformations referred for features of either complicated or uncomplicated HSP (table S1), we performed WES on 93 individuals typically from two affected siblings or cousins where possible, for multiplex families, or one affected and one unaffected sibling or both parents, for simplex families. We prioritized predicted protein frame shift, stop codon, splice defects, and conserved nonsynonymous amino acid substitution mutations [Genomic Evolutionary Rate Profile (GERP) score > 4 or phastCons (genome conservation) score > 0.9]. We excluded variants with an allele frequency of greater than 0.2% in our internal exome database of over 2000 individuals. We genotyped each informative member from the majority of families with a 5000 single-nucleotide polymorphism (SNP) panel and generated genome-wide parametric multipoint linkage plots or used WES data to generate homozygosity plots (2). We excluded variants falling outside of homozygous intervals <2.0 Mb threshold (fig. S1).
We tested segregation of every variant meeting these criteria (table S2). We report a candidate HSP gene only if there was a single deleterious variant that segregated in the family or if the gene was identified as mutated in multiple families (3). For 15 families, a single genetic cause could not be identified. We identified mutations in 13 genes known to be mutated in HSP (33% of the cases in our cohort) (table S3 and fig. S2), supporting the methodology. These include EIF2B5, associated with vanishing white-matter disease [Online Mendelian Inheritance in Man (OMIM) no. 603896]; CLN8, associated with ceroid lipofuscinosis (OMIM 600143); and ARG1, which causes arginase deficiency (OMIM 207800). The diversity of genes identified speaks to the heterogeneity of HSP presentations. ALS2 (OMIM 205100) was mutated in four different families presenting with uncomplicated HSP, and ATL1 (OMIM 182600) was mutated in three different families, some displaying partial penetrance (4).
We identified 14 candidate genes not previously implicated in disease (Table 1), accounting for 42% of the cases in our cohort. We also evaluated five non-consanguineous families by WES, implicating one additional candidate gene. We estimated, on the basis of our false discovery rate (FDR), that fewer than 0.1 alleles per family should pass this threshold randomly, dependent on the number of informative meiosis, suggesting that fewer than 1:10 genes identified with this method should prove false positive (i.e., identify by chance) (3).
Table 1.
Novel candidate HSP genes. List of novel candidate HSP genes identified through WES, divided into major functional modules (ERAD, etc.). OMIM nomenclature refers to established or new (beginning with SPG58) locus. Position refers to the Genome Browser release 19 map. Family 787 has a compound heterozygous mutation in the MARS gene. C, complicated; U, uncomplicated forms of HSP. ADP, adenosine diphosphate; IMP, inosine monophosphate; ATP, adenosine triphosphate; DDHD, Asp-Asp-His domain; GPI, glycosyl phosphatidylinositol; GTPase, guanosine triphosphatase; tRNA, transfer RNA. N/A, not applicable. Single-letter amino acid abbreviations are as follows: C, Cys; E, Glu; F, Phe; G, Gly; I, Ile; K, Lys; L, Leu; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; and X, termination.
| Gene Symbol | Locus Nomenclature |
Entrez Gene ID |
Gene Name | Putative Biological Function | Family # | Position (hg19) | Nucleotide Change | Deduced Protein Change |
Effect on Protein | Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|
| Endoplasmic reticulum and ER-associated degradation (ERAD) | ||||||||||
| ARL6IP1 | SPG61 | 23204 | ADP-ribosylation factor- like 6 interacting protein 1 |
Protein transport | 827 | chr16:18804609 TGTTT>T | c.576_579delAAAC | p.K193Ffs36X | Frameshift | C |
| ERLIN1 | SPG62 | 10613 | ER lipid raft associated 1 | ER-associated degradation | 786 1098 1598 |
chr10:101914679 G>A chr10:101943559 C>A chr10:101912067 CCTGGTA>C |
c.C763T c.G149T c.862_868delACCAGG |
p. R255X p.G50V p.del203-204YQ |
Stop codon Missense Frameshift |
U U |
| Endosome and membrane trafficking | ||||||||||
| KIF1C | SPG58 | 10749 | Kinesin family member 1C | Retrograde Golgi to ER transport | 789 803 RBT53 |
chr17:4904143 G>A chr17:4925567 C>T Del |
c.G183A c.C2191T - |
Splice p.R731X - |
N/A Stop codon Absent protein |
C U C |
| USPS | SPG59 | 9101 | Ubiquitin specific peptidase 8 |
Deubiquitinating enyzme | 882 | chr15:50769124 C>A | c.C928A | p.Q310K | Missense | C |
| WDR48 | SPG60 | 57599 | WD repeat domain 48 | Regulator of deubiquitination | 910 | chr3:39135498 AAAG>A | c.1879_1881delAAG | p.del628E | Amino acid del | C |
| Nucleotide metabolism | ||||||||||
| AMPD2 | SPG63 | 271 | Adenosine monophosphate deaminase 2 |
Deaminates AMP to IMP in purine nucleotide metabolism |
1526 | chr1:110167989 CT>C | c.318delT | p.C107Afs365X | Frameshift | C |
| ENTPD1 | SPG64 | 953 | Ectonucleoside triphosphate diphosphohydrolase 1 |
Hydrolyzes ATP and other nucleotides to regulate purinergic transmission |
1242 1800 |
chr10:97605168 G>A chr10:97604339 G>T |
c.G649A c.G719T |
p.G217R p.E181X |
Missense Stop codon |
C C |
| NT5C2 | SPG65 | 22978 | 5′-nucleotidase, cytosolic II | Preferentially hydrolyzes IMP, in both purine/pyrimidine nucleotide metabolism |
659 1290 1549 NGT2 1829 |
chr10:104899162 C>T chr10:104850739 CT>C chr10:104853067 C>A chr10:104934653 C>T chr10:104861028 T> A |
c.175+1G>A c.1225delA c.988-1G>T c.G86A c.A445T |
Splice p.S409Vfs436X Splice p.R29X p.R149X |
N/A Frameshift N/A Stop codon Stop codon |
U C U U C |
| Lipid/Myelination related | ||||||||||
| ARS | SPG66 | 340075 | Arylsulfatase family, member I |
Hydrolyze sulfate esters Hormone biosynthesis |
1349 | chr5:149676845 A>AT | c.1641insA | p.C548Mfs559X | Frameshift | C |
| DDHD2 | SPG54 | 23259 | DDHD domain-containing protein 2 |
Phospholipase preferentially hydrolyzes phosphatidic acid |
1675 1314 |
chr8:38103473 C>G chr8:38103270 C>T |
c.1057+5C>G c.C859T |
Splice p.R287X |
N/A Stop codon |
C C |
| PGAP1 | SPG67 | 80055 | Post-GPI attachment to proteins 1 |
GPI biosynthesis | 1241 | chr2:197712670 C>A | c.1952+1G>T | Splice | N/A | C |
| Axon guidance/synapse related | ||||||||||
| FLRT1 | SPG68 | 23769 | Fibronectin leucine rich transmembrane protein 1 |
Cell adhesion and receptor signaling |
709 | chr11:63885762 T>C | c.T2023C | Stop-loss | Stop-loss | C |
| RAB3GAP2 | SPG69 | 25782 | RAB3 GTPase activating protein subunit 2 |
Exocytosis of neurotransmitters and hormones |
738 | chr1:220357421 A>C | c.T1955G | p.L252X | Stop codon | C |
| Other | ||||||||||
| MARS | SPG70 | 4141 | Methionyl-tRNA synthetase |
Cytosolic methionyl-tRNA synthetase |
787 | chr12:57881886 G>A chr12:57908741 C>T |
c.G13A;c.C2104T | p.V5M;p.R702W | Missense Missense |
C |
| ZFR | SPG71 | 51663 | Zinc finger RNA binding protein |
RNA localization? | 1611 | chr5:32406955 A>G | c.T956C | p.L319P | Missense | U |
The mutations in the 15 novel genes were identified in patients presenting with a spectrum of HSP phenotypes. Three of these genes, ERLIN1, KIF1C, and NT5C2, were found independently mutated in more than one family, and all mutations were predicted to be highly deleterious. All but one was homozygous, whereas the non-consanguineous family 787, with four affected and six healthy children, displayed a compound heterozygous mutation. This approach thus identified a host of novel candidate genes for further investigation.
Extending Results to Larger HSP Cohort
An additional cohort of 200 patients diagnosed with HSP (5) were screened for mutations in these genes with exome sequencing or microfluidic polymerase chain reaction (PCR) followed by sequencing (3). Additional mutations in ERLIN1, ENTPD1, KIF1C, NT5C2, and DDHD2 were identified (Table 1), thus validating these in the pathogenesis of HSP. Microfluidic PCR provided threshold coverage of only 68% of the targeted exons, suggesting that improved methods will be required to fully evaluate this second cohort. While this paper was in preparation, DDHD2 was published as mutated in complicated HSP, cross-validating results (6).
Functional Testing Candidates with Expression and Zebrafish
To understand the potential role of these disease genes in HSP, we profiled their expression across multiple human tissues with reverse transcription PCR. Expression was specific to neural tissue for the genes FLRT1 and ZFR, suggesting a neuronal function (fig. S3). For most, however, we noted broadly distributed expression patterns, suggesting functions in other tissues but that neurons show increased susceptibility to genetic mutations.
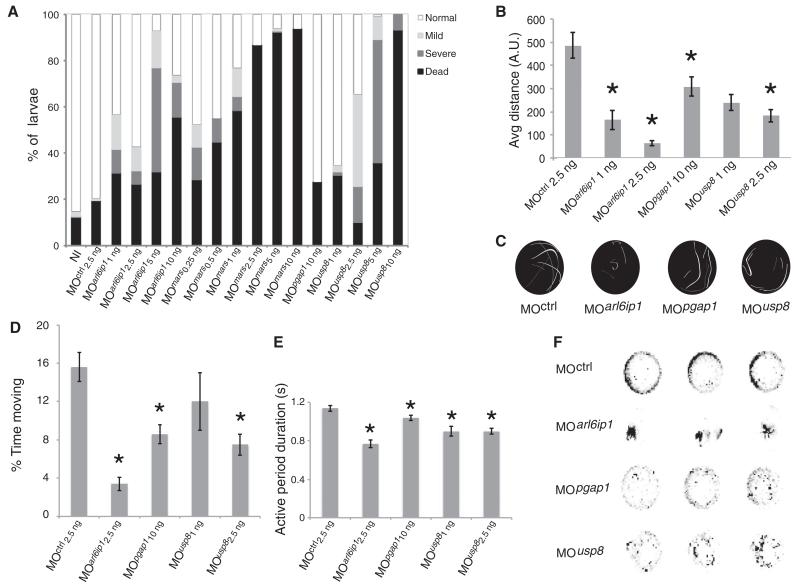
To functionally validate the private genes (i.e., those mutated in a single family), we performed knockdown modeling in zebrafish. Phylogenetic analysis indicated a single zebrafish ortholog for the private genes ARL6IP1, MARS, PGAP1, and USP8. Morphants were phenotyped for lethality and defects in body axis (Fig. 1 and fig. S4), motor neuron morphology (fig. S5), and evoked and spontaneous swimming behavior, all relevant to HSP. Except for mars morphants, which were too severe to be analyzed completely, we identified phenotypes for all morphants in both touch-induced and spontaneous locomotion behavior, as previously reported for other HSP candidate genes (7). Although more work is warranted to conclusively uncover the role of the tested genes in CST degeneration, our in vivo functional validation supports the genetic data.
Fig. 1. Functional validation of private HSP genes in zebrafish.
(A) Quantification of 24-hours-post-fertilization (hpf) embryos mortality (black) and curly-tail (gray) phenotypes for noninjected (NI), scrambled, and morphants (MO) at stated nanogram concentrations. Overt phenotypes were observed for all MOs except MOpgap1. (B) Average touch-response distance (in arbitrary units, A.U.) in 72-hpf larvae, showing blunted response for all MOs. (C) Immediate touch-response trajectory of example larvae, each shaded uniquely. Mars2 MO was too severe to be tested, whereas others showed reduced response. (D to F) Spontaneous locomotion at 6 days post fertilization. (D) Average percent of time spent moving over a 30-min window showed a reduction for all for at least one dose. (E) Average active period duration, showing reduction for all. (F) Representative kymographs recording fish position (black dot) over 30-min recording. MOs showed either short distance traveled (MOarl6ip1) or reduced movements per recording (MOpgap1 and MOusp8). *P < 0.01 (t test). N > 2 experiments with n > 20 animals per experiment. Error bars indicate standard error.
HSP-Related Proteins Interact Within a Network
To generate an HSPome containing all known and candidate genes as well as proximal interactors, we first created a protein network of all known human genes and/or proteins. We then extracted the subnetwork containing all previously published HSP mutated genes (seeds, table S4) to derive the HSP seeds network and then extracted the subnetwork containing all seed genes plus candidate HSP genes (from Table 1) to derive the HSP seed + candidates network (Fig. 2A).
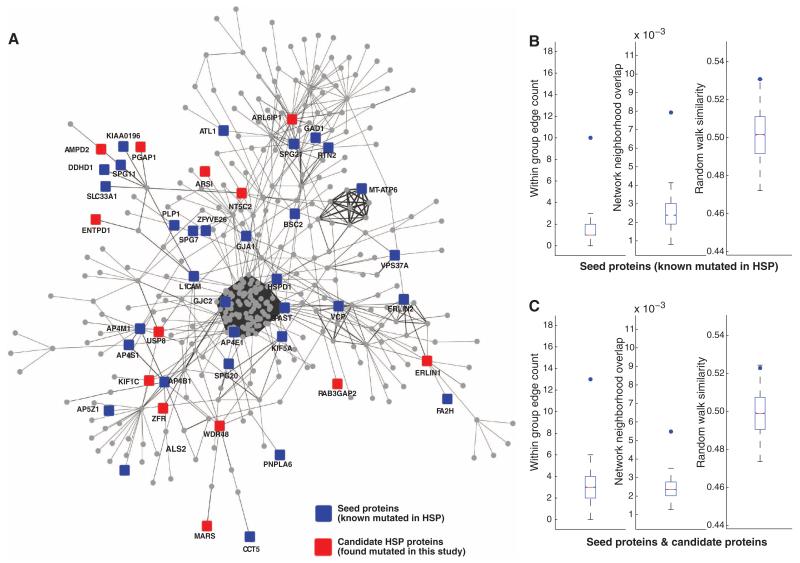
Fig. 2. Hereditary spastic paraplegia interactome.
(A) HSP seeds + candidate network (edge-weighted force-directed layout), demonstrating many of the genes known to be mutated in HSP (seeds, blue) and new HSP candidates (red), along with others (circles) constituting the network. (B and C) Comparison of statistical strength of HSP subnetworks with 10,000 permutations of randomly selected proteins. Dots denote the value of the metric on the true set (i.e., seeds or seeds + candidates). Box and whisker plots denote matched null distributions (i.e., 10,000 permutations). (B) Seed (known mutated in HSP) versus random proteins drawn with the same degree distribution. (C) Seed plus candidate HSP versus a matching set of proteins. (Left) Within group edge count (i.e., number of edges between members of the query set). (Middle) Interaction neighborhood overlap (i.e., Jaccard similarity). (Right) Network random walk similarity.
We tested whether the HSP seeds network was more highly connected than expected by chance. We compared the connectivity of the network comparing the 43 seeds to a background network generated by 10,000 permutations of randomly selected sets of 43 seeds from the global network using three different measures of connectivity: (i) the number of edges within the query set (within group edge count), (ii) the mean overlap in interaction neighborhoods between pairs of proteins in the query set (Jaccard similarity), and (iii) the mean random walk similarity (i.e., the expected “time” it takes to get from one protein to another when performing a random walk on the network) (8). By all three measures, we found the HSP seed proteins were more cohesive than expected at random (P = 2.0 × 10−04, P = 1.3 × 10−03, and P = 1.5 × 10−05) (Fig. 2B and supplementary data 1).
We also examined whether the HSP seed + candidates network, containing 43 seeds plus 15 candidates, was more highly connected than expected in a background of 10,000 random permutations (Fig. 2C). The addition of the candidates to the HSP seeds network resulted in a set significantly more highly connected than expected by chance (P = 3.1 × 10−02, P = 1.2 × 10−03, P = 4.8 × 10−04, respectively). We conclude that these newly identified genes are more cohesive than would be expected with candidates selected at random.
To identify proximal interactors, we expanded the global network by including HumanNet protein interaction database (9) and literature-curated interactions from STRING (10) to derive an expanded global network (fig. S6). This network propagation method assigns a priority score to each protein within the network (11). From this expanded network, we extracted the expanded HSP seeds network and found that 7 of the 15 newly identified candidates have significant support in the network (ARS1, DDHD2, ERLIN1, FLRT1, KIF1C, PGAP1, and RAB3GAP2, FDR < 0.1). Genes involved in biochemical pathways, such as NT5C2, AMPD2, and ENTPD1, did not emerge from this analysis, probably because of a lack of metabolic network edges in the input networks. Proteins that were not well characterized or represented in public databases also did not show enrichment.
We next expanded the HSP seed + candidate network to derive the HSPome (i.e., HSP seeds + candidates + proximal interactors network), allowing a global view of HSP and flagging other potential genes that may be mutated in HSP patients. The HSPome contains 589 proteins (i.e., potential HSP candidates) (supplementary data 2 and table S5).
Implicated Causal Genes Suggest Modules of HSP Pathology
Studies in HSP consistently report an ascending axonal CST degeneration (12), but the processes modulating this degeneration are not well defined. Supporting the hypothesis that individual rare mutations in distinct genes may converge on specific biological pathways, we identified major modules involved in the pathophysiology of HSP. Several HSP genes have previously implicated endoplasmic reticulum (ER) biology (i.e., ATL1, REEP1, RTN2, and SPAST) and the ER-associated degradation (ERAD) pathway (i.e., ERLIN2) (13-15). From the HSPome, we focused on this ER subnetwork containing the newly identified genes ARL6IP1 and ERLIN1 (fig. S7). ARL6IP1 encodes a tetraspan membrane protein localized to the ER, composed of highly conserved hydrophobic hairpin domains implicated in the formation of ER tubules (16). We overexpressed ARL6IP1 in cells and noted dramatically altered ER shape (fig. S7). The ERAD system controls protein quality control, critical for cellular adaptation to stress and survival. ERLIN1 encodes a prohibin-domain-containing protein localized to the ER that forms a ring-shaped complex with ERLIN2, further implicating defective ERAD in HSP etiology.
We identified an endosomal and membrane-trafficking subnetwork composed of seeds and candidates KIF1C, USP8, and WDR48, implicating the endosomal sorting complexes required for transport (ESCRT) pathway (fig. S8). USP8 encodes a deubiquitinating enzyme (DUB) in the ESCRT pathway (17). The WDR48-encoded protein forms stable complexes with multiple DUBs, such as USP1, USP12, and USP46, and is required for enzymatic activity and linked to lysosomal trafficking (18, 19). KIF1C encodes a motor protein localized to the ER/Golgi complex, suggesting a role in trafficking (20). To validate the effect of the putative splicing mutation in family 789, we obtained fibroblasts and confirmed skipping of exon 4 (fig. S9). Defects in ESCRTare linked to neurodegenerative disorders such as frontotemporal dementia, Charcot Marie Tooth disease, and recently AR-HSP (21-23). Additionally, the HSP gene products SPG20, SPAST, and ZYFVE26 interact with components of this complex (24-26). Taken together, this suggests that disruptions in ESCRT and endosomal function can lead to HSP and other forms of neurodegeneration.
AMPD2, ENTPD1, and NT5C2 are involved in purine nucleotide metabolism (fig. S10). Nucleotide metabolism is linked to the neurological disorder Lesch-Nyhan disease, among others (27), but was not previously implicated in HSP. AMPD2 encodes one of three adenosine monophosophate (AMP) deaminase enzymes involved in balancing purine levels (28). Mutations in AMPD2 have been recently linked to a neurodegenerative brainstem disorder (28). In addition, the deletion we have identified in this study affects just the longest of the three AMPD2 isoforms, indicating that the most N-terminal domain of AMPD2 is important to prevent motor neuron degeneration. ENTPD1 encodes an extracellular ectonuclease hydrolyzing adenosine nucleotides in the synaptic cleft (29). NT5C2 encodes a downstream cytosolic purine nucleotide 5′ phosphatase. Purine nucleotides are neuroprotective and play a critical role in the ischemic and developing brain (29); thus, alterations in their levels could sensitize neurons to stress and insult. ENTPD1 was recently identified as a candidate gene in a family with nonsyndromic intellectual disability, but HSP was not evaluated (30).
Candidate HSP Genes Identified by Network Analysis
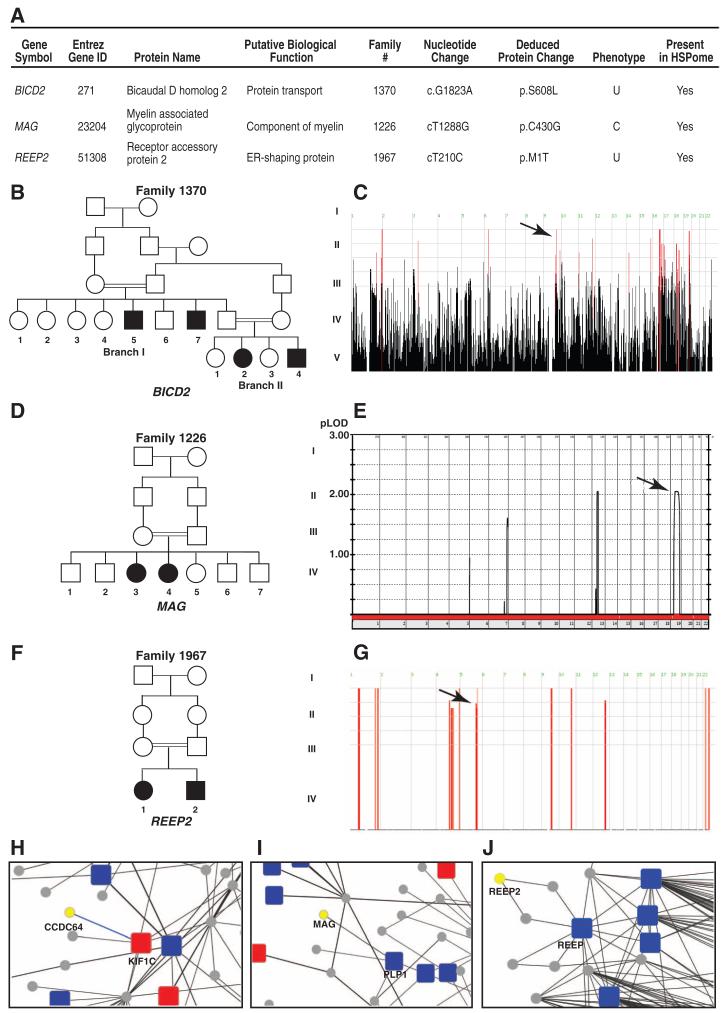
For families that were not included in our initial analysis, we interrogated our exome database for variants in genes emerging from the extended HSPome network. By using this method, we identified potentially pathogenic variants in MAG, BICD2, and REEP2, found in homozygous intervals in three families (Fig. 3), validating the usefulness of the HSPome to identify new HSP genes. Interacting with KIF1C in the HSPome is CCDC64, encoding a member of the Bicaudal family (31), a paralog of the BIC2 gene that emerged in the HSPome (FDR < 0.05, table S5). Family 1370 displays a homozygous Ser608→Leu608 missense change in the BIC2 gene within a homozygous haplotype. The Drosophila bicaudal-D protein is associated with Golgi-to-ER transport and potentially regulates the rate of synaptic vesicle recycling (32). Coimmunoprecipitation confirmed that BICD2 physically interacts with KIF1C (fig. S11). Recently, a mutation in BICD2 was implicated in a dominant form of HSP (33).
Fig. 3. Genes from HSP networks found mutated in HSP.
(A) HSP candidate genes predicted from the HSPome found mutated in the HSP cohort. BICD2, MAP, and REEP2 were subsequently found mutated in HSP families 1370 (B), 1226 (D), and 1967 (F), respectively. (C) Homozygosity plot from family 1370. Red bars, regions of homozygosity; arrow, homozygous block containing BICD2. (E) Linkage plot of family 1226; arrow, MAG locus. (G) Homozygosity plot; arrow, REEP2 locus. (H to J) Zoom in from HSPome for specific interaction identifying candidates CCDC64 (a paralog of BIC2D), MAG, and REEP2 (yellow) with previously published (blue) and newly identified (red) genes mutated in HSP. Blue lines denote manually curated interactions.
MAG was identified as a significant potential HSP candidate (FDR < 0.05) from the HSPome, interacting with PLP1, the gene product mutated in SPG2. MAG is a membrane-bound adhesion protein implicated in myelin function, and knockout mice display defects of the periaxonal cytoplasmic collar in the spinal cord with later oligodendrocyte degeneration (34). MAG was found mutated in family 1226, displaying a homozygous Cys430→Gly430 missense mutation.
REEP2 encodes the receptor expression-enhancing protein 2, a paralog of REEP1, mutated in SPG31 (35). Family 1967 displays a homozygous Met1→Thr1 mutation in REEP2 removing the canonical start codon and is mutated in a second recessive HSP family in an independent cohort (36). All of these gene mutations segregated with the phenotype in the family according to recessive inheritance and were not encountered in our exome database, consistent with pathogenicity. Although further validation of these three candidates is necessary in larger cohorts, the data suggest the HSPome can be useful to identify HSP-relevant pathways and genes.
Link Between HSP and Neurodegenerative Disease Genes
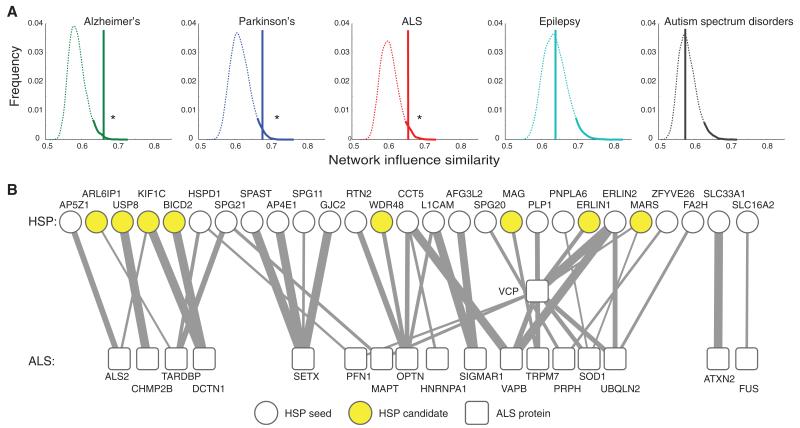
Some of the genes we identified in this cohort have been previously associated with other neurodegenerative disorders (e.g., CLN8, EIF2B5, and AMPD2) primarily affecting areas of the nervous system other than the corticospinal tract. Prompted by this observation, we used the network to examine the similarity of HSP genes (seed + candidates) to other common neurological disorders. By using the random walk distance, we found that the set of HSP seeds plus candidates is significantly overlapping with sets of genes previously implicated in three neurodegenerative disorders, amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Parkinson’s disease (P = 1.1 × 10−02, P = 7.6 × 10−03, P = 1.6 × 10−02, respectively) (Fig. 4). In contrast, we did not find a similar association with sets for representative neurodevelopmental disorders such as autism spectrum disorders and epilepsy (P = 0.49 and P = 0.51, respectively; fig. S12), nor with nonneurological disorders represented by heart and pulmonary disorders.
Fig. 4. Functional link between HSP genes and genes of other neurodegenerative conditions.
(A) Density distribution representing random walk distances of OMIM-derived neurodegeneration gene networks along with 10,000 permutations of randomly selected protein pools compared with the HSP seeds plus candidates pool. The top 5%ile distance is shaded. Only for Parkinson’s, Alzheimer’s, and ALS do the HSP seeds plus candidates fall within this 5%, whereas epilepsy and autism spectrum disorder show no statistical overlap. (B) Bipartite network showing the top links between the set of HSP and ALS proteins. Clear circles, HSP seeds; yellow circles, HSP candidates; boxes, ALS genes (VCP and ALS2 are implicated as causative of both HSP and ALS); line thickness, diffusion similarity between the two proteins.
Discussion
By using WES, we identified 18 previously unknown candidates for AR-HSP (fig. S13), three of which (ERLIN1, KIF1C, and NT5C2) alone explain almost 20% of this cohort. These new candidates are predicted to display near 100% risk of HSP when mutated (37). All mutations were predicted as damaging to protein function, probably resulting in null or severely reduced function, consistent with the recessive mode of inheritance. In about 25% of the families a single candidate gene mutation could not be identified, probably a result of two factors: (i) Some mutations are in noncoding regions. (ii) Some causative mutations within the exome do not stand out more than other variants.
Four of our candidate HSP genes are located within previously identified loci for AR-HSP for which genes were not known: ENTPD1, NT5C2, ERLIN1, and MARS. Both ERLIN1 and NT5C2 are in the SPG45 locus (38) and ENTPD1 resides in SPG27 (39). Recently, the MARS2 gene, encoding a methionyl-tRNA synthetase, was implicated in the spastic ataxia 3 (SPAX3) phenotype (40). KIF1C is within the spastic ataxia 2 (SAX2) locus (41). On the basis of our findings, we returned to the original SPAX2 family and identified a homozygous deletion of exons 14 to 18, confirming KIF1C as the SPAX2 gene (fig. S14).
Our data support the idea that rare genetic mutations may converge on a few key biological processes, and our HSP interactome demonstrates that many of the known and candidate HSP genes are highly connected. This highlights important biological processes, such as cellular transport, nucleotide metabolism, and synapse and axon development. Some of the HSP gene modules suggest potential points of treatment; for example, the nucleotide metabolism module or the lipid metabolism module could be targeted by bypassing specific metabolic blocks. Our HSPome ranked list of genes also provides candidates for unsolved cases of HSP. In addition to our analysis, we were able to link HSP with more common neurodegenerative disorders, indicating that the study of one disorder might advance the understanding of other neurodegenerative disorders as well.
Our study supports the principle that integrating family-based gene discovery together with prior knowledge (represented here as known causative genes and pathways) can facilitate the identification of biological pathways and processes disrupted in disease. Furthermore, this mode of analysis should be highly useful in the future to aid in the validation of private mutations in genes found in single families, to identify novel candidate genes and pathways, and for the discovery of potential therapeutic targets.
Supplementary Material
Acknowledgments
We are grateful to the participating families. Supported by the Deutsche Forschungsgemeinschaft (G.N.); the Brain and Behavior Research Foundation (A.G.F.); NIH R01NS041537, R01NS048453, R01NS052455, P01HD070494, and P30NS047101 (J.G.G.); French National Agency for Research (G.S., A.D.); the Verum Foundation (A.B.); the European Union (Omics call, “Neuromics” A.B.); Fondation Roger de Spoelberch (A.B.); P41GM103504 (T.I.); “Investissements d’avenir” ANR-10-IAIHU-06 (to the Brain and Spine Institute, Paris); and Princess Al Jawhara Center of Excellence in Research of Hereditary Disorders. Genotyping services provided in part by Center for Inherited Disease Research contract numbers HHSN268200782096C, HHSN268201100011I, and N01-CO-12400. We thank the Broad Institute (U54HG003067 to E. Lander), the Yale Center for Mendelian Disorders (U54HG006504 to R. Lifton and M. Gunel), M. Liv for technical expertise, and E. N. Smith, N. Schork, M. Yahyaoui, F. Santorelli, and F. Darios for discussion. Data available at dbGaP (accession number phs000288). UCSD Institutional Review Board (070870) supervised the study. The authors declare no competing financial interests.
Footnotes
Supplementary Materials
www.sciencemag.org/content/343/6170/506/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S15
Tables S1 to S6
References (42-52)
References and Notes
- 1.Blackstone C. Annu. Rev. Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.See supplementary text available on Science Online.
- 4.D’Amico A, et al. Neurology. 2004;62:2138–2139. doi: 10.1212/01.wnl.0000127698.88895.85. [DOI] [PubMed] [Google Scholar]
- 5.Tesson C, et al. Am. J. Hum. Genet. 2012;91:1051–1064. doi: 10.1016/j.ajhg.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuurs-Hoeijmakers JH, et al. Am. J. Hum. Genet. 2012;91:1073–1081. doi: 10.1016/j.ajhg.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fassier C, et al. Nat. Neurosci. 2010;13:1380–1387. doi: 10.1038/nn.2662. [DOI] [PubMed] [Google Scholar]
- 8.Vandin F, Upfal E, Raphael BJ. J. Comput. Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 9.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, et al. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. PLOS Comput. Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLuca GC, Ebers GC, Esiri MM. Neuropathol. Appl. Neurobiol. 2004;30:576–584. doi: 10.1111/j.1365-2990.2004.00587.x. [DOI] [PubMed] [Google Scholar]
- 13.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Hum. Mol. Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montenegro G, et al. J. Clin. Invest. 2012;122:538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildirim Y, et al. Hum. Mol. Genet. 2011;20:1886–1892. doi: 10.1093/hmg/ddr070. [DOI] [PubMed] [Google Scholar]
- 16.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 17.Wright MH, Berlin I, Nash PD. Cell Biochem. Biophys. 2011;60:39–46. doi: 10.1007/s12013-011-9181-9. [DOI] [PubMed] [Google Scholar]
- 18.Park J, et al. Immunity. 2002;17:221–233. doi: 10.1016/s1074-7613(02)00368-0. [DOI] [PubMed] [Google Scholar]
- 19.Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. J. Biol. Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorner C, et al. J. Biol. Chem. 1998;273:20267–20275. doi: 10.1074/jbc.273.32.20267. [DOI] [PubMed] [Google Scholar]
- 21.Lee SM, Chin LS, Li L. J. Cell Biol. 2012;199:799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skibinski G, et al. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 23.Zivony-Elboum Y, et al. J. Med. Genet. 2012;49:462–472. doi: 10.1136/jmedgenet-2012-100742. [DOI] [PubMed] [Google Scholar]
- 24.Renvoisé B, et al. Mol. Biol. Cell. 2010;21:3293–3303. doi: 10.1091/mbc.E09-10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lumb JH, Connell JW, Allison R, Reid E. Biochim. Biophys. Acta. 2012;1823:192–197. doi: 10.1016/j.bbamcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Sagona AP, et al. Nat. Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 27.Jinnah HA, Sabina RL, Van Den Berghe G. Handb. Clin. Neurol. 2013;113:1827–1836. doi: 10.1016/B978-0-444-59565-2.00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akizu N, et al. Cell. 2013;154:505–517. doi: 10.1016/j.cell.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thauerer B, Zur Nedden S, Baier-Bitterlich G. J. Neurochem. 2012;121:329–342. doi: 10.1111/j.1471-4159.2012.07692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Najmabadi H, et al. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 31.Schlager MA, et al. EMBO J. 2010;29:1637–1651. doi: 10.1038/emboj.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, et al. EMBO J. 2010;29:992–1006. doi: 10.1038/emboj.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oates EC, et al. Am. J. Hum. Genet. 2013;92:965–973. doi: 10.1016/j.ajhg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Z, et al. J. Peripher. Nerv. Syst. 2002;7:181–189. doi: 10.1046/j.1529-8027.2002.02023.x. [DOI] [PubMed] [Google Scholar]
- 35.Züchner S, et al. Am. J. Hum. Genet. 2006;79:365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteves T, et al. Am. J. Hum. Genet. published online 31 December 2013 (10.1016/j.ajhg.2013.12.005) [Google Scholar]
- 37.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dursun U, Koroglu C, Kocasoy Orhan E, Ugur SA, Tolun A. Neurogenetics. 2009;10:325–331. doi: 10.1007/s10048-009-0191-3. [DOI] [PubMed] [Google Scholar]
- 39.Meijer IA, et al. Ann. Neurol. 2004;56:579–582. doi: 10.1002/ana.20239. [DOI] [PubMed] [Google Scholar]
- 40.Bayat V, et al. PLOS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouslam N, et al. Hum. Genet. 2007;121:413–420. doi: 10.1007/s00439-007-0328-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.