Abstract
The night and day cycle governs the circadian (24 hourly) rhythm of activity and rest in animals and humans. This is reflected in daily changes of the global gene expression pattern and metabolism, but also in the local physiology of various tissues. A central clock in the brain co-ordinates the rhythmic locomotion behaviour, as well as synchronizing various local oscillators, such as those found in the musculoskeletal system. It has become increasingly recognized that the internal molecular clocks in cells allow a tissue to anticipate the rhythmic changes in their local environment and the specific demands of that tissue. Consequently, the majority of the rhythmic clock controlled genes and pathways are tissue specific. The concept of the tissue-specific function of circadian clocks is further supported by the diverse musculoskeletal phenotypes in mice with deletions or mutations of various core clock components, ranging from increased bone mass, dwarfism, arthropathy, reduced muscle strength and tendon calcification. The present review summarizes the current understanding of the circadian clocks in muscle, bone, cartilage and tendon tissues, with particular focus on the evidence of circadian rhythms in tissue physiology, their entrainment mechanisms and disease links, and the tissue-specific clock target genes/pathways. Research in this area holds strong potential to advance our understanding of how circadian rhythms control the health and disease of the musculoskeletal tissues, which has major implications in diseases associated with advancing age. It could also have potential implications in sports performance and sports medicine.
Keywords: cartilage entrainment, circadian rhythm, homoeostasis, muscle
Abbreviations: ADAM, a disintegrin and metalloproteinase; Adrb2, β2-adrenergic receptor; Bmal1, brain and muscle ARNT-like 1; BMP, bone morphogenetic protein; BMSC, bone marrow-derived stem cell; CCG, clock-controlled gene; Creb/CREB, cAMP-responsive-element-binding protein; Cry, cryptochrome circadian clock; ECM, extracellular matrix; Fbxo32, F-box protein 32; Gilz, glucocorticoid-induced leucine zipper; Grem2, gremlin 2; HBM, high bone mass; Ihh, Indian hedgehog; KO, knockout; LD, light–dark; Mmp, matrix metalloproteinase; Mrf4, muscle-specific regulatory factor 4; MSC, mesenchymal stem cell; Myf, myogenic factor; MyoD, myogenic differentiation; Noc, nocturnin; Nr1d1, nuclear receptor subfamily 1, group D, member 1; Per, period circadian clock; Pgc1, Pparg co-activator 1; Pparg, peroxisome-proliferator-activated receptor γ; PTH, parathyroid hormone; RORE, ROR/REV-ERB-binding element; SCN, suprachiasmatic nuclei; Sirt1, sirtuin 1; TGF-β, transforming growth factor-β; WT, wild-type
INTRODUCTION
The mammalian circadian system is organized in a hierarchical manner. The central clock [SCN (suprachiasmatic nuclei)] in the hypothalamus controls rest/activity rhythms, and is entrained on a daily basis by the LD (light–dark) cycle, whereas clocks in peripheral tissues were thought to be slave oscillators, subject to the neuronal or hormonal control by the SCN. However, the emerging paradigm is that SCN synchronizes peripheral tissue clocks, but is not an absolute master [1]. The autonomous peripheral clocks contain identical clock genes/proteins and can be entrained by external time cues, such as feeding/fasting and physical exercise, independently of the SCN and the LD cycle. This means that the LD cycle can be overridden in peripheral tissues by other zeitgebers (time cues), allowing animals greater flexibility in responding to demands of the environment [2–4].
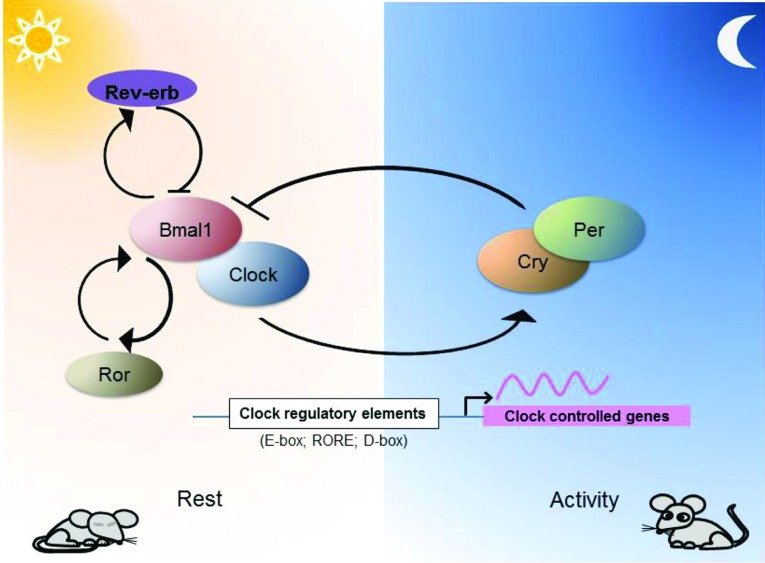
In mammals the circadian clock relies on a transcriptional/translational feedback loop consisting of the transcriptional activators Bmal1 (brain and muscle ARNT-like 1)/Clock and the repressor complex Per1–Per2 (period circadian clock 1/2) and Cry1/Cry2 (cryptochrome circadian clock 1/2). The Bmal1/Clock complex activates the expression of the Per and Cry genes. The Per/Cry complex periodically feeds back to suppress the activity of Bmal1/Clock, inhibiting their own transcription. Nuclear hormone receptors Rev-Erb (repressor) and Ror (activator) form an additional stabilizing loop with Bmal1 to fine-tune the precision of the clock (Figure 1). The interaction of these core components and modulation by additional regulators results in an approximately 24-h period. In addition to the autoregulation, the core clock transcription factors rhythmically control the expression of other CCGs (clock-controlled genes) through specific regulatory elements [E-box, D-box and RORE (ROR/REV-ERB-binding element)] in their promoters [5]. Despite the same core clock mechanism operating in different tissues, the circadian transcriptome of peripheral tissues is strikingly different with only a small overlap [6]. Many of the oscillating genes are key tissue-specific transcription factors, characteristic structural proteins or are involved in metabolic pathways fundamental for that particular tissue [7–10]. Conditional ablation of the peripheral clocks in liver, pancreas, adipose tissue and skin revealed profound and diverse disorders and pathologies [11–13], highlighting the importance of the local clocks in tissue physiology and diseases. In this regard, the musculoskeletal system is particularly relevant to the daily rest/activity cycles. Coupling of the local gene expression and physiology to the daily loading/unloading and related metabolic changes could be an important part of the musculoskeletal biology. Here we review the current developments in the field of circadian biology with respect to the musculoskeletal system and the implications of the circadian rhythms in tissue homoeostasis and disease.
Figure 1. A simplified model of the core clock components of the mammalian circadian oscillator.
Bmal1/Clock complex activates transcription of genes containing E-box sequences in their promoters. Among these are Per1/2 and Cry1/2, which following dimerization can inhibit the activity of the Bmal1/Clock complex. Rev-Erb and Ror provide an additional stabilizing loop to fine-tune the expression of Bmal1. In addition to E-boxes, other clock regulatory elements (such as ROREs and D-boxes) are commonly found in the promoters of CCGs.
MUSCLE CLOCK
Skeletal muscle is the most abundant tissue in the human body comprising approximately 40% of body weight. Accompanied by bones, cartilage and tendons it provides the means of locomotion, but also plays an important role in metabolic homoeostasis of the whole organism. The muscle is a malleable organ with the ability to adapt its mass to the different pathophysiological conditions through regulation of pathways responsible for protein and cellular turnover. External stimuli, such as exercise and nutrients, can increase the rate of muscle protein synthesis and result in hypertrophy (increase in muscle mass). Conversely starvation, immobilization and aging increase the rate of protein degradation and result in muscle atrophy (reduction in mass) [14].
The daily pace of light and dark, rest and activity, is reflected in the physiology of the muscle tissue. There has been ample evidence for the time of day-dependent variation in muscle physiology and neuromuscular performance [15–18]. Early evidence for the diurnal expression of clock genes in skeletal muscle came from studies in humans [19]. That study also showed that exercise up-regulated the expression of Bmal1, Per1 and Per2. However, the first comprehensive studies of the muscle circadian transcriptome were performed by Miller et al. [20] and McCarthy et al. in 2007 [7]. More than 200 rhythmic genes were identified in skeletal muscle. Interestingly, both studies reported a large cluster of genes to be peaking at ZT18 (zeitgeber time 18), corresponding to the middle of the active phase in mice. These findings further reinforce the notion that locomotor activity may play a role in entraining the rhythmic expression of genes in skeletal muscle. The largest Gene Ontology clusters consisted of genes involved in biosynthesis and metabolism (18%) and regulation of gene transcription (17%). The specificity of the muscle circadian transcriptome was underscored by the presence of skeletal muscle characteristic genes such as MyoD (myogenic differentiation), Ucp3 (uncoupling protein 3), Fbxo32 (F-box protein 32; Atrogin1), Pdk4 (pyruvate dehydrogenase kinase 4) and Myh1 (myosin heavy chain I). Comparison with arrhythmic ClockΔ19 mutant mice showed that a large proportion of the muscle circadian genes either lost their rhythmicity or shifted their phase. The majority was found to be down-regulated, including genes involved in muscle contraction (such as actin, dystrophin, titin and myosin heavy chain IIb), protein synthesis and energy metabolism [7].
Consistent with the key pathways controlled by the circadian clock, various muscle phenotypes were reported in Clock- or Bmal1-deficient mice (summarized in Table 1). Bmal1−/− mice exhibit reduced muscle mass accompanied by decreased diameter and number of muscle fibres [21,22]. Myoblasts isolated from Bmal1−/− mice exhibited impaired formation of organized myotubes and a considerably lower percentage of MHC-positive myonuclei. Lower expression levels of myogenic markers such as Myf5 (myogenic factor 5), Mrf4 (muscle-specific regulatory factor 4/Myf6), MyoD and Myf4 (myogenin), as well as MHC3, may partially explain the phenotype. Further, expression of several genes involved in canonical Wnt signalling was markedly reduced in isolated myotubes from Bmal1−/− mice, including Wnt10a, Dvl2 (dishevelled segment polarity protein 2), β-catenin and Tcf3 (transcription factor 3). The KO (knockout) cells also remained unresponsive to Wnt3a stimulation in vitro as evidenced by the lack of nuclear accumulation of β-catenin after treatment. Overexpression of MyoD and Wnt3a in the Bmal1−/− myoblasts resulted in improved myogenic differentiation, but failed to fully rescue the myotube formation defect, indicating involvement of other Bmal1-controlled pathways in the process [22].
Table 1. Summary of musculoskeletal phenotypes found in mice deficient in various core circadian clock components.
| Tissue phenotype | ||||
|---|---|---|---|---|
| Circadian mutant | Muscle | Bone | Cartilage | Tendon |
| Cry KO or mutants | Not reported | High bone mass [41,78] | Not reported | Not reported |
| Increase in osteoblast number [41] | ||||
| Normal osteoblast number, but decreased osteoclast activity [78] | ||||
| Per KO or mutants | Not reported | High bone mass and increase in osteoblast number [41,78] | Not reported | Not reported |
| Per−/−/Cry−/− | Not reported | Normal phenotype [78] | Not reported | Not reported |
| Bmal1−/− | Reduced diameter and number of muscle fibres at 40 weeks [21,22] | Thicker bones [41] | Calcification of ribcage cartilage [62] | Ectopic calcification [62] |
| Disorganized myofilaments [24] | Increase in osteoblast number [41] | Affected growth plate (shorter bones) [62] | ||
| Abnormal mitochondria [24] | Decreased expression of Col XIhh and Alp in the growth plate [61] | |||
| Rev-Erbα−/− | Impaired myogenic differentiation [22] | Not reported | Not reported | Not reported |
| Misalignment of Z lines [29] | ||||
| Abnormal mitochondria [29] | ||||
| Impaired muscle regeneration [29] | ||||
| Increased autophagy [29] | ||||
| Clock mutant | Disorganized myofilaments [24] | Not reported | Not reported | Ectopic calcification [77] |
| Abnormal mitochondria [24] | ||||
Both Clock mutant and Bmal1−/− mice exhibit decreased muscle force and disorganized arrangement of the thin and thick filaments. Interestingly, a similar phenotype was found in MyoD−/− mice. MyoD is a muscle specific basic helix–loop–helix transcription factor which, along with other myogenic regulatory factors such as Myf5 and Mrf4, controls the expression of a myriad of genes during myogenesis [23]. MyoD was one of the rhythmic genes identified in the muscle which is a direct target of Clock/Bmal1 through a functional E-box in its promoter [24,25]. Also of interest is the muscle-specific rescue model of the global Bmal1−/− mice. Although the activity level and longevity of the global mutants were restored by muscle Bmal1 overexpression, the behavioural rhythmicity remains abolished, and the tendon calcification phenotype is still present. These findings further support the concept of tissue-specific functions of clocks in driving a diverse range of physiology [26].
Clock mutant and Bmal1-KO mice were found to have a dramatically smaller number and volume of mitochondria in muscles with aberrant morphology. The mitochondrial abnormalities are possibly due to decreased expression of Pgc1a [Pparg (peroxisome-proliferator-activated receptor γ) co-activator 1α] and Pgc1b [7,27]. Pgc1 genes are a family of co-activators that activate the expression of mitochondrial genes and are major regulators that determine the type of muscle fibre [28]. Interestingly, the Rev-Erb-α [Nr1d1 (nuclear receptor subfamily 1, group D, member 1)]-KO mouse also shows decreased mitochondrial content and lower respiratory chain function in isolated organelles, and lower muscle endurance. The mitochondria also displayed abnormal morphology and the muscles showed a slight misalignment of Z lines and vacuolated fibres. The ATP, NAD+ and NADH concentrations were lower in skeletal muscle from Nr1d1−/− mice, revealing dysregulation of muscle energy metabolism. Notably, the expression of the Sirt1 (sirtuin 1) gene was reduced in the Nr1d1−/− muscle, associated with heavy acetylation (and activation) of Pgc1a [29]. SIRT1 is an NAD+-dependent deacetylase involved in the control of muscle mitochondrial function and Pgc1a is one of its targets [30].
Feeding/fasting is considered as one of the zeitgebers (time cues) to synchronize the skeletal muscle clock. It was found that although the core clock components are unaffected in mice fasted for 24 h, the circadian rhythm of MyoD and Myf4 was disrupted and their expression was reduced. Furthermore, the amplitude of rhythmic expression of MuRF1 [Trim63 (tripartite motif-containing 63)] and Fbxo32 genes, E3 ubiquitin ligases involved in muscle wasting, was increased [31]. This suggests that, at least in the muscle, it is not the food consumption, but rather the fasting which exhibits an inhibitory effect on muscle specific rhythmic genes, leading to resetting of the muscle clock. Another candidate zeitgeber is locomotor activity. Scheduled exercise entrains the muscle peripheral clock, while leaving the SCN cycle unaffected [32–34]. Further research is clearly needed to define the entrainment mechanisms by physical activity.
BONE CLOCK
Bones provide the scaffold for skeletal muscles and bear the weight of the organism. They also play an important role in mineral balance as a reservoir of calcium and phosphorus. Bone homoeostasis depends on two opposing processes, resorption and synthesis. Normal bone remodelling requires strict temporal control of these two processes to guarantee maintenance of the bone mass and structural quality [35]. These processes are regulated by two distinct types of cells, the osteoclasts and osteoblasts. First, there is a rapid phase of resorption of old mineralized bone by osteoclasts which is followed by a relatively slow phase of new bone formation by osteoblasts [36,37]. Diurnal variations in serum markers of bone metabolism, such as calcium, C-telopeptide, osteocalcin, skeletal ALP (alkaline phosphatase), PTH (parathyroid hormone), tartrate-resistant acid phosphatase and calcitonin have long been recognized [38–40].
Characterization of bone diurnal transcriptome from mice kept in 12:12 h LD cycle revealed that 26% of genes expressed in calvarial bone exhibit near 24-h oscillations. Among the rhythmic genes found were core clock genes as well as genes involved in BMP (bone morphogenetic protein) and FGF (fibroblast growth factor) signalling, Wnt signalling, multiple ADAM (a disintegrin and metalloproteinase) proteases and various pro-collagen isoforms, underscoring the scope of clock influence on bone physiology [8].
Studies of the skeletal phenotypes of various circadian gene KO mice (summarized in Table 1) established the circadian system as an important regulator of bone homoeostasis. Mice lacking the Per1 gene or the PAS domain (Per/Arnt/Sim domain) of Per2 show normal bone morphology up to 6 weeks of age. After this time increase in bone mass starts to be evident and progresses with age. Similar HBM (high bone mass) phenotype can be seen in Per1/Per2 and Cry1/Cry2 double-KO mice. Biochemical and histomorphometric analyses found a significant increase in the number of osteoblasts in bones of these mutant mice, which correlated with increased mineral apposition rate and bone-formation rate [41]. Somewhat surprisingly, Bmal1-KO mice also exhibit increased bone-formation rate, mineral apposition rate and osteoblast numbers similar to Per- and Cry-KO mice. These phenotypes point to a mechanism in which the circadian clock negatively regulates bone formation by inhibition of osteoblast proliferation.
Another mechanism through which the circadian clock could affect bone formation is the differentiation of osteoblasts from BMSCs (bone marrow-derived stem cells). Noc (nocturnin), a deadenylase that regulates gene expression post-transcriptionally [42], has been shown to be a direct rhythmic target of Bmal1–Clock heterodimer [43]. Noc is down-regulated during differentiation of osteoblasts in vitro and its overexpression inhibits osteoblastogenesis. Noc-KO mice display a HBM phenotype and reduced numbers of adipocytes in the bone marrow, suggesting a shift in BMSC differentiation favouring the osteoblastic route [44,45]. Pparg2 has a well-established role in MSC (mesenchymal stem cell) fate determination. Pparg2 activation favours MSC differentiation along the adipogenic lineage over osteogenic differentiation, and suppresses osteogenic signalling pathways such as BMP, TGF-β (transforming growth factor-β), Wnt, and osteoblast-specific transcription factors such as Runx2 (runt-related transcription factor 2), Sox9 (SRY-box 9) and Sp7 (osterix) [46]. In vivo silencing of Pparg2 in bone marrow was found to result in reduction in intramarrow adiposity and increased trabecular bone formation [47]. Interestingly, Noc can interact with Pparg2 and promote its nuclear translocation [44]. Expression of Pparg2 was significantly decreased in Noc-KO brown adipocytes [44]. Further in vivo studies are clearly warranted to test this interesting hypothesis.
Another potential mechanistic link between circadian disruption and bone phenotype could be Gilz (glucocorticoid-induced leucine zipper/TSC22D3), a glucocorticoid-responsive transcriptional regulator. Gilz displays a robust circadian rhythm in brown and white adipose tissue, liver, cartilage, tendon and calvarial bone [8,10]. Shi and colleagues found that Gilz expression can be induced by dexamethasone in 2T3 osteoblasts and 3T3-L1 pre-adipocytes. Gilz was shown to block transcription of Pparg2 by competing with C/ebpα (CCAAT/enhancer-binding protein α) [48,49]. Overexpression of Gilz in MSCs in vitro leads to inhibition of adipogenesis and enhancement of osteogenesis [46,50].
The output signals from the SCN can be transmitted to the local bone clocks through β-adrenergic (sympathetic), glucocorticoid signalling and feeding/fasting [51]. It has been shown that leptin regulates the bone clock through sympathetic nervous system and Adrb2 (β2-adrenergic receptor) present on the osteoblasts, thus linking global metabolism and the bone peripheral clock. Adrb2 acts through Creb1 (cAMP-responsive-element-binding protein 1; a cell-cycle regulator) to stimulate the expression of clock genes such as Bmal1 and Clock [36]. This implies that osteoblast activity is coordinated by both the external stimuli and the internal osteoblast clock to maintain the diurnal rhythms in bone formation and resorption, and consequently the maintenance of bone mass. In osteoclasts, glucocorticoids were shown to be a more potent time cue than sympathetic signalling. Circadian expression of genes in osteoclasts can be stimulated in vitro with dexamethasone but not with isoprenaline [52]. Dexamethasone was also shown to induce rhythmic bone resorption in an in vitro pit assay [52]. Adrenalectomy of mice reduces the amplitude of Per1 and Per2 genes in cancellous bone, and completely abolishes rhythmic expression of osteoclast specific genes Ctsk (cathepsin K) and Nfatc1 (nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1). Glucocorticoid injection was able to restart the rhythmic expression of these two osteoclast markers in vivo [52]. Similarly to the muscle, the diurnal variations in the bone metabolism seem to be governed by timing of food intake and fasting, providing an additional entrainment mechanism [53,54].
CARTILAGE CLOCK
Cartilage in the growth plate or the end of long bones (articular cartilage) consists of abundant ECM (extracellular matrix), sparsely populated by chondrocytes. The growth plate is responsible for the longitudinal growth of bones whereas the arti-cular cartilage provides a smooth, resilient surface for the movement of joints. Isolated from the vasculature and lacking innervations, the fine balance between anabolic and catabolic activities is critical to maintain cartilage tissue homoeostasis. Disruptions to this balance are implicit in the degeneration and destruction of the articular cartilage tissue, which is frequently seen in diseases such as osteoarthritis and rheumatoid arthritis [55].
Evidence for diurnal variations in cartilage metabolism has been well described. Based on measurements of phosphorus and calcium fractions from rat tibial growth plate it was shown that cartilage mineralization occurs at night [56]. Moreover, measurements of chondrocyte mitotic index and height of the growth plate showed that chondrocytes proliferate most actively in the early morning. The proliferation phase is followed by expansion of the growth plate, reaching peak height at noon in rats [57]. The peak height of growth plate coincides with the highest collagen matrix synthesis as evidenced by incorporation of [3H]proline [58]. Diurnal variations in serum levels of COMP (cartilage oligomeric matrix protein), hialuronic acid, keratan sulphate, aggrecan, collagen type II and TGF-β have also been reported [59,60].
Deletion of the core clock gene Bmal1 in mice provided genetic evidence for the role of the molecular clock in cartilage physiology. Although the Bmal1-KO mice are shorter than the WT (wild-type) and the overall weight of the skeleton is lower, Bmal1 appears to have opposing effect on osteoblasts and chondrocytes. Closer inspection of the skeleton reveals that the KO mice have higher bone formation rate, mineral apposition rate and osteoblast number, but their bones are significantly shorter [21,41,61,62]. It was proposed that the Bmal1−/− phenotype in bone length is caused, at least partially, by the altered circadian expression of Ihh (Indian hedgehog) in the growth plate, a master regulator of endochondral ossification [61].
In contrast, the circadian clock in articular cartilage has only recently been described. Long-term imaging of the long bone from clock gene reporter mouse revealed strong rhythmic bioluminescence signals from the articular cartilage and growth plate, with limited signal in the ossified bones [63]. More recently, autonomous circadian rhythms in mouse articular cartilage tissues (and human chondrocyte-derived cell lines) have been unequivocally demonstrated using real-time bioluminescence photon counting of explant cultures of xiphoid and femoral head cartilage [9]. Both types of cartilage display robust circadian oscillations. Dexamethasone and temperature cycles were able to entrain the cartilage circadian rhythm [9]. The temperature response could provide a mechanism by which the central clock can synchronize cartilage rhythms. This is particularly interesting because cartilage is avascular and not innervated. Cartilage clock could also potentially be entrained by the daily rhythm of activity as the Clock gene was identified as a mechanosensitive gene that is down-regulated in cartilage by mechanical stress [64].
Time series microarrays revealed hundreds of rhythmic genes in murine cartilage, including catabolic extracellular proteases [Adamts4/9 and Mmp14 (matrix metalloproteinase 14)], anabolic or ECM genes (e.g. Timp4, elastin and fibrillins), and genes involved in apoptosis [Xiap (X-linked inhibitor of apoptosis)] and oxidative stress pathways [9]. Adamts4 is one of the main aggrecanases responsible for remodelling the aggrecan network in articular cartilage and has been implicated in progression of osteoarthritis [65]. Mmp14 is a membrane-tethered protease with multiple roles within cartilage, including activation of other matrix proteases, such as Mmp13 and Mmp9 [66,67], modulation of TGF-β signalling [68,69], articular cartilage homoeostasis and chondrogenic differentiation [70,71]. The circadian rhythmicity of many cartilage specific genes found in mouse is supported by microarray studies in rat cartilage under diurnal (LD cycle) conditions [72].
PTH, which exhibits diurnal variation in serum concentration [38], could be another factor with the ability to entrain the circadian clock in growth plate chondrocytes. PTH, which signals through the PPR (PTH-related protein receptor), was shown to induce Per1 expression in osteoblasts and chondrocytes. It appears that PTH can induce Per1 expression by a mechanism related to cAMP/PKA (protein kinase A)/CREB signalling in both types of cells [73,74]. Interestingly, Takarada et al. [61] showed that overexpression of Per1 in vitro suppresses chondrocyte differentiation by binding and blocking a functional E-box in the Ihh promoter. Therefore the circadian clock could serve as a modulator of the interplay between Ihh and PTHrP (PTH-related protein) signalling which regulates differentiation of chondrocytes in the growth plate [75].
Interestingly, in aged cartilage tissue, the amplitude of circadian oscillations is significantly reduced [9]. This reduction could have a substantial impact on the susceptibility to joint diseases such as osteoarthritis, due to the loss of rhythmic expression of the downstream cartilage genes controlling tissue homoeostasis. Expression of core clock genes was found to be affected early on in an experimental mouse model of osteoarthritis, further supporting a role of clock genes in the initiation and progression of osteoarthritis [9]. Future studies should address the functional significance of the cartilage rhythm in relation to joint degeneration and repair.
TENDON CLOCK
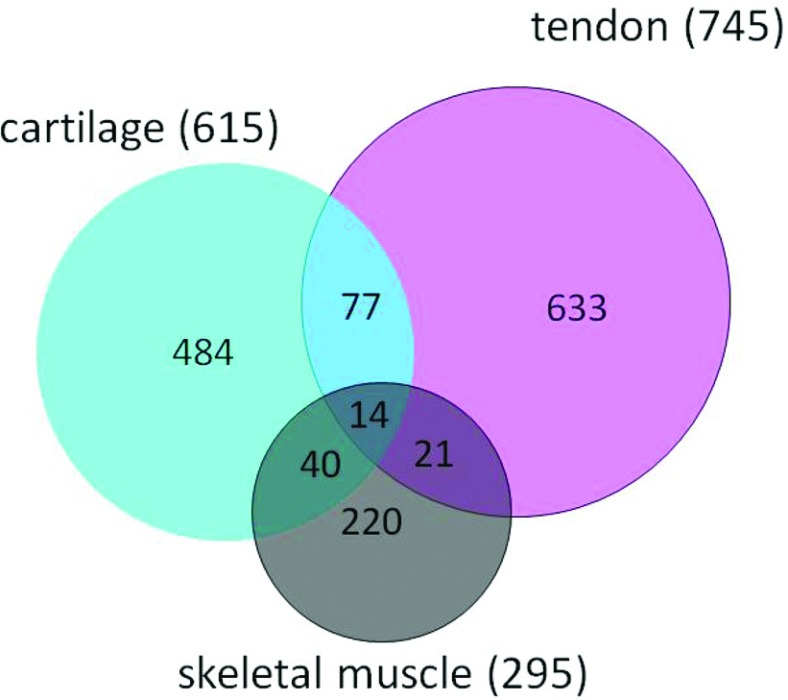
Tendons are an essential component of the musculoskeletal system that provides transmission of forces and an attachment point of muscles to bones. Uniform collagen fibrils arranged parallel to each other are the main component of the tissue with a small population of fibroblasts called tenocytes responsible for the formation and maintenance of the vast ECM [76]. The daily loading/unloading of tendons during the rest/activity cycle suggests potential circadian control of tissue physiology. In fact, the Bmal1−/− mice showed an extensive ectopic tendon calcification phenotype, although the underlying mechanisms remain unknown [62]. Recently, Yeung et al. [77] described an autonomous circadian clock in mouse tendon tissue and human tenocytes. The authors reported the first tendon circadian transcriptome and the age-associated changes in the expression of clock genes in mouse tendon. Importantly, the tendon transcriptome study used tissue samples from the same animals used for the cartilage studies [9], and performed the analysis using identical algorithms and stringency criteria. Therefore it is possible to compare appropriately the clock target genes between two skeletal compartments in the joint, i.e. the cartilage and tendon. This comparison revealed only ~15% overlap between the two closely related tissues, highlighting the tissue-specific adaptations of the circadian system (see Figure 2). Glucocorticoid signalling was shown to be capable of entraining the tendon clock [77]. However, additional mechanisms, such as mechanical loading, feeding/fasting or temperature cycles may well be at play for synchronizing this particular local clock.
Figure 2. Venn diagram comparing the circadian transcriptome of mouse cartilage, tendon and skeletal muscle.
Although all the tissues share the same core molecular clock mechanism, there is limited overlap in the rhythmic CCGs between tissues. Circadian time series microarrays of the musculoskeletal system showed that the circadian clock controls tissue specific sets of target genes, many of which were hallmark transcription factors, key signalling pathways or structural components of the particular tissue. Please note that the transcriptome study in calvarial bone [4] was performed on diurnal animals (kept under LD conditions instead of constant darkness), and therefore is not directly comparable with the circadian transcriptomes listed in the present review. Adapted from Yeung, C.Y., Gossan, N., Lu, Y., Hughes, A., Hensman, J.J., Bayer, M.L., Kjaer, M., Kadler, K.E. and Meng, Q.J. (2014) Gremlin 2 is a BMP antagonist that is regulated by the circadian clock. Sci. Rep. 4, 5183 [77].
Ectopic calcification of the tendons in Clock mutant mice could be observed at as early as 18 weeks of age. Because the calcified tendon phenotype was observed in both global Bmal1-KO mice [62] and ClockΔ19 mutant mice [77], these data strongly suggest a role of the core circadian clock complex (Bmal1–Clock) in regulating tendon physiology. Grem2 (gremlin 2), an antagonist of BMP signalling, was among the rhythmic genes identified by the microarray study and was shown to oscillate in antiphase to that of the BMP signalling activities. Importantly, BMP signalling is known to play a role in calcification of tendons. Recombinant Grem2 protein was demonstrated to inhibit the activation of Smad signalling by BMP2 and prevent deposition of calcium in human tenocytes cultured in an osteogenic medium. The importance of the circadian clock in tendon is further supported by the finding that aged WT mice show dampened circadian rhythm in their tendons, with disturbed expression of Grem2, and spontaneous calcification [77]. Together, these data support a hypothesis that the tendon circadian rhythm plays a critical role in tissue homeostasis. Future studies should be directed towards understanding the contribution of the local tendon oscillator to the tissue homeostasis, and to explore whether a similar clock-controlled pathway (BMP signalling) also operates in human tendinopathies.
SUMMARY
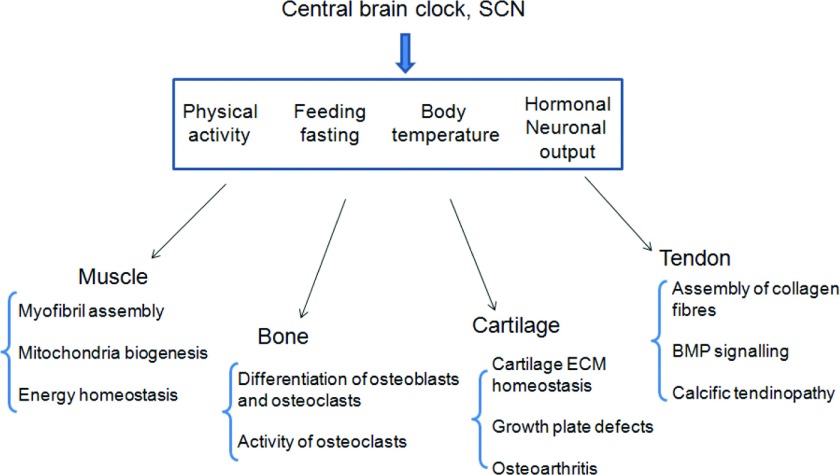
Our understanding of the influence of the circadian clock on the physiology of the musculoskeletal system is still in an early phase (see Figure 3). Although diurnal variations in serum markers of bone and cartilage metabolism and time of day dependent neuromuscular performance were known to exist for a long time, only several studies in the last decade addressed the roles of the peripheral clocks in homoeostasis of the musculoskeletal system. In tissues that are under heavy load the structural components deteriorate with use and age. It may provide an advantage to temporally divide the clean-up and the rebuilding phases so that the two opposing processes, degradation and synthesis, do not interfere with each other. Loss of rhythm could make both phases less efficient and more costly for the organism, but could also result in structurally inferior ECM in bone, cartilage and tendon or myofibrils in muscle. This view is supported by the phenotype of circadian mutant mice which display disorganized ultrastructure of myofibrils in the muscle. The coupling of the circadian phase in gene expression and metabolism with the animal's activity rhythm may also be beneficial. For example, in cartilage, the majority of catabolic genes peaked early in the day, following the night time activity of mice. Meanwhile the anabolic genes peaked 12 h later, early at night. Similar peaking time of anabolic genes was observed in the muscle. Chronic decoupling of the metabolic phases and the animal's daily activity could lead to degenerative changes in cartilage and overall reduced performance in muscles. On the other hand, synchrony among different tissues could play critical roles in physiology. In long bones, the rhythm of growth plate cartilage expansion needs to be in tune with the activity of osteoclasts and osteoblasts. Indeed, loss of circadian rhythm in cartilage results in shorter bones in Bmal1-KO mice.
Figure 3. The circadian control of tissue homoeostasis within the musculoskeletal system.
Light and other environmental zeitgebers entrain the master clock in the SCN. The SCN then generate endogenous circadian time cues (e.g. hormonal, neuronal and body temperature cycles) to synchronize the peripheral oscillators. The musculoskeletal clocks can also be entrained by other time cues, such as food intake and physical activity, either directly or indirectly (through the SCN). The circadian rhythms control a variety of genes and pathways crucial for the correct functioning of the particular musculoskeletal tissue. Mutations of the core clock genes can result in a range of pathologies affecting structural components of the tissues, energy metabolism and differentiation along the tissue specific lineages.
So far, studies of the circadian clocks in the musculoskeletal system provide a glimpse into the rhythmicity of the physiology of tissues involved. Nevertheless, the findings point to an essential role of the molecular clock in the homoeostasis of the muscle, bone, cartilage and tendon. Future studies should test the hypothesis that chronic circadian disruption (during ageing or in shift workers) contributes to the increased susceptibility to diseases of the musculoskeletal system.
ACKNOWLEDGEMENTS
Limited by the space constraints, we apologize to those researchers whose work we could not cite. We thank Professor Karl Kadler (University of Manchester) for his critical reading and advice on this review before submission.
FUNDING
This work has been funded by the Medical Research Council (MRC) Career Development Award [G0900414] and an MRC project grant [grant number MR/K019392/1 (to Q.-J.M.)].
References
- 1.Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Roenneberg T., Merrow M. Circadian clocks: the fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 2005;6:965–971. doi: 10.1038/nrm1766. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert S. M., Weaver D. R. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Yan J., Wang H., Liu Y., Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy J. J., Andrews J. L., McDearmon E. L., Campbell K. S., Barber B. K., Miller B. H., Walker J. R., Hogenesch J. B., Takahashi J. S., Esser K. A. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zvonic S., Ptitsyn A. A., Kilroy G., Wu X., Conrad S. A., Scott L. K., Guilak F., Pelled G., Gazit D., Gimble J. M. Circadian oscillation of gene expression in murine calvarial bone. J. Bone Miner. Res. 2007;22:357–365. doi: 10.1359/jbmr.061114. [DOI] [PubMed] [Google Scholar]
- 9.Gossan N., Zeef L., Hensman J., Hughes A., Bateman J. F., Rowley L., Little C. B., Piggins H. D., Rattray M., Boot-Handford R. P., Meng Q. J. The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis Rheum. 2013;65:2334–2345. doi: 10.1002/art.38035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zvonic S., Ptitsyn A. A., Conrad S. A., Scott L. K., Floyd Z. E., Kilroy G., Wu X., Goh B. C., Mynatt R. L., Gimble J. M. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 11.Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamia K. A., Storch K. F., Weitz C. J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janich P., Pascual G., Merlos-Suarez A., Batlle E., Ripperger J., Albrecht U., Cheng H. Y., Obrietan K., Di Croce L., Benitah S. A. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 14.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology. 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 15.Callard D., Davenne D., Gauthier A., Lagarde D., Van Hoecke J. Circadian rhythms in human muscular efficiency: continuous physical exercise versus continuous rest. A crossover study. Chronobiol. Int. 2000;17:693–704. doi: 10.1081/CBI-100101075. [DOI] [PubMed] [Google Scholar]
- 16.Bessot N., Moussay S., Clarys J. P., Gauthier A., Sesboue B., Davenne D. The influence of circadian rhythm on muscle activity and efficient force production during cycling at different pedal rates. J. Electromyogr. Kinesiol. 2007;17:176–183. doi: 10.1016/j.jelekin.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Sargent C., Ferguson S. A., Darwent D., Kennaway D. J., Roach G. D. The influence of circadian phase and prior wake on neuromuscular function. Chronobiol. Int. 2010;27:911–921. doi: 10.3109/07420528.2010.488901. [DOI] [PubMed] [Google Scholar]
- 18.Teo W., Newton M. J., McGuigan M. R. Circadian rhythms in exercise performance: implications for hormonal and muscular adaptation. J. Sports Sci. Med. 2011;10:600–606. [PMC free article] [PubMed] [Google Scholar]
- 19.Zambon A. C., McDearmon E. L., Salomonis N., Vranizan K. M., Johansen K. L., Adey D., Takahashi J. S., Schambelan M., Conklin B. R. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4:R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller B. H., McDearmon E. L., Panda S., Hayes K. R., Zhang J., Andrews J. L., Antoch M. P., Walker J. R., Esser K. A., Hogenesch J. B., Takahashi J. S. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee S., Nam D., Guo B., Kim J. M., Winnier G. E., Lee J., Berdeaux R., Yechoor V. K., Ma K. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. J. Cell Sci. 2013;126:2213–2224. doi: 10.1242/jcs.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkes C. A., Tapscott S. J. MyoD and the transcriptional control of myogenesis. Semin. Cell. Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Andrews J. L., Zhang X., McCarthy J. J., McDearmon E. L., Hornberger T. A., Russell B., Campbell K. S., Arbogast S., Reid M. B., Walker J. R., et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Patel S. P., McCarthy J. J., Rabchevsky A. G., Goldhamer D. J., Esser K. A. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 2012;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDearmon E. L., Patel K. N., Ko C. H., Walisser J. A., Schook A. C., Chong J. L., Wilsbacher L. D., Song E. J., Hong H. K., Bradfield C. A., Takahashi J. S. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Li S., Liu T., Borjigin J., Lin J. D. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 28.Lin J., Handschin C., Spiegelman B. M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Woldt E., Sebti Y., Solt L. A., Duhem C., Lancel S., Eeckhoute J., Hesselink M. K., Paquet C., Delhaye S., Shin Y., et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shavlakadze T., Anwari T., Soffe Z., Cozens G., Mark P. J., Gondro C., Grounds M. D. Impact of fasting on the rhythmic expression of myogenic and metabolic factors in skeletal muscle of adult mice. Am. J. Physiol. Cell Physiol. 2013;305:C26–C35. doi: 10.1152/ajpcell.00027.2013. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G. D., Sakaki Y., Menaker M., Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka Y., Honma S., Honma K. Scheduled exposures to a novel environment with a running-wheel differentially accelerate re-entrainment of mice peripheral clocks to new light-dark cycles. Genes Cells. 2008;13:497–507. doi: 10.1111/j.1365-2443.2008.01183.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolff G., Esser K. A. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med. Sci. Sports Exerc. 2012;44:1663–1670. doi: 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng X., McDonald J. M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011;6:121–145. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downey P. A., Siegel M. I. Bone biology and the clinical implications for osteoporosis. Phys. Ther. 2006;86:77–91. doi: 10.1093/ptj/86.1.77. [DOI] [PubMed] [Google Scholar]
- 37.Sommerfeldt D. W., Rubin C. T. Biology of bone and how it orchestrates the form and function of the skeleton. Eur. Spine J. 2001;10:S86–S95. doi: 10.1007/s005860100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenspan S. L., Dresner-Pollak R., Parker R. A., London D., Ferguson L. Diurnal variation of bone mineral turnover in elderly men and women. Calcif. Tissue Int. 1997;60:419–423. doi: 10.1007/s002239900256. [DOI] [PubMed] [Google Scholar]
- 39.Shao P., Ohtsuka-Isoya M., Shinoda H. Circadian rhythms in serum bone markers and their relation to the effect of etidronate in rats. Chronobiol. Int. 2003;20:325–336. doi: 10.1081/CBI-120019343. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava A. K., Bhattacharyya S., Li X., Mohan S., Baylink D. J. Circadian and longitudinal variation of serum C-telopeptide, osteocalcin, and skeletal alkaline phosphatase in C3H/HeJ mice. Bone. 2001;29:361–367. doi: 10.1016/S8756-3282(01)00581-6. [DOI] [PubMed] [Google Scholar]
- 41.Fu L., Patel M. S., Bradley A., Wagner E. F., Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Goldstrohm A. C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 43.Li R., Yue J., Zhang Y., Zhou L., Hao W., Yuan J., Qiang B., Ding J. M., Peng X., Cao J. M. CLOCK/BMAL1 regulates human nocturnin transcription through binding to the E-box of nocturnin promoter. Mol. Cell Biochem. 2008;317:169–177. doi: 10.1007/s11010-008-9846-x. [DOI] [PubMed] [Google Scholar]
- 44.Kawai M., Green C. B., Lecka-Czernik B., Douris N., Gilbert M. R., Kojima S., Ackert-Bicknell C., Garg N., Horowitz M. C., Adamo M. L., et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPARγ nuclear translocation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guntur A. R., Kawai M., Le P., Bouxsein M. L., Bornstein S., Green C. B., Rosen C. J. An essential role for the circadian-regulated gene nocturnin in osteogenesis: the importance of local timekeeping in skeletal homeostasis. Ann. N.Y. Acad. Sci. 2011;1237:58–63. doi: 10.1111/j.1749-6632.2011.06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shockley K. R., Lazarenko O. P., Czernik P. J., Rosen C. J., Churchill G. A., Lecka-Czernik B. PPARγ2 nuclear receptor controls multiple regulatory pathways of osteoblast differentiation from marrow mesenchymal stem cells. J. Cell Biochem. 2009;106:232–246. doi: 10.1002/jcb.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James A. W., Shen J., Khadarian K. A., Pang S., Chung G., Goyal R., Asatrian G., Velasco O., Kim J., Zhang X., et al. Lentiviral delivery of PPARγshRNAalters the balance of osteogenesis and adipogenesis, improving bone microarchitecture. Tissue Eng. A. 2014 doi: 10.1089/ten.TEA.2013.0736. doi:10.1089/ten.TEA.2013.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi X., Shi W., Li Q., Song B., Wan M., Bai S., Cao X. A glucocorticoid-induced leucine-zipper protein, GILZ, inhibits adipogenesis of mesenchymal cells. EMBO Rep. 2003;4:374–380. doi: 10.1038/sj.embor.embor805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W., Yang N., Shi X. M. Regulation of mesenchymal stem cell osteogenic differentiation by glucocorticoid-induced leucine zipper (GILZ) J. Biol. Chem. 2008;283:4723–4729. doi: 10.1074/jbc.M704147200. [DOI] [PubMed] [Google Scholar]
- 50.Lecka-Czernik B., Gubrij I., Moerman E. J., Kajkenova O., Lipschitz D. A., Manolagas S. C., Jilka R. L. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARγ2. J. Cell. Biochem. 1999;74:357–371. doi: 10.1002/(SICI)1097-4644(19990901)74:3<357::AID-JCB5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 51.Komoto S., Kondo H., Fukuta O., Togari A. Comparison of β-adrenergic and glucocorticoid signaling on clock gene and osteoblast-related gene expressions in human osteoblast. Chronobiol. Int. 2012;29:66–74. doi: 10.3109/07420528.2011.636496. [DOI] [PubMed] [Google Scholar]
- 52.Fujihara Y., Kondo H., Noguchi T., Togari A. Glucocorticoids mediate circadian timing in peripheral osteoclasts resulting in the circadian expression rhythm of osteoclast-related genes. Bone. 2014;61:1–9. doi: 10.1016/j.bone.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 53.Bjarnason N. H., Henriksen E. E., Alexandersen P., Christgau S., Henriksen D. B., Christiansen C. Mechanism of circadian variation in bone resorption. Bone. 2002;30:307–313. doi: 10.1016/S8756-3282(01)00662-7. [DOI] [PubMed] [Google Scholar]
- 54.Karsdal M. A., Byrjalsen I., Riis B. J., Christiansen C. Investigation of the diurnal variation in bone resorption for optimal drug delivery and efficacy in osteoporosis with oral calcitonin. BMC Clin. Pharmacol. 2008;8:12. doi: 10.1186/1472-6904-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martel-Pelletier J., Boileau C., Pelletier J. P., Roughley P. J. Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol. 2008;22:351–384. doi: 10.1016/j.berh.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Russell J. E., Grazman B., Simmons D. J. Mineralization in rat metaphyseal bone exhibits a circadian stage dependency. Proc. Soc. Exp. Biol. Med. 1984;176:342–345. doi: 10.3181/00379727-176-41880. [DOI] [PubMed] [Google Scholar]
- 57.Stevenson S., Hunziker E. B., Herrmann W., Schenk R. K. Is longitudinal bone growth influenced by diurnal variation in the mitotic activity of chondrocytes of the growth plate? J. Orthop. Res. 1990;8:132–135. doi: 10.1002/jor.1100080117. [DOI] [PubMed] [Google Scholar]
- 58.Igarashi K., Saeki S., Shinoda H. Diurnal rhythms in the incorporation and secretion of 3H-proline and 3H-galactose by cartilage cells and osteoblasts in various bone-forming sites in growing rats. Orthod. Waves. 2013;72:11–15. doi: 10.1016/j.odw.2012.09.001. [DOI] [Google Scholar]
- 59.Andersson M. L., Petersson I. F., Karlsson K. E., Jonsson E. N., Mansson B., Heinegard D., Saxne T. Diurnal variation in serum levels of cartilage oligomeric matrix protein in patients with knee osteoarthritis or rheumatoid arthritis. Ann. Rheum. Dis. 2006;65:1490–1494. doi: 10.1136/ard.2005.051292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong S. Y., Stabler T. V., Criscione L. G., Elliott A. L., Jordan J. M., Kraus V. B. Diurnal variation of serum and urine biomarkers in patients with radiographic knee osteoarthritis. Arthritis Rheum. 2006;54:2496–2504. doi: 10.1002/art.21977. [DOI] [PubMed] [Google Scholar]
- 61.Takarada T., Kodama A., Hotta S., Mieda M., Shimba S., Hinoi E., Yoneda Y. Clock genes influence gene expression in growth plate and endochondral ossification in mice. J. Biol. Chem. 2012;287:36081–36095. doi: 10.1074/jbc.M112.408963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bunger M. K., Walisser J. A., Sullivan R., Manley P. A., Moran S. M., Kalscheur V. L., Colman R. J., Bradfield C. A. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 63.Okubo N., Minami Y., Fujiwara H., Umemura Y., Tsuchiya Y., Shirai T., Oda R., Inokawa H., Kubo T., Yagita K. Prolonged bioluminescence monitoring in mouse ex vivo bone culture revealed persistent circadian rhythms in articular cartilages and growth plates. PLoS ONE. 2013;8:e78306. doi: 10.1371/journal.pone.0078306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanbe K., Inoue K., Xiang C., Chen Q. Identification of clock as a mechanosensitive gene by large-scale DNA microarray analysis: downregulation in osteoarthritic cartilage. Mod. Rheumatol. 2006;16:131–136. doi: 10.3109/s10165-006-0469-3. [DOI] [PubMed] [Google Scholar]
- 65.Verma P., Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem. 2011;112:3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- 66.Knauper V., Bailey L., Worley J. R., Soloway P., Patterson M. L., Murphy G. Cellular activation of proMMP-13 by MT1-MMP depends on the C-terminal domain of MMP-13. FEBS Lett. 2002;532:127–130. doi: 10.1016/S0014-5793(02)03654-2. [DOI] [PubMed] [Google Scholar]
- 67.Chellaiah M. A., Ma T. Membrane localization of membrane type 1 matrix metalloproteinase by CD44 regulates the activation of pro-matrix metalloproteinase 9 in osteoclasts. Biomed. Res. Int. 2013;2013:302392. doi: 10.1155/2013/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velasco-Loyden G., Arribas J., Lopez-Casillas F. The shedding of betaglycan is regulated by pervanadate and mediated by membrane type matrix metalloprotease-1. J. Biol. Chem. 2004;279:7721–7733. doi: 10.1074/jbc.M306499200. [DOI] [PubMed] [Google Scholar]
- 69.Urena J. M., Merlos-Suarez A., Baselga J., Arribas J. The cytoplasmic carboxy-terminal amino acid determines the subcellular localization of proTGF-α and membrane type matrix metalloprotease (MT1-MMP) J. Cell Sci. 1999;112:773–784. doi: 10.1242/jcs.112.6.773. [DOI] [PubMed] [Google Scholar]
- 70.Szabova L., Yamada S. S., Wimer H., Chrysovergis K., Ingvarsen S., Behrendt N., Engelholm L. H., Holmbeck K. MT1-MMP and type II collagen specify skeletal stem cells and their bone and cartilage progeny. J. Bone Miner. Res. 2009;24:1905–1916. doi: 10.1359/jbmr.090510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/S0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 72.Honda K. K., Kawamoto T., Ueda H. R., Nakashima A., Ueshima T., Yamada R. G., Nishimura M., Oda R., Nakamura S., Kojima T., et al. Different circadian expression of major matrix-related genes in various types of cartilage: modulation by light-dark conditions. J. Biochem. 2013;154:373–381. doi: 10.1093/jb/mvt068. [DOI] [PubMed] [Google Scholar]
- 73.Hanyu R., Hayata T., Nagao M., Saita Y., Hemmi H., Notomi T., Nakamoto T., Schipani E., Knonenbery H., Kaneko K., et al. Per-1 is a specific clock gene regulated by parathyroid hormone (PTH) signaling in osteoblasts and is functional for the transcriptional events induced by PTH. J. Cell Biochem. 2011;112:433–438. doi: 10.1002/jcb.22957. [DOI] [PubMed] [Google Scholar]
- 74.Hinoi E., Ueshima T., Hojo H., Iemata M., Takarada T., Yoneda Y. Up-regulation of per mRNA expression by parathyroid hormone through a protein kinase A-CREB-dependent mechanism in chondrocytes. J. Biol. Chem. 2006;281:23632–23642. doi: 10.1074/jbc.M512362200. [DOI] [PubMed] [Google Scholar]
- 75.Lanske B., Karaplis A. C., Lee K., Luz A., Vortkamp A., Pirro A., Karperien M., Defize L. H., Ho C., Mulligan R. C., et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- 76.Kadler K. E., Hill A., Canty-Laird E. G. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 2008;20:495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeung C. Y., Gossan N., Lu Y., Hughes A., Hensman J. J., Bayer M. L., Kjaer M., Kadler K. E., Meng Q. J. Gremlin-2 is a BMP antagonist that is regulated by the circadian clock. Sci. Rep. 2014;4:5183. doi: 10.1038/srep05183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maronde E., Schilling A. F., Seitz S., Schinke T., Schmutz I., van der Horst G., Amling M., Albrecht U. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS ONE. 2010;5:e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]