Abstract
Archaeal RNA polymerases (RNAPs) are closely related to eukaryotic RNAPs, and in Euryarchaea, genomic DNA is wrapped and compacted by histones into archaeal nucleosomes. In eukaryotes, transcription of DNA bound into nucleosomes is facilitated by histone tail modifications and chromatin remodeling complexes, but archaeal histones do not have histone tails and archaeal genome sequences provide no evidence for archaeal homologs of eukaryotic chromatin remodeling complexes. We have therefore investigated the ability of an archaeal RNAP, purified from Methanothermobacter thermautotrophicus, to transcribe DNA bound into an archaeal nucleosome by HMtA2, an archaeal histone from M. thermautotrophicus. To do so, we constructed a template that allows transcript elongation to be separated from transcription initiation, on which archaeal nucleosome assembly is positioned downstream from the site of transcription initiation. At 58°C, in the absence of an archaeal nucleosome, M. thermautotrophicus RNAP transcribed this template DNA at a rate of ∼20 nucleotides per second. With an archaeal nucleosome present, transcript elongation was slowed but not blocked, with transcription pausing at sites before and within the archaeal nucleosome. With additional HMtA2 binding, complexes were obtained that also incorporated the upstream regulatory region. This inhibited transcription presumably by preventing archaeal TATA-box binding protein, general transcription factor TFB, and RNAP access and thus inhibiting transcription initiation.
In almost all eukaryotes, two copies of essentially the same four histones, H2A, H2B, H3, and H4, wrap and compact nuclear DNA into nucleosomes (55), and three related RNA polymerases (RNAPs), Pol I, II, and III, transcribe primarily rRNAs, mRNAs, and tRNAs, respectively (15). Logically, therefore, a histone-based DNA wrapping mechanism compacted the DNA, and Pol I, II, and III, or their common RNAP ancestor, were present in the first eukaryotic nucleus. These basic components of genome compaction and gene expression were presumably also present in the immediate prokaryotic ancestor of the eukaryotic nucleus, and this apparently belonged to the lineage that gave rise to contemporary Archaea (32, 45) but not to Bacteria. There are no histone-fold-containing proteins in Bacteria (12), but histones are present in members of the euryarchaeal branch of the Archaea. Archaeal histones have sequences conserved in all four eukaryotic histones and form histone folds almost identical to those that constitute the globular core of the eukaryotic nucleosome (17, 31). Archaeal histones similarly bind, wrap, and compact DNA forming complexes designated archaeal nucleosomes (41). These most closely resemble the tetrasome (2), the structure formed at the center of the eukaryotic nucleosome where ∼90 bp of DNA are circularized around a histone (H3 + H4)2 tetramer (3, 29, 58). Archaeal RNAPs have subunits and subunit complexities similar to those of all three eukaryotic RNAPs but most closely resemble Pol II (62). Consistent with this, archaeal promoters have TATA boxes and TFB-responsive elements (BRE), and archaeal homologs of the eukaryotic TATA-box binding protein (TBP) and general transcription factor TFIIB, designated TFB in Archaea, recruit archaeal RNAP to the promoter (9, 45). Archaea also have TFE, a homolog of the α subunit of TFIIE (8, 20) that stimulates transcription from some promoters, and TFS, a homolog of TFIIS, that facilitates stalled transcript elongation (21). Archaea do not have homologs of the β subunit of TFIIE, of TFIIA or TFIIH, or of Pol II-specific transcription elongation factors (42, 56).
In cells that employ histones for genome compaction, mechanisms must exist to access histone-bound DNA, and determining how eukaryotic nuclear DNA is transcribed within the constraints of nucleosomes and chromatin is currently an area of intense investigation. Most of these studies focus on posttranslation histone modifications and the roles of large, multisubunit complexes in facilitating eukaryotic nucleosome assembly, restructuring, and movement (10, 11, 23, 32, 38, 42, 44, 53, 56, 65). Archaeal genome sequences provide no evidence for archaeal relatives of the subunits of the chromatin remodeling complexes, but both crenarachaeal and euryarchaeal genomes do encode proteins with sequences related to the Elp3 histone acetylases and Sir2 and Rpd3 deacetylases (26, 43). Crenarchaea do not, however, have histones, and the histones present in Euryarchaea do not have histone tails (45) and therefore lack the lysine targets acetylated in eukaryotic histones by the Elp3 complex (11, 23, 64). These archaeal proteins presumably therefore have other substrates, and a Sir2 family member from the crenarchaeon Sulfolobus solfataricus has been shown to deacetylate Alba, an abundant but nonhistone DNA binding protein (7). Given this situation, it seemed possible that archaeal RNAPs might have the inherent ability to transcribe DNA bound into an archaeal nucleosome, without the aid of other factors. To determine if this is the case, a transcription template was constructed that allows transcription initiation and elongation to be separated, on which archaeal nucleosome assembly is positioned at a site downstream from the site of transcription initiation. The results obtained using this template demonstrate that transcript elongation by the archaeal RNAP purified from the histone-containing euryarchaeaon Methanothermobacter thermautotrophicus (16) is slowed, but is not blocked, on encountering an archaeal nucleosome. When archaeal histone-containing complexes were assembled that also incorporated the upstream regulatory region, transcription was inhibited.
MATERIALS AND METHODS
Protein purifications.
The HMtA2-encoding hmtA2 gene (MTH1696) (49) was amplified from M. thermautotrophicus genomic DNA with flanking HindIII and EcoRI sites added by using oligonucleotides with the sequences 5′-TAATAAGCTTACTTAAACAAGGAGGG and 5′-GTGATATAGAATTCTGATATGGATGC. Following HindIII-plus-EcoRI digestion, this DNA molecule was ligated with HindIII-plus-EcoRI-digested pRAT4 (39), and the resulting plasmid was transformed into Escherichia coli BL21. Recombinant HMtA2 was purified and quantitated after isopropyl-β-d-thiogalactopyranoside-induced synthesis in E. coli BL21(DE3), using the procedures described for HMfB (46). For relevance to the in vivo conditions (40, 41), archaeal histone concentrations are stated in terms of dimers per 100 bp, with an accuracy of ±25%. The preparation of His6-tagged M. thermautotrophicus TBP and TFB and native RNAP and their use in in vitro transcription have been described previously (16).
Template construction.
Figure 1A shows the sequence of the 284-bp DNA template constructed for this study. The promoter-containing region from upstream of the hmtB gene in M. thermautotrophicus (MTH0254) (16) and the 24-bp U-less cassette were assembled in vitro by using overlapping oligonucleotides (sequences available on request; Ransom Hill Biosciences, Ramona, Calif.). This assembly was cloned into NsiI-plus-EcoRI-digested pLitmus28 (New England Biolabs, Beverly, Mass.), and the resulting plasmid was digested with EcoRI plus HindIII and ligated with an EcoRI-HindIII restriction fragment from pKS564 that contained the 60-bp Selex1 sequence (4, 5). Template DNA preparations were generated by PCR amplification from the resulting plasmid and purified by passage through a Qiaquik spin column (QIAGEN, Valencia, Calif.). For ternary complex isolation, a primer with a biotin molecule attached to the 5′ nucleotide was used to amplify the template DNA.
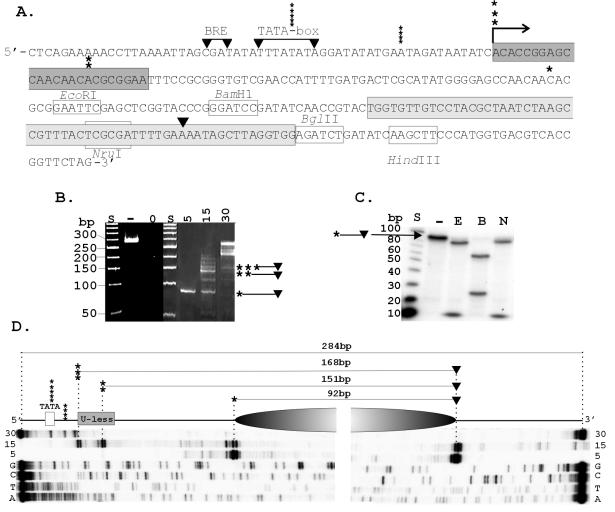
FIG. 1.
Transcription template and position of HMtA2 assembly. (A) The sequence of the transcription template is shown with the BRE and TATA-box sequence from the HMtB promoter (16), the U-less cassette (dark gray), Selex1 sequence (light gray) (4), site of transcription initiation (→), and 3′ (▾) and 5′ (asterisks) boundaries of the MN-protected fragments indicated. (B) Electrophoretic separation of the fragments of the template DNA protected from MN digestion by assembly into complexes at the HMtA2 dimer/100-bp ratios indicated above the corresponding lane. Control lanes contained size standards (S), untreated template DNA (−) and template DNA exposed to MN for 1 min in the absence of HMtA2 (0). (C) Autoradiogram of the electrophoretic separation of the restriction fragments generated from a population of the ∼90-bp MN-protected DNA fragments after 32P end labeling and digestion with EcoRI (E), BamHI (B), or NruI (N). Control lanes contained size standards (S) and an aliquot of the ∼90-bp molecules not exposed to restriction enzymes (−). (D) Diagram showing the position of the archaeal nucleosome (shaded oval), the 5′ and 3′ boundaries, and the precise lengths of the fragments of the template DNA protected from MN digestion as determined by the lengths of the primer extension products indicated below. The extension products were generated from 32P-labeled primers that hybridized to the MN-protected fragments bound by 5, 15, or 30 HMtA2 dimers per 100 bp of template DNA. Adjacent lanes contained the products of sequencing reactions (G, C, T, A) generated from the template DNA, using the same primers.
EMSAs.
For electrophoretic mobility polyacrylamide gel shift assays (EMSAs), aliquots of 32P-labeled template DNA (50 ng) were incubated with increasing amounts of HMtA2 for 20 min at 25°C in 50 μl of transcription buffer (20 mM Tris HCl [pH 8], 120 mM KCl, 10 mM MgCl2, 2 mM dithiothreitol). The products were separated by electrophoresis through 6% polyacrylamide gels run in Tris-borate-EDTA buffer at 8V/cm and visualized by autoradiography (16). Aliquots of ternary complexes containing a 32P-labeled 24-nucleotide (nt) transcript, generated as described below, were incubated with increasing amounts of HMtA2 in transcription buffer for 20 min at 25°C. The products were subjected to electrophoresis through 5% polyacrylamide gels and visualized by phosphorimaging (Storm model 840; Amersham Biosciences, Piscataway, N.J.).
MN and primer extension footprinting.
Aliquots of template DNA (50 ng) were incubated for 20 min at 25°C with increasing amounts of HMtA2 in 50 μl of transcription buffer. One microliter of 100 mM CaCl2 and 1 U of micrococcol nuclease (MN) (Sigma, St. Louis, Mo.) were added, and the reaction mixtures were placed at 37°C for 1 min. One microliter of 100 mM EDTA, 20 mg of proteinase K (Invitrogen, San Diego, Calif.)/ml, and 10% (wt/vol) sodium dodecyl sulfate were then added, and incubation was continued at 37°C for 30 min. Undigested DNA molecules were separated by electrophoresis through 10% polyacrylamide gels and visualized by staining using SYBR-Gold (Molecular Probes, Eugene, Oreg.). Preparations of these MN-protected fragments were gel purified, 32P end labeled using [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen, Gaithersburg, Md.), and subjected to restriction enzyme digestions. The fragments generated were separated by electrophoresis through 12% denaturing polyacrylamide gels and visualized by autoradiography. The 5′ and 3′ boundaries of MN-protected fragments were also determined from the lengths of primer extension products generated using 32P-labeled primers that hybridized within the MN-protected regions.
In vitro transcription.
Increasing amounts of HMtA2 were incubated for 20 min with 50 ng of the template DNA in transcription buffer. TBP (50 ng), TFB (300 ng), 5 μl of RNAP, 200 μM ATP, 200 μM UTP, 200 μM GTP, 20 μM CTP (Roche Applied Science, Indianapolis, Ind.), and 5 μCi of [α-32P]CTP (3 kCi/mmol; ICN, Costa Mesa, Calif.) were added, and the mixtures (50-μl final volume) were incubated at 58°C for 30 min. The transcripts synthesized were separated by electrophoresis through 6% denaturing polyacrylamide gels (16), visualized by autoradiography, and quantitated by phosphorimaging.
Ternary complex isolation and stalled transcript elongation.
Ternary complexes that contained a 32P-labeled 24-nt U-less transcript were generated by incubation of biotin-labeled template DNA (50 ng), TBP (50 ng), TFB (300 ng), 5 μl of RNAP, 200 μM ATP, 200 μM GTP, 20 μM CTP, and 20 μCi of [α-32P]CTP (3 kCi/mmol) in 50 μl of transcription buffer for 20 min at 58°C. After the mixture was cooled to room temperature, 10 μg of streptavidin-coated Dynabeads (Dynal Biotech, Lake Success, N.Y.) was added and the mixture was incubated at room temperature for 5 min. The beads were then captured by attraction to a magnet, the supernatant was removed, and after being washed four times with 25 μl of transcription buffer, the beads were resuspended in transcription buffer and incubated with or without HMtA2 for 20 min at 25°C. The reaction mixture was then placed at 58°C, and ATP, GTP, CTP, and UTP (final concentration of 200 μM each) were added. Aliquots (10 μl) were removed at increasing times and mixed with 10 μl of 95% deionized formamide-20 mM EDTA. The transcripts synthesized were separated by electrophoresis through 6% polyacrylamide sequencing gels or through gels in which the upper two-thirds were 6% polyacrylamide and the lower third was 18% polyacrylamide. 32P-labeled transcripts were then visualized and quantitated by phosphorimaging.
RESULTS
Template construction and positioning of HMtA2 assembly.
M. thermautotrophicus RNAP initiates transcription accurately in vitro from the promoter of the M. thermautotrophicus histone-encoding hmtB gene (16). We therefore constructed a template that had the BRE and TATA box from this promoter positioned appropriately to direct transcription initiation at the start of a 24-bp sequence (a U-less cassette) that requires only ATP, CTP, and GTP for transcription. To direct and localize archaeal nucleosome assembly away from this regulatory region, the 60-bp Selex1 sequence was incorporated ∼100 bp downstream from the U-less cassette (Fig. 1A). This sequence was SELEX selected previously from a large population of random sequence molecules based on inherently high affinity for HMfB, an archaeal histone from Methanothermus fervidus (4). It was known to direct the positioned assembly of HMfB (5), and as shown in Fig. 1B to D, it positioned HMtA2 assembly with base-pair accuracy when incorporated into the template DNA. Archaeal histone dimers polymerize to bind and wrap DNA (3-5), and at histone dimer-to-DNA ratios similar to those in vivo (two to three dimers per 100 bp) (40, 41), archaeal nucleosomes are formed in vitro that have ∼90 bp circularized around an archaeal histone tetramer core (3, 33). At higher histone-to-DNA ratios, additional histone binding occurs, resulting in larger complexes that incorporate longer lengths of DNA (3, 45). As this predicts, complexes assembled in reaction mixtures that contained up to five histone dimers per 100 bp protected only ∼90-bp fragments of the template DNA from MN digestion (Fig. 1B) whereas complexes formed at higher histone-to-DNA ratios protected longer regions of the template DNA. These increased in length in multiples of ∼30 bp, consistent with protection by the addition of histone dimers (29, 30). Restriction enzyme digestions of the ∼90-bp MN-protected fragments revealed that >95% of these molecules had essentially the same sequence (Fig. 1C), and the primer extension products generated using primers that hybridized within the MN-protected region confirmed this result (Fig. 1D). HMtA2 assembly on the template DNA resulted in a precisely positioned archaeal nucleosome that protected a 92- ± 2-bp sequence that extended from ∼50 bp downstream of the U-less cassette through most of the Selex1 sequence. Primer extension products were also generated using the longer MN-protected fragments as the template DNA. These demonstrated that with additional HMtA2 polymerization, DNA was protected from MN digestion that extended from the 92- ± 2-bp sequence at first only in the 5′ direction towards the promoter. As illustrated in Fig. 1D, at a ratio of 15 HMtA2 dimers per 100 bp, the predominant MN-protected DNA fragment was 151 ± 2 bp, and this contained the 92- ± 2-bp region plus ∼60 bp that extended 5′ from this region and included part of the U-less cassette (Fig. 1D). At a ratio of 30 dimers per 100 bp, HMtA2 assembly incorporated template DNA both 5′ and 3′ to the 92- ± 2-bp region, and the complexes formed protected almost the entire template from MN digestion (Fig. 1D).
Reduction of transcript accumulation by HMtA2 binding.
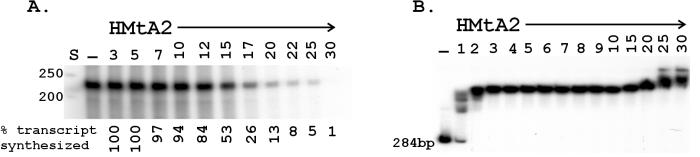
As previously documented (16), transcription by the M. thermautotrophicus in vitro system resulted in the linear accumulation of runoff transcripts for >30 min at 58° C, the growth temperature of M. thermautotrophicus. When HMtA2 was incubated with the template DNA before the addition of TBP, TFB, RNAP, and nucleoside triphosphates, there was no reduction in transcript accumulation at histone-to-DNA ratios up to five dimers per 100 bp. Consistent with an earlier report (50), at higher histone-to-DNA ratios transcript accumulation was reduced, and there was no detectable transcription in reaction mixtures that contained >30 HMtA2 dimers per 100 bp (Fig. 2A). EMSAs confirmed that HMtA2 bound to the template DNA under the in vitro transcription reaction conditions, and as shown in Fig. 2B, HMtA2-containing complexes were assembled on every template DNA molecule present in reaction mixtures where HMtA addition (three to six dimers per 100 bp) (Fig. 2A) had no detectable effect on transcript accumulation.
FIG. 2.
Runoff transcription and EMSA of HMtA2 binding. (A) Electrophoresis of the 225-nt 32P-labeled runoff transcripts synthesized in 30 min at 58°C on templates preincubated with HMtA2 as indicated (dimers per 100 bp) above each lane. The amount of transcript, as a percentage of that synthesized in the absence of HMtA2 (−) is listed below each lane. Lane S contained size standards. (B) Autoradiogram of the electrophoretic separation of the complexes formed by incubation of 32P-labeled template DNA (50 ng) without (−) and with HMtA2 at the histone dimer/100-bp ratio indicated above each lane.
HMtA2 binding does not prevent transcript elongation.
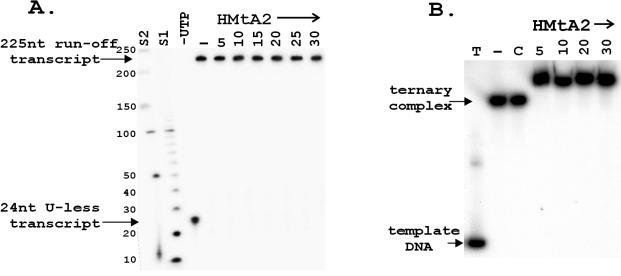
M. thermautotrophicus RNAP initiated transcription in reaction mixtures that contained ATP, GTP, and [32P]CTP but no UTP, but stalled after transcribing the U-less cassette. The resulting ternary complexes were removed from the reaction mixture by streptavidin affinity, and aliquots were incubated with increasing amounts of HMtA2. These complexes were then added to complete but nonradioactive in vitro transcription reaction mixtures. At all HMtA2-to-ternary complex ratios, the 32P-labeled 24-nt transcript present in the ternary complex was extended into a 225-nt runoff transcript. Given this result, it seemed possible that HMtA2 might not be able to bind to DNA already bound into a ternary complex, but EMSA proved that this was not the case. Incubation with HMtA2 reduced the electrophoretic mobility of the ternary complexes (Fig. 3B). To confirm that this resulted from histone-DNA binding rather than nonspecific protein aggregation, control experiments were undertaken using an archaeal histone variant, HMfB K13T+R19S+T54K, that has no DNA binding ability (51). Even at very high histone-to-ternary complex ratios, incubation with the variant resulted in no decrease in the electrophoretic mobility of the ternary complexes (Fig. 3B).
FIG. 3.
Stalled-transcript elongation and EMSA of ternary complexes. (A) Ternary complexes that contained the 32P-labeled 24-nt U-less transcript were incubated without (−) or with HMtA2 and then added to a complete transcription reaction mixture and incubated at 58°C for 20 min. A control aliquot of the ternary complexes was incubated in a reaction mixture that lacked UTP (−UTP). The transcripts synthesized were separated by electrophoresis, and the 32P-labeled transcripts were detected by autoradiography and quantitated by β-decay measurement. The HMtA2 dimer/100-bp ratio in the reaction mixture is indicated above the corresponding lane. The control lanes contained size standards (S1 and S2). (B) Aliquots of ternary complexes that contained a 32P-labeled 24-nt U-less transcript were incubated with HMtA2 at the histone dimer/100-bp ratio indicated above each lane. The products were separated by electrophoresis and visualized by autoradiography. Control lanes contained a sample of 32P-end-labeled template DNA (T), an aliquot of the ternary complexes incubated without HMtA2 addition (−), and an aliquot incubated with the HMfB K13T+R19S+T54K variant which lacks DNA binding ability (51) at a ratio of 30 histone dimers per 100 bp (C).
The presence of an archaeal nucleosome slows but does not prevent transcript elongation.
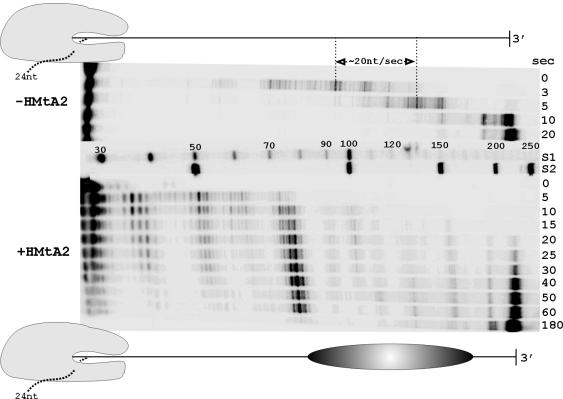
Although HMtA2 binding to the template DNA did not prevent stalled-transcript elongation (Fig. 3A), it was still possible that it reduced the rate of transcription. To determine if this was the case, HMtA2 was incubated with ternary complexes containing a 32P-labeled 24-nt stalled transcript to allow assembly of an archaeal nucleosome at the Selex1 site, and these complexes were then added to a complete in vitro transcription reaction mixture. The lengths of the 32P-labeled transcripts synthesized after increasing times of incubation at 58°C were determined. On control templates lacking an archaeal nucleosome, transcript elongation occurred at a rate of ∼20 nt/s and almost all of the 24-nt transcripts that were extended reached full length (225 nt) in 20 s. With the archaeal nucleosome present, transcript elongation was slowed and 225-nt transcripts first became evident only after ∼30 s (Fig. 4). The M. thermautotrophicus RNAP paused at several locations, both before and within the archaeal nucleosome, and the predominant pause site coincided precisely with the 5′ boundary of the archaeal nucleosome (Fig. 4). When a longer template that had 15 bp inserted between the U-less cassette and Selex1 sequence was used, transcription pausing occurred at the same sites relative to the location of the archaeal nucleosome (result not shown), again with a predominant pause occurring at the 5′ boundary of the nucleosome.
FIG. 4.
Transcription in the absence and presence of an archaeal nucleosome. Ternary complexes containing a 32P-labeled 24-nt U-less transcript incubated with HMtA2 (+HMtA2) or without HMtA2 (−HMtA2) were added to a complete reaction mixture and placed at 58°C. Aliquots were taken at the times indicated (seconds), and the transcripts synthesized were separated by electrophoresis and visualized by phosphorimaging. Control lanes contained nucleotide size standards (S1 and S2). In the diagrams, the template DNA is shown to scale, with or without the positioned archaeal nucleosome. Footprinting studies indicate that an archaeal RNAP in a ternary complex extends ∼12 bp downstream from the site of nucleoside triphosphate polymerization (52). The rate of transcription was estimated, as indicated, from the increase in length of transcripts during a 2-s incubation at 58°C.
DISCUSSION
M. thermautotrophicus RNAP transcription through an archaeal nucleosome.
The results obtained with M. thermautotrophicus RNAP and DNA assembled into an archaeal nucleosome are consistent with those reported for transcription in vitro of eukaryotic nucleosome-containing templates by phage T7 and SP6 RNAPs (14, 25, 37, 61) and by eukaryotic Pol III (54). In all these cases, progress of the RNAP in vitro is slowed but not blocked on encountering the histone-DNA complex and RNAP pausing occurs at ∼10-bp intervals during transcription through the complex (Fig. 4). To explain these observations, it has been proposed that the advancing RNAP causes short regions of DNA to detach sequentially from the histone core and so moves forward, stepwise through the nucleosome, transcribing each detached region. The pausing observed then occurs, once per helical turn, where the advancing RNAP and histone core must most closely interact (6, 54). After transcription, the detached DNA sequence is rebound by the histone core, and so the nucleosome remains intact and is not disassembled by passage of the RNAP. Based on the pausing observed (Fig. 4), the archaeal nucleosome remained intact during transcription by M. thermautotrophicus RNAP, but this is difficult to substantiate further experimentally. Eukaryotic nucleosome assembly in vitro requires an assembly protocol (30), and the presence of a nucleosome on the template DNA after transcription is therefore de facto evidence that transcription did not disassemble the nucleosome. In contrast, archaeal nucleosome assembly occurs spontaneously under in vitro transcription conditions (Fig. 2), and therefore it remains to be demonstrated conclusively that transcription does not result in archaeal nucleosome disassembly followed by immediate reassembly.
In contrast to the phage RNAPs (14, 25, 37, 61), Pol III (54), archaeal RNAP (Fig. 4), eukaryotic Pol II (10, 13, 22, 24), and E. coli bacterial RNAP (60) cannot extend a promoter-initiated transcript through a nucleosome in vitro under physiological salt conditions. They can do so if the salt concentration is increased (60, 61) or, in the case of Pol II, if transcription is end initiated rather than promoter initiated (27). The basis for this higher sensitivity to a nucleosome barrier is unknown, but it has been suggested that this difference between Pol II and Pol III has evolved to provide an additional opportunity for Pol II-specific regulation (24, 27). Transcription elongation factors, such as SWI/SNF and FACT (10, 23, 36, 38, 56, 65), can then be used to regulate gene expression by facilitating Pol II transcription through a nucleosome. Consistent with this evolutionary scenario, archaeal genomes do not encode relatives of the Pol II elongation factors (26, 43), but this suggestion does not address the inability of E. coli RNAP to transcribe through a nucleosome (60).
Rate of archaeal transcription in vitro.
M. thermautotrophicus RNAP elongated transcripts in vitro at a rate of ∼20 nt/s at 58°C (Fig. 4). This is the first report of the rate of transcription by an archaeal RNAP, and although the comparisons are very tenuous given the differences in assays, templates, and temperatures used, this is consistent with the rates reported for transcription in vitro by other multisubunit RNAPs. Transcription rates in vitro appear to correlate inversely with RNAP complexity (59), with single-subunit phage enzymes incorporating nucleotides in vitro at rates of 200 to 400 nt/s whereas the rates reported for multisubunit bacterial and eukaryotic RNAPs range from 10 to 35 nt/s (1, 19, 34, 59).
Archaeal nucleosome positioning.
Positioned nucleosomes regulate the expression of many eukaryotic genes, and from the sequences of DNA molecules that localize eukaryotic nucleosome assembly (28, 57, 63), nucleosome positioning rules have been established (28). Consistent with archaeal and eukaryotic nucleosomes having structural homology, the DNA molecules SELEX selected from a random population based on their preferential binding by the archaeal histone HMfB also have sequences that conform to these rules (4). As a representative, the SELEX-selected molecule that contained the 60-bp Selex1 sequence was shown to have high affinity for HMfB (apparent Kd of 20 nM) and to direct HMfB assembly into an archaeal nucleosome with preferred rotational and translational positioning (5). The portability and the extension of these properties to other archaeal histones were not however known. As shown in Fig. 1, the 60-bp Selex1 sequence also directed the positioned assembly of an archaeal nucleosome when flanked by sequences entirely different from those in the original SELEX-selected molecule, and Selex1 also positioned the assembly of HMtA2, an archaeal histone from a different archaeon with a sequence ∼73% identical to that of HMfB (45, 49).
Regulation of gene expression by archaeal nucleosome assembly.
Different histones from the same archaeon have different affinities for the same DNA sequence (5, 33) and are synthesized differentially depending on growth conditions (18, 47). Given these observations and that a relatively short DNA sequence can precisely position archaeal nucleosome assembly (Fig. 1), archaeal nucleosome positioning could be used in vivo to regulate gene expression, replication, and/or recombination. Consistent with this, electron microscopy of archaeal genomic DNAs has revealed that archaeal nucleosomes are not as regularly packed as nucleosomes in eukaryotic chromatin but rather are interspersed between histone-free regions (41, 48). Transcription regulation could then be based on a binding competition between archaeal histones and transcription factors. Most archaeal transcription regulators identified to date do, in fact, function by competing with TBP and TFB for binding to the TATA-BRE region or with RNAP for the site of transcription initiation (9, 45). In this regard, when HMtA2 assembly incorporated the upstream regulatory region (Fig. 1), transcription in vitro was inhibited (Fig. 2). The almost universal presence of histones in eukaryotes has led to the argument that it was the evolution of the histone fold-based mechanism of DNA compaction that facilitated genome expansion and eukaryote evolution (35). Accommodating much larger genomes within the confines of a nucleus may however also have required that essentially all the genomic DNA was bound by histones. Under such conditions, regulation based on a simple competition between histone and transcription factor would have been compromised, and histone tails and histone-modifying and chromatin remodeling complexes may then have been needed to access and regulate the expression of histone-bound DNA. As eukaryotic genome sequences accumulate, it may become possible to identify when the sophistication of eukaryotic chromatin expression arose. From the archaeal genome sequences now available, we can conclude that it was after the divergence of the archaeal and eukaryotic lineages (32, 45).
Acknowledgments
We thank K. Sandman for the construction and gift of plasmid pKS564 and D. Soares for the HMfB K13T+R19S+T54K variant.
This research was supported by grants from the Department of Energy (DE-FGO2-87ER13731) and the National Institutes of Health (GM53185).
REFERENCES
- 1.Adelman, K., A. La Porta, T. J. Santangelo, J. T. Lis, J. W. Roberts, and M. D. Wang. 2002. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc. Natl. Acad. Sci. USA 99:13538-13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alilat, M., A. Sivolob, B. Révet, and A. Prunell. 1999. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J. Mol. Biol. 291:815-841. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, K. A., C. S. Chow, and J. N. Reeve. 1999. Histone stoichiometry and DNA circularization in archaeal nucleosomes. Nucleic Acids Res. 27:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, K. A., S. L. Pereira, J. Widom, and J. N. Reeve. 2000. Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution. J. Mol. Biol. 303:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, K. A., F. Marc, K. Sandman, and J. N. Reeve. 2002. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J. Biol. Chem. 277:9293-9301. [DOI] [PubMed] [Google Scholar]
- 6.Bednar, J., V. M. Studitsky, S. A. Grigoryev, G. Felsenfeld, and C. L. Woodcock. 1999. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol. Cell 4:377-386. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. D., A. B. Brinkman, J. van der Oost, and S. P. Jackson. 2001. The archaeal TFIIE alpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. [DOI] [PubMed] [Google Scholar]
- 10.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studisky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1096. [DOI] [PubMed] [Google Scholar]
- 11.Berger, S. L. 2002. Histone modifications in transcription regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 12.Caetano-Anollés, G., and C. Caetano-Anollés. 2003. An evolutionarily structured universe of protein architecture. Genome Res. 13:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C.-H., and D. S. Luse. 1997. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 272:23427-23434. [DOI] [PubMed] [Google Scholar]
- 14.Chirinos, M., F. Hernández, and E. Palacián. 1999. Transcription of DNA templates associated with histone (H3 + H4)2 tetramers. Arch. Biochem. Biophys. 370:222-230. [DOI] [PubMed] [Google Scholar]
- 15.Cramer, P. 2002. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 12:89-97. [DOI] [PubMed] [Google Scholar]
- 16.Darcy, T. J., W. Hausner, D. E. Awery, A. M. Edwards, M. Thomm, and J. N. Reeve. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181:4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decanniere, K., A. M. Babu, K. Sandman, J. N. Reeve, and H. Heinemann. 2000. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 303:35-47. [DOI] [PubMed] [Google Scholar]
- 18.Dinger, M. E., G. J. Baillie, and D. R. Musgrave. 2000. Growth phase- dependent expression and degradation of histones in the thermophilic archaeon Thermococcus zilligii. Mol. Microbiol. 36:876-885. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, A. M. C. M. Kane, R. A. Young, and R. D. Kornberg. 1991. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266:71-75. [PubMed] [Google Scholar]
- 20.Hanzelka, B. L., T. J. Darcy, and J. N. Reeve. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausner, W., U. Lange, and M. Musfeld. 2000. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 275:12393-12399. [DOI] [PubMed] [Google Scholar]
- 22.Izban, M. G., and D. S. Luse. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates J. Biol. Chem. 267:13647-13655. [PubMed] [Google Scholar]
- 23.Kim, J.-H., W. S. Lane, and D. Reinberg. 2002. Human elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99:1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 25.Kirov, N., I. Tsaneva, E. Einbinder, and R. Tsanev. 1992. In vitro transcription through nucleosomes by T7 RNA polymerase. EMBO J. 11:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krypides, N. C., and C. A. Ouzounis. 1999. Transcription in Archaea. Proc. Natl. Acad. Sci. USA 96:8545-8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y. V., D. J. Clark, V. Tchernajenko, M. E. Dahmus, and V. M. Studitsky. 2003. Role of C-terminal domain phosphorylation in RNA polymerase II transcription through the nucleosome. Biopolymers 68:528-538. [DOI] [PubMed] [Google Scholar]
- 28.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 29.Luger, K., A. M. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 30.Luger, K, T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 31.Luger, K., and T. J. Richmond. 1998. DNA binding within the nucleosome. Curr. Opin. Struct. Biol. 8:33-40. [DOI] [PubMed] [Google Scholar]
- 32.Malik, H. S., and S. Henikoff. 2003. Phylogenetics of the nucleosome. Nat. Struct. Biol. 10:882-891. [DOI] [PubMed] [Google Scholar]
- 33.Marc, F., K. Sandman, R. Lurz, and J. N. Reeve. 2002. Archaeal histone tetramer formation determines DNA affinity, and the direction of DNA supercoiling. J. Biol. Chem. 277:30879-30886. [DOI] [PubMed] [Google Scholar]
- 34.Matsuzaki, H., G. A. Kassavetis, and E. P. Geiduschek. 1994. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 235:1173-1192. [DOI] [PubMed] [Google Scholar]
- 35.Minsky, A., R. Ghirlando, and Z. Reich. 1997. Nucleosomes: a solution to a crowded intracellular environment? J. Theor. Biol. 188:379-385. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar, G. J., H.-Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill, T. E., J. G. Smith, and E. M. Bradbury. 1993. Histone octamer dissociation is not required for transcript elongation through arrays of nucleosome cores by T7 RNA polymerase in vitro. Proc. Natl. Acad. Sci. USA 90:6203-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 39.Peränen, J., M. Rikkonen, M. Hyvönen, and L. Kääriäinen. 1996. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236:371-373. [DOI] [PubMed] [Google Scholar]
- 40.Pereira, S. L. 1997. An in vivo study of the archaeal histone HMf from the hyperthermophilic methanogen, Methanothermus fervidus. Ph.D. thesis. The Ohio State University, Columbus.
- 41.Pereira, S. L., R. A. Grayling, R. Lurz, and J. N. Reeve. 1997. Archaeal nucleosomes. Proc. Natl. Acad. Sci. USA 94:12633-12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 43.Ponting, C. P. 2002. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30:3643-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Protacio, R. U., G. Li, P. T. Lowary, and J. Widom. 2000. Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol. Cell. Biol. 20:8866-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeve, J. N. 2003. Archaeal chromatin and transcription. Mol. Microbiol. 48:587-598. [DOI] [PubMed] [Google Scholar]
- 46.Sandman, K., K. Bailey, S. L. Pereira, D. Soares, W.-T. Li, and J. N. Reeve. 2001. Archaeal histones and nucleosomes. Methods Enzymol. 334:116-129. [DOI] [PubMed] [Google Scholar]
- 47.Sandman, K., R. A. Grayling, B. Dobrinski, R. Lurz, and J. N. Reeve. 1994. Growth phase dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc. Natl. Acad. Sci. USA 91:12624-12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shioda, M., K. Sugimori, T. Shiroya, and S. Takayanagi. 1989. Nucleosome-like structures associated with chromosomes of the archaebacterium Halobacterium salinarium. J. Bacteriol. 171:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, D. R., L. A. Doucette-Stamm, C. DeLoughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. The complete genome sequence of Methanobacterium thermoautotrophicum strain ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soares, D., I. Dahlke, W.-T. Li, K. Sandman, C. Hethke, M. Thomm, and J. N. Reeve. 1998. Archaeal histone stability, DNA binding, and transcription inhibition above 90°C. Extremophiles 2:75-81. [DOI] [PubMed] [Google Scholar]
- 51.Soares, D. J., K. Sandman, and J. N. Reeve. 2000. Mutational analysis of archaeal histone-DNA interactions. J. Mol. Biol. 297:39-47. [DOI] [PubMed] [Google Scholar]
- 52.Spitalny, P., and M. Thomm. 2003. Analysis of the open region and of DNA- protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 278:30497-30505. [DOI] [PubMed] [Google Scholar]
- 53.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 405:41-45. [DOI] [PubMed] [Google Scholar]
- 54.Studitsky, V. M., G. A. Kassavetis, E. P. Geiduschek, and G. Felsenfeld. 1997. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science 278:1960-1963. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan, S., D. W. Sink, K. L. Trout, I. Makalowska, P. M. Taylor, A. D. Baxevanis, and D. Landsman. 2002. The histone database. Nucleic Acids. Res. 30:341-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svejstrup, J. Q. 2002. Chromatin elongation factors. Curr. Opin. Genet. Dev. 12:156-161. [DOI] [PubMed] [Google Scholar]
- 57.Thåström, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non- natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 58.Tomschik, M., M. A. Karymov, J. Zlatanova, and S. H. Leuba. 2001. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure 9:1201-1211. [DOI] [PubMed] [Google Scholar]
- 59.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 60.Walter, W., M. L. Kireeva, V. M. Studitsky, and M. Kashlev. 2003. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J. Biol. Chem. 278:36148-36156. [DOI] [PubMed] [Google Scholar]
- 61.Walter, W., and V. M. Studitsky. 2001. Facilitated transcription through the nucleosome at high ionic strength occurs via a histone octamer transfer mechanism. J. Biol. Chem. 276:29104-29110. [DOI] [PubMed] [Google Scholar]
- 62.Werner, F., and R. O. J. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10:635-646. [DOI] [PubMed] [Google Scholar]
- 63.Widlund, H. R., H. Cao, S. Simonsson, E. Magnusson, T. Simonsson, P. E. Nielsen, J. D. Kahn, D. M. Crothers, and M. Kubista. 1997. Identification and characterization of genomic nucleosome-positioning sequences. J. Mol. Biol. 267:807-817. [DOI] [PubMed] [Google Scholar]
- 64.Winkler, G. S., A. Kristjuhan, H. Erdjument-Bromage, P. Tempst, and J. Q. Svetjstrup. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99:3517-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]