Abstract
We report here the transcriptome analyses of highly expressed genes that are subject to catabolite repression or activation mediated by the cyclic AMP receptor protein (Crp). The results reveal that many operons encoding enzymes of central carbon metabolic pathways (e.g., Krebs cycle enzymes), as well as transporters and enzymes that initiate carbon metabolism, are subject to direct Crp-mediated catabolite repression. By contrast, few enzyme-encoding genes (direct regulation) but many ribosomal protein- and tRNA-encoding genes (indirect regulation) are subject to Crp-dependent glucose activation. Additionally, Crp mediates strong indirect catabolite repression of many cytoplasmic stress response proteins, including the major chaperone proteins, five ATP-dependent protease complexes, and several cold and heat shock proteins. These results were confirmed by (i) phenotypic analyses, (ii) real-time PCR studies, (iii) reporter gene fusion assays, and (iv) previously published reports about representative genes. The results serve to define and extend our appreciation of the Crp regulon.
A dominant mechanism by which Escherichia coli and other related bacteria sense carbon sufficiency involves cyclic AMP and its receptor protein, Crp (15, 35). The mechanisms by which Crp regulates gene expression in response to variable cytoplasmic levels of cyclic AMP have been extensively investigated with primary emphasis on E. coli and Salmonella strains (5, 34, 35, 41). Dozens of operons have been shown to be subject to Crp-mediated control by using classical approaches (17). Transcriptome and proteome approaches have been used to study the control of various regulons, as well as to facilitate glucose flux analyses (10, 39). Indeed, the transcriptome approach has allowed investigators to challenge established paradigms, and the technology has enjoyed rapid adoption among researchers (7). However, no report has focused on genomewide analyses of Crp-mediated catabolite regulation in E. coli. In this study, we corrected this deficiency by conducting combined transcriptome, phenotypic, and bioinformatic analyses of the Crp-mediated responses of E. coli to exogenous glucose availability. We also tabulated comparative data derived from the classical literature and confirmed representative regulatory responses by using alternative approaches. We show here that a variety of stress-related genes, encoding chaperones (1, 4, 38), ATP-dependent proteases (9, 11, 20), and certain temperature shock proteins (12, 31, 40), are regulated in response to the presence of Crp, apparently by an indirect mechanism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study were BW25113 and an isogenic crp mutant derivative, LJ3017, constructed as described by Zhang et al. (43). Strains were grown at 37°C with agitation at 250 rpm in Luria-Bertani (LB) broth containing 50 mM potassium phosphate, pH 7.4, and 0.2 mM l-cysteine with or without 0.4% glucose. Cells were grown in 25 ml of medium in 250-ml shake flasks starting at an optical density at 600 nm (OD600) of 0.05. Each determination was performed three times, and the results were averaged (6).
Cell harvesting and preparation of RNA.
Cells from each triplicate experiment were harvested in the exponential growth phase when cultures reached an OD600 of 0.5. The total contents of each shake flask (25 ml) were poured into a Millipore vacuum filtration apparatus (catalog no. 1004700) with a Millipore 0.8-μm-pore-size filter (catalog no. AAWPO4700). The collected cells with filter were immediately transferred to a 200-ml glass beaker containing liquid nitrogen. RNA was extracted from each sample as described by Caldwell et al. (6).
Target preparation.
RNA harvested from a given E. coli strain and at a given time point was reverse transcribed into biotin-labeled cDNA by the method of de Saizieu et al. (8). Total RNA (18 μg) was incubated at 37°C overnight in an 80-μl reaction mixture consisting of 1× GIBCO first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2); 10 mM dithiothreitol; 40 μM random hexamer; 0.3 mM concentrations (each) of dCTP, dGTP, and dTTP; 0.12 mM dATP; 0.3 mM biotin-dATP (NEN catalog no. NEL999); and 1,800 U of SuperScript II reverse transcriptase. To remove RNA, the reaction mixture was brought to 0.25 M NaOH and incubated at 65°C for 30 min. The reaction mixture was neutralized with HCl, and the nucleic acid was precipitated at −20°C in ethanol with 2.5 M ammonium acetate and 20 μg of glycogen. The pellet was washed, air dried, resuspended in water, and quantitated by UV spectroscopy. The yield was approximately 10 to 12 μg of biotin-labeled cDNA. This cDNA (10 μg) was fragmented in 33 μl of 1× One-Phor-All buffer (Amersham-Pharmacia no. 27-0901-02) with 3.75 mU of DNase I at 37°C for 10 min. After the DNase had been heat killed, fragmentation was validated by running 1.5 μg of the fragmented cDNA on a 1.2% agarose gel. Biotin-containing cDNA routinely ranged in size from 25 to 400 nucleotides. The remaining 8 μg of cDNA was hybridized to an Affymetrix (Santa Clara, Calif.) E. coli GeneChip array.
Array description.
The Affymetrix E. coli array has been described in detail by Selinger et al. (37). Briefly, each microarray contains 295,936 25-mer oligonucleotide probes. Half of the probes are a perfect match to the corresponding E. coli chromosomal sequences, while the other half have a single mismatch at the 13th base position. The array includes 4,327 genes and intergenic regions (2). The complete set of averaged data is provided at our website (www-biology.ucsd.edu/∼msaier/supmat; see Table S1).
Hybridization, scanning, and data collection.
The hybridization, scanning, and data collection procedures described by Caldwell et al. (6) were followed.
RT-PCR studies.
Bacterial cells were cultured in LB broth with or without 0.4% glucose exactly as for the microarray analyses. When the OD600 reached 0.5, cells were collected by centrifugation at 4°C. Total RNA was subsequently isolated with the RNeasy Mini Kit (Qiagen) in accordance with the manufacturer's protocol. Residual DNA present in the RNA preparations was removed by RNase-free DNase (Stratagene). cDNAs were synthesized with the superscript first-strand synthesis kit (Invitrogen) in accordance with the manufacturer's instructions and stored at −20°C prior to use. Real-time PCR (RT-PCR) was carried out on the LightCycler instrument (Roche Diagnosis Corporation) in accordance with the manufacturer's recommended protocol. Primers used for the RT-PCR were as follows: for aceE, GAA GAA GGT GTT GAG CGT GC and TTG CGG AAGA CTG GAA GGAC; for clpB, CGAC ATC CTG AAA GCA GCAG and CAG ACC TTC AAC GAT GGCAG; for ibpA (hslT), CG CTT TAC CGT TCT GCT ATT GG and TGC GTT CAA AGT TGC GTT CAG.
Data analysis.
Array images were analyzed with Affymetrix Microarray Suite 5.0 software to determine raw expression intensity values, which then were scaled and normalized to a target value of 1,000. These data were imported into a Microsoft Excel spreadsheet for further analysis. The intensity values in the data sets ranged from about 3 × 10−1 to 8 × 104. Three data sets were obtained for each of the following experimental conditions: wild type in LB, wild type in LB plus glucose, crp mutant in LB, and crp mutant in LB plus glucose. The data sets for each strain and condition were compared pairwise to determine the Pearson correlation coefficient with the formula r = [n(ΣXY) − (ΣX)(ΣY)/{[nΣX2 − (ΣX)2][nΣY2 − (ΣY)2]}1/2. For each triplicate data set, the two sets with the highest Pearson correlation coefficient were retained for further analysis. The statistical significance of fold changes between data sets of different strains and conditions was determined as described by Caldwell et al. (6) and also in Results. Briefly, for each pair of replicate data sets, a table was generated from the average of the logs of the two replicate signals (ALS) and their log ratio (LR). From this table, with a series of sliding windows with a size of 201 ALS values, the mean ALS values and the standard deviation values of the corresponding LR were calculated (SDLR).
RESULTS
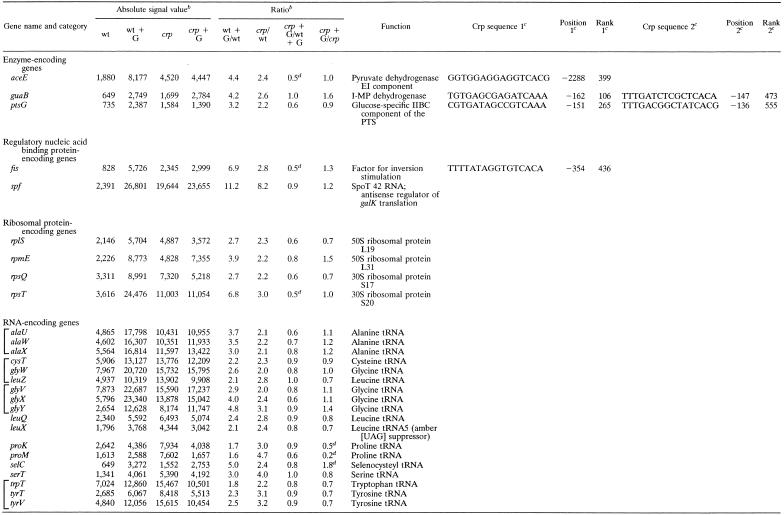
Glucose-activated Crp-dependent genes. Table 1 summarizes the genes that gave (i) a statistically significant change (the fold change was at least 4.37 times higher than the SDLR for the ALS of the gene on the wild-type and crp strains), (ii) consistently large signals (signal intensity, ≥1,000), (iii) a crp/wild-type ratio of ≥2.0 following growth in LB broth, and (iv) activation by glucose in a Crp-dependent fashion. Very few enzyme-encoding genes met these criteria, although several genes encoding ribosomal proteins and tRNAs did. Enzyme-encoding genes showing Crp-mediated glucose activation were the aceE gene, encoding the pyruvate dehydrogenase E1 component; the guaB gene, encoding inosine 5′ monophosphate dehydrogenase; and the ptsG gene, encoding the glucose-specific IIBC permease of the phosphotransferase system (PTS). These genes may be subject to repression by the cyclic AMP-Crp complex, although ptsG is subject to glucose activation by a distinct Mlc-dependent mechanism (16, 28, 29). A few other enzyme-encoding genes exhibited Crp-mediated glucose activation but did not satisfy all of the criteria cited above (see Table S1 at our website). These genes included, for example, aceF, encoding the pyruvate dehydrogenase E2 component; accC, encoding acetyl coenzyme A (CoA) carboxylase; ack, encoding acetate kinase; and adk, encoding adenylate kinase. It is surprising that so few highly expressed enzyme-encoding genes are directly activated by the cyclic AMP-Crp complex compared to enzyme-encoding genes that are subject to repression (see below).
TABLE 1.
Genes subject to Crp-dependent glucose activationa
Genes tabulated gave a high level of reproducibility for replicate values, large signals, and crp/wt ratios greater than 2.0. Brackets indicate genes included within a single operon or regulon.
Abbreviations: wt, wild type; crp, the isogenic crp-null mutant; G, grown in LB medium plus glucose. When G is not indicated, cells were grown in LB medium.
Crp binding sites were identified by using the GRASP-DNA program (36). The position relative to the binding site of the translational start site of the gene and the rank of that site relative to all other sites in the genome are provided as detailed by Schilling et al. (36).
Crp-independent glucose repression (proK and proM) and activation (selC) are presumably mediated by another transcription factor. The two-fold decrease observed for three genes, aceE, fis, and rpsT, when the crp mutant is compared with the wild type in the presence of glucose could be due to Crp-mediated activation of gene expression in the presence of glucose in the wild-type strain.
Two other genes, fis, encoding the Fis (factor for inversion stimulation) protein, a basic, histone-like, chromatin-binding protein, and the spf gene, encoding an antisense regulator of galK translation, were subject to strong Crp-dependent glucose activation. Crp-binding sites could be found in or near most of the enzyme-encoding and fis operons. On this basis, we assume that these genes are directly regulated by Crp.
Many genes concerned with transcription and translation proved to be subject to Crp-dependent glucose activation. These genes encode tRNAs, ribosomal proteins, elongation factors, and RNA polymerase subunits (see Table S1 at our E. coli transcriptome website). Those satisfying the criteria cited above are listed in Table 1. We could not find Crp-binding sites in or near these genes or operons, suggesting that they are indirectly regulated, possibly in response to changes in growth rate due to the presence or absence of Crp and/or glucose availability (14). This suggestion is in agreement with previous reports on select ribosomal protein and tRNA-encoding genes (14).
Glucose-repressed Crp-dependent genes.
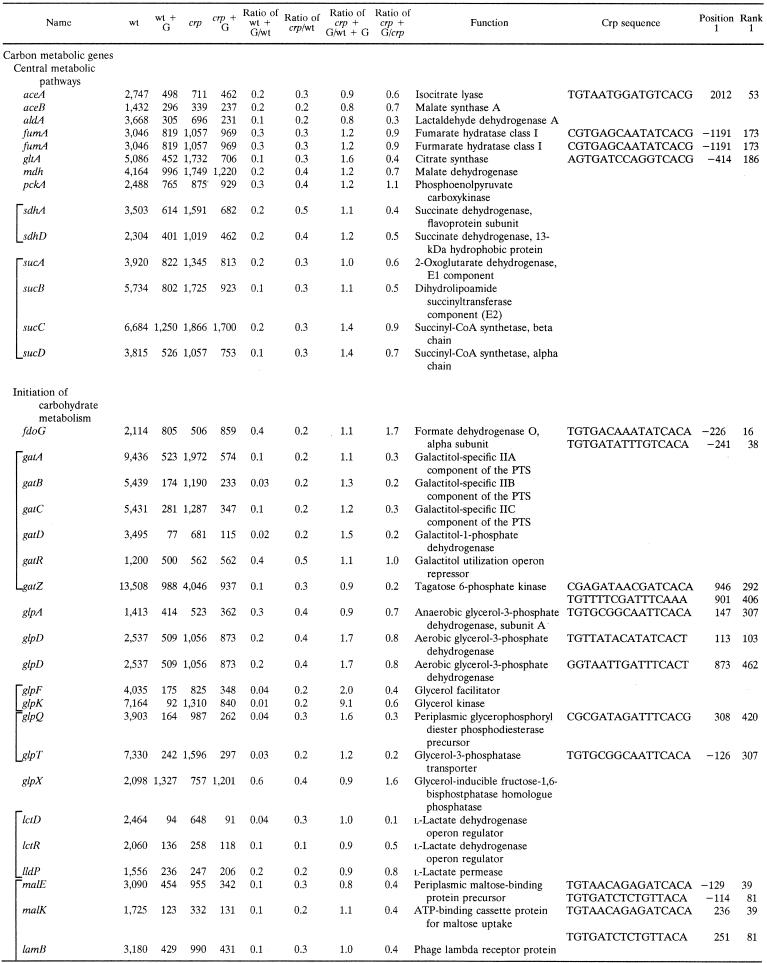
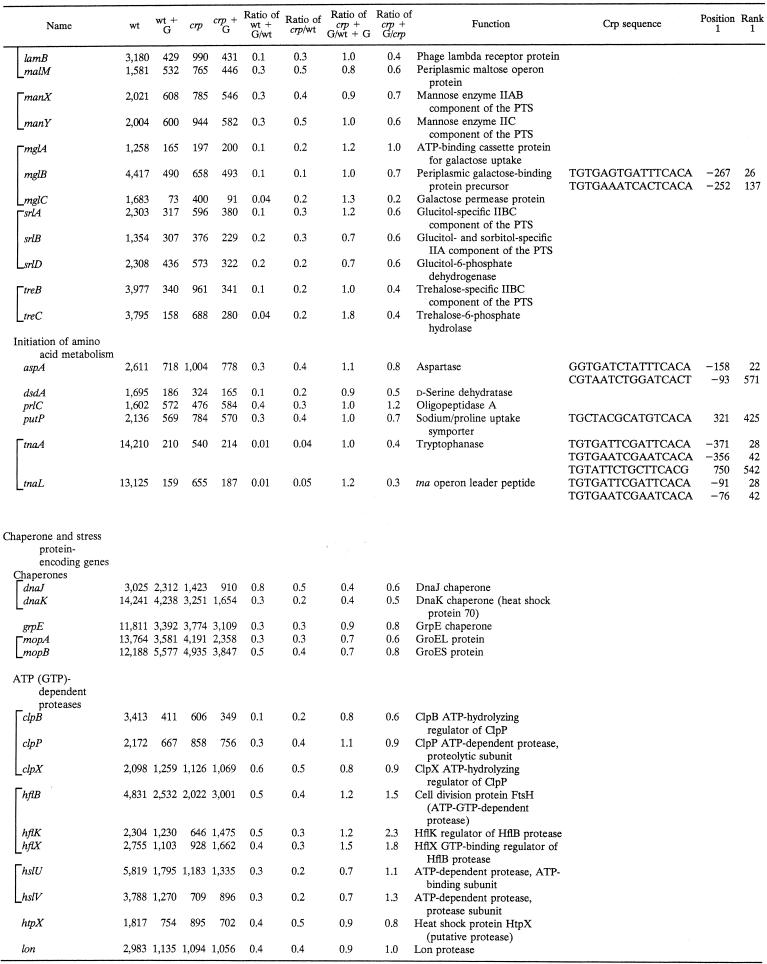
Table 2 tabulates genes subject to Crp-dependent glucose repression identified by the same criteria cited above for Table 1, except that the crp/wild-type strain signal ratio had to be ≤0.5. The majority of these genes encode enzymes and transporters involved in carbon catabolism. These proteins include (i) enzymes of central carbon metabolism (e.g., the Krebs cycle), (ii) transporters and enzymes that initiate the metabolism of an exogenous carbon compound (e.g., galactitol), and (iii) transporters and enzymes that initiate the metabolism of an exogenous amino acid or peptide that can be used as a primary source of both carbon and nitrogen (e.g., aspartate). The second largest class encodes stress response proteins primarily concerned with protein folding and degradation. Thus, 51 of the 72 genes listed in Table 2 are carbon catabolic genes while 18 of these genes are stress response genes. Only three genes fell into the “miscellaneous” category. For all of these genes, the crp mutation eliminated or decreased the magnitude of catabolite repression. Residual glucose repression in the crp mutant strain is presumably due to effects of other transcription factors.
TABLE 2.
Genes subject to CRP-dependent glucose repressiona
Brackets indicate genes included within a single operon or regulon. Other conventions are as described in the footnotes to Table 1.
The prominent carbon catabolic genes include several genes encoding enzymes in central pathways of carbon catabolism. These enzymes include (i) the glyoxalate shunt enzymes (AceA and AceB); (ii) lactaldehyde dehydrogenase A (AldA); (iii) most of the Krebs cycle enzymes, including fumarase (FumA), citrate synthase (GltA), malate dehydrogenase (Mdh), succinate dehydrogenase (Sdh), 2-oxoglutarate dehydrogenase (SucAB), and succinyl-CoA synthetase (SucCD); and (iv) the central gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PckA). However, nearly half of the genes exhibiting Crp-dependent catabolite repression are concerned with the initiation of carbon metabolism. These genes include the enzymes catalyzing catabolism of formate (Fdo), galactitol (Gat), glycerol and α-glycerophosphate (Glp) lactate (Lct and Lld), maltose (Mal), mannose (Man), galactose (Mgl), glucitol (or sorbitol) (Srl), and trehalose (Tre). Four of these carbohydrates, galactitol, mannose, glucitol, and trehalose, are substrates of the E. coli PTS, while six of these compounds, formate, glycerol, α-glycerol phosphate, lactate, maltose, and galactose, are non-PTS carbon and energy sources. Many of these genes (i.e., gat, glp, mgl, lld, mal, and tre) showed residual glucose repression in the crp mutant. In several of these cases, the fruR mutation by itself diminished the repressive effect of glucose while the crp fruR double mutant exhibited full resistance to glucose repression. The genes that most clearly showed dual control of catabolite repression by Crp and Cra were gat, mgl, tre, and dsd (data not shown). These observations suggest that in several cases, catabolite repression depends on both Crp and Cra (32, 33). The fact that many genes show residual glucose repression in the crp mutant is not surprising since multiple mechanisms of catabolite repression are recognized in E. coli (35).
Crp also regulates genes encoding enzymes, permeases, and regulators that initiate the catabolism of amino acids as both carbon and nitrogen sources. Representative genes encode (i) aspartase (AspA), (ii) d-serine dehydratase (DsdA), (iii) the primary proline uptake permease (PutP), (iv) tryptophanase (TnaA), and (v) the tna operon leader peptide (TnaL). One gene encoding an oligopeptidase (PrlC) is also listed. Many additional carbon metabolic genes not meeting our rigorous criteria for inclusion in Table 2 were similarly found to be subject to Crp-dependent catabolite repression, as expected on the basis of published results, as well as our phenotypic data (see below; see also Table S1 at our website).
Several additional interesting enzyme- or transporter-encoding genes proved to be subject to strong glucose repression mediated at least partially by Crp (see our website). For example, the nmpC and tsx outer membrane porin-encoding genes fell into this category. Both exhibited only partial relief from catabolite repression in the crp mutant, but a crp-fruR double mutant (32, 33) showed no glucose repression (unpublished results). The same behavior was observed for the dadA and dadX genes encoding d-amino acid dehydrogenase and alanine racemase, respectively, as well as the genes encoding the dicarboxylate transporters DctA and DcuA. Crp-binding sites could be identified in the control regions of some of these genes but not others. It therefore appears that Crp and the fruR gene product, Cra, cooperate in the regulation of many operons in E. coli.
Phenotypic analyses.
We conducted phenotypic analyses, measuring the oxidation of various carbon and nitrogen sources with 96-well Biolog microtiter plates (3). For carbon sources of interest that were not included in the Biolog plates, or where the responses were ambiguous, we conducted studies of growth on minimal agar plates containing the carbon source at 0.2%. Table 3 summarizes some of the results. For a more complete compilation of the phenotypic data for carbon and nitrogen sources found to be under Crp control, see our website (Table S2).
TABLE 3.
Oxidation or utilization of various carbon sources by isogenic wild-type and crp mutant strains of E. coli
| Carbon source | Strain
|
|
|---|---|---|
| wta | crp | |
| Acetate | − | |
| Fumarate | + | − |
| d,l-Malate | + | − |
| Succinate | + | − |
| Formateb | + | − |
| Galactitolb | + | − |
| d-α-Ketoglutarate | + | − |
| d,l-Lactate | + | − |
| Glycerol | + | − |
| d,l-α-Glycerol phosphate | + | − |
| Maltose | + | − |
| Maltotriose | + | − |
| Mannoseb | + | ± |
| Galactoseb | + | ± |
| d-Glucitol | + | − |
| Trenaloseb | + | ± |
| l-Aspartate | + | − |
| Glycyl-l-aspartate | + | − |
| Glycyl-l-glutamate | + | − |
| l-Proline | ± | − |
| Tryptophanb | + | − |
| d-Serine | + | ± |
wt, wild type.
Compound studied by substrate utilization on minimal agar plates. All others were determined with Biolog 96-well plates, which measure substrate oxidation.
Almost all of the carbon sources tested (e.g., acetate, fumarate, d- and l-malate, succinate, α-ketoglutarate, lactate, glycerol, d,l-α-glycerol phosphate, maltose, maltotriose, and glucitol) gave positive wild-type responses but negative crp mutant responses. Of the potential nitrogen sources, l-aspartate, several peptides, and proline also gave positive wild-type responses but negative crp mutant responses. d-Serine showed a decreased but appreciable response in the mutant relative to that in the wild type. Oxidation of galactitol, galactose, mannose, trehalose, and tryptophan did not show differences between the wild type and the crp mutant on the Biolog plates, but when these compounds were assayed for growth on minimal plates, the wild type, but not the crp mutant, proved to use galactitol, trehalose, and tryptophan while both strains utilized galactose and mannose, with the mutant growing less well than the wild type. Thus, the transcriptome data are in agreement with the phenotypic data in all of the cases tested.
Correlation between transcriptome and classical approaches.
Table 4 summarizes the results of our transcriptome analyses and compares them with corresponding published data obtained by classical approaches. In most cases, the fold effects of glucose on gene expression in a wild-type background were examined. In every such case, the direction of the effect (repression or activation) was the same. Similar agreement has been reported for transcriptome analyses versus traditional methods conducted with Bacillus subtilis (21). Quantitative differences may be within the range of experimental error considering that in all cases different strains were used and that in several cases different media were used. The different genetic backgrounds could have contributed to the observed differences. However, for all but one of the glucose-repressed genes, the fold repression by glucose was greater when assayed by microarray technology than when lacZ reporter gene fusions were used. The lacZ fusion technology may introduce artifacts affecting gene expression, as discussed previously (27).
TABLE 4.
Correlation between the transcriptome results of this study and gene expression analyses reported in the literature
| Method of assay | Condition | Reference | Fold change | Array result |
|---|---|---|---|---|
| ptsG-lacZ | Minimal medium + glucose/minimal medium + glycerol | 30 | 7.5 | 3.2 |
| fis (Northern blotting) | crp/wt | 22 | 2.0 | 2.8 |
| sucA-lacZ | LB + glucose/LB | 23 | 0.6 | 0.2 |
| mdh-lacZ | LB + glucose/LB | 24 | 0.6 | 0.2 |
| fumA-lacZ | LB + glucose/LB | 25 | 0.2 | 0.3 |
| gltA-lacZ | LB + glucose/LB | 26 | 0.4 | 0.1 |
| tnaA (tryptophanase sp act) | Minimal medium + tryptophan + glucose/minimal medium + tryptophan | 13 | 0.2 | 0.01 |
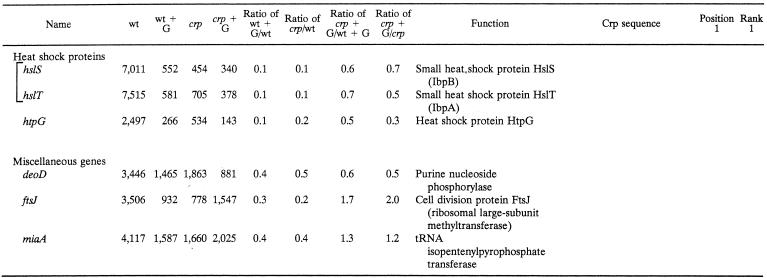
Crp-mediated control of stress response genes.
The stress response proteins whose synthesis proved to be under Crp control include the major chaperone proteins DnaJK, GrpE, and GroEL/ES (MopAB) (Table 2). Five cytoplasmic ATP-dependent proteases or protease complexes (ClpPBX, HflBKX, HslUV, HtpX, and Lon) also proved to be subject to strong Crp-mediated catabolite repression. In the case of the hfl genes, the crp mutation reversed glucose repression (Table 2) while the fruR mutation abolished it (data not shown). Finally, three biochemically ill-defined heat shock genes (hstS, hstT, and htpG) are listed in Table 2. We also identified putative stationary-phase- or carbon starvation-inducible genes (e.g., cspD, and yjiY) that are subject to strong Crp-dependent glucose repression (42). Crp-binding sites could be found upstream of both the cspD and yjiY genes. It is therefore clear that many stress response genes in E. coli are subject to Crp-dependent catabolite repression, in agreement with the fact that crp mutants are sensitive to certain types of stress and starvation conditions (18, 19).
From a mechanistic standpoint, it is important to note that we were not able to find Crp-binding sites in the control regions of most of these stress genes and operons. The regulatory consequences of inclusion of glucose in the medium and of the loss of the Crp protein are therefore presumed to be indirect in most cases. Sigma-32 is known to be under Crp control, but its contribution to the rates of gene transcription should be minimal under the conditions used in our studies (19). Of the three miscellaneous genes (Table 2), one is a purine nucleoside phosphorylase while two are involved in translational regulation.
Confirmation by RT-PCR.
Three of the genes exhibiting Crp-dependent glucose regulation were selected for confirmation by RT-PCR. Table 5 summarizes the results and compares them with the transcriptome data reported in Tables 1 and 2. The three genes analyzed are the aceE gene, which is subject to catabolite activation (Table 1), and the stress response genes clpB and hslT/ibpA (Table 2). Excellent agreement between the two methods is apparent. The only minor discrepancies are observed for the clpB and hslT/ibpA genes after growth of the crp mutant in LB plus glucose. In these two instances, the differences between the two methods are only twofold. Since the repressive effect of glucose was about 10-fold in both cases, this difference may be within the range of experimental error.
TABLE 5.
Comparison of RT-PCR and microarray analyses of gene expression in response to glucose and a crp mutationa
| Gene | RT-PCR
|
Microarray
|
||||||
|---|---|---|---|---|---|---|---|---|
| (wt + G)/wt | crp/wt | crp + G/wt + G | crp + G/crp | (wt + G)/wt | crp/wt | crp + G/wt + G | crp + G/crp | |
| aceE | 2.5 | 1.6 | 0.6 | 0.9 | 4.4 | 2.4 | 0.5 | 1.0 |
| clpB | 0.1 | 0.2 | 1.8 | 0.8 | 0.1 | 0.2 | 0.8 | 0.6 |
| hslT/ibpA | 0.1 | 0.1 | 1.5 | 0.7 | 0.1 | 0.1 | 0.7 | 0.5 |
The values shown are signal ratios. wt, wild type; G, growth in LB medium plus glucose.
DISCUSSION
In this paper, we have provided a comprehensive analysis of Crp-mediated catabolite control in E. coli. The transcriptome data (Tables 1 and 2, as well as Table S1 at our website) were confirmed and extended by (i) citing published data for representative genes (Table 4), (ii) conducting phenotypic analyses that reflect gene expression control (Tables 3 and S2), and (iii) conducting RT-PCR experiments (Table 5). The agreement between the different methods was striking, allowing us to conclude that the transcriptome data are reliable.
Far more highly expressed genes proved to be subject to direct catabolite repression (Table 2) than catabolite activation (Table 1) (see also Table S1 at our website). Most of the carbon metabolic genes subject to catabolite control showed responses to glucose and the loss of Crp as expected. Thus, many genes that function in the initiation of carbon utilization proved to be strongly repressed, as were several central carbon metabolic genes (Table 2). By contrast, genes that showed glucose activation included the glucose transporter gene ptsG, those that encode specific metabolic enzymes that are required for growth under glucose fermentative conditions such as aceE and guaB (Table 1), a few regulatory nucleic acid-binding protein-encoding genes such as the fis and spf genes, and many genes subject to a positive growth rate response. Only in the former genes were Crp-binding sites identified in the control regions (Table 1).
Perhaps of greatest interest was the surprising number of stress genes that are subject to catabolite repression (Tables 2 and 5). These genes encode many chaperone proteins (1, 4, 38), all of the important cytoplasm ATP-dependent protease complexes that can function both as general chaperones and for protein degradation (9, 11, 20), and both heat shock and cold shock genes (12, 31, 40). Sometimes the repressive effects were very large (Table 2), even though a Crp-binding site could not be identified in the control regions of the encoding operons. Indirect mechanisms mediated by transcription factors under Crp control are therefore likely. Further experiments are required to ascertain what these mechanisms are.
Acknowledgments
We thank Ron Sapolsky for assistance in the data analysis and Mary Beth Hiller for assistance in the preparation of the manuscript.
This work was supported by National Institutes of Health grant GM64368 from the National Institute of General Medical Sciences. G.G. acknowledges a sabbatical fellowship from the Dirección General de Asuntos del Personal Académico de la Universidad Nacional Autónoma de México.
REFERENCES
- 1.Agashe, V. R., and F.-U. Hartl. 2000. Roles of molecular chaperones in cytoplasm protein folding. Semin. Cell Dev. Biol. 11:15-25. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 4.Bukau, B., T. Hesterkamp, and J. Luirink. 1996. Growing up in a dangerous environment: a network of multiple targeting and folding pathways for nascent polypeptides in the cytosol. Trends Cell Biol. 6:480-486. [DOI] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, R. J., and M. J. Campbell. 2000. Transcription, genomes, function. Trends Genet. 16:409-415. [DOI] [PubMed] [Google Scholar]
- 8.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan, D. A., A. Mogk, and B. Bukau. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol. Life Sci. 59:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, R., H. Tao, K. T. Shanmugam, S. W. York, and L. O. Ingram. 2002. Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol. Prog. 18:6-20. [DOI] [PubMed] [Google Scholar]
- 11.Hengge, R., and B. Bukau. 2003. Proteolysis in prokaryotes: protein quality control and regulatory principles. Mol. Microbiol. 49:1451-1462. [DOI] [PubMed] [Google Scholar]
- 12.Inouye, M. 1999. Cold-shock response and adaptation. J. Mol. Microbiol. Biotechnol. 1:191. [PubMed] [Google Scholar]
- 13.Isaacs, H., Jr., D. Chao, C. Yanofsky, and M. H. Saier, Jr. 1994. Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli. Microbiology 140:2125-2134. [DOI] [PubMed] [Google Scholar]
- 14.Jinks-Robertson, S., and M. Nomura. 1987. Ribosomes and tRNA, p. 1358-1385. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 15.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S.-J., W. Boos, J.-P. Bouché, and J. Plumbridge. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 19:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magasanik, B., and F. C. Neidhardt. 1987. Regulation of carbon and nitrogen utilization, p. 1318-1325. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 18.Matin, A. 1991. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 5:3-10. [DOI] [PubMed] [Google Scholar]
- 19.Matin, A., M. Baetens, S. Pandza, C. H. Park, and S. Waggoner. 1999. Survival strategies in the stationary phase, p. 30-48. In E. Rosenberg (ed.), Microbial ecology and infectious disease. American Society for Microbiology, Washington, D.C.
- 20.Matouschek, A. 2003. Protein unfolding—an important process in vivo? Curr. Opin. Struct. Biol. 13:98-109. [DOI] [PubMed] [Google Scholar]
- 21.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 22.Nasser, W., R. Schneider, A. Travers, and G. Muskhelishvili. 2001. CRP modulates fis transcription by alternate formation of activating and repressing nucleoprotein complexes. J. Biol. Chem. 276:17878-17886. [DOI] [PubMed] [Google Scholar]
- 23.Park, S. J., G. Chao, and R. P. Gunsalus. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode α-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 179:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, S. J., P. A. Cotter, and R. P. Gunsalus. 1995. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon and heme availability. J. Bacteriol. 177:6652-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S. J., and R. P. Gunsalus. 1995. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J. Bacteriol. 177:6255-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, S. J., J. McCabe, J. Turna, and R. P. Gunsalus. 1994. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J. Bacteriol. 176:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pessi, G., C. Blumer, and D. Haas. 2001. lacZ fusions report gene expression, don't they? Microbiology 147:1993-1995. [DOI] [PubMed] [Google Scholar]
- 28.Plumbridge, J. 1998. Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol. 29:1053-1063. [DOI] [PubMed] [Google Scholar]
- 29.Plumbridge, J. 1999. Expression of the phosphotransferase system both mediates and is mediated by Mlc regulation in Escherichia coli. Mol. Microbiol. 33:260-273. [DOI] [PubMed] [Google Scholar]
- 30.Plumbridge, J. 2001. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:371-380. [PubMed] [Google Scholar]
- 31.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154:573-580. [DOI] [PubMed] [Google Scholar]
- 32.Ramseier, T. M., S. Bledig, V. Michotey, R. Feghali, and M. H. Saier, Jr. 1995. The global regulatory protein, FruR, modulates the direction of carbon flow in enteric bacteria. Mol. Microbiol. 16:1157-1169. [DOI] [PubMed] [Google Scholar]
- 33.Ramseier, T. M., D. Nègre, J.-C. Cortay, M. Scarabel, A. J. Cozzone, and M. H. Saier, Jr. 1993. In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, pts, icd and ace operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol. 234:28-44. [DOI] [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr., S. Chauvaux, J. Deutscher, J. Reizer, and J.-J. Ye. 1995. Protein phosphorylation and the regulation of carbon metabolism: comparisons in gram-negative versus gram-positive bacteria. Trends Biochem. Sci. 20:267-271. [DOI] [PubMed] [Google Scholar]
- 35.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 36.Schilling, C. H., L. Held, M. Torre, and M. H. Saier, Jr. 2000. GRASP-DNA: a web application to screen prokaryotic genomes for specific DNA-binding sites and repeat motifs. J. Mol. Microbiol. Biotechnol. 2:495-500. [PubMed] [Google Scholar]
- 37.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 38.Shinde, U., and M. Inouye. 2000. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11:35-44. [DOI] [PubMed] [Google Scholar]
- 39.Snapyan, M., M. Lecocq, L. Guevel, M. C. Arnaud, A. Ghochikyan, and V. Sakanyan. 2003. Dissecting DNA-protein and protein-protein interactions involved in bacterial transcriptional regulation by a sensitive protein array method combining a near-infrared fluorescence detection. Proteomics 3:647-657. [DOI] [PubMed] [Google Scholar]
- 40.Weber, M. H., and M. A. Marahiel. 2003. Bacterial cold shock responses. Sci. Prog. 86:9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, J., and R. C. Johnson. 1997. Cyclic AMP receptor protein functions as a repressor of the osmotically inducible promoter proP P1 in Escherichia coli. J. Bacteriol. 179:2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamanaka, K., and M. Inouye. 1997. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Z., M. Aboulwafa, M. H. Smith, and M. H. Saier, Jr. 2003. The ascorbate transporter of Escherichia coli. J. Bacteriol. 185:2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]