Summary
Cyclic dinucleotides (CDNs) play central roles in bacterial pathogenesis and innate immunity. The mammalian enzyme cGAS synthesizes a unique cyclic dinucleotide (cGAMP) containing a 2′-5′ phosphodiester linkage essential for optimal immune stimulation, but the molecular basis for linkage specificity is unknown. Here we show that the Vibrio cholerae pathogenicity factor DncV is a prokaryotic cGAS-like enzyme whose activity provides a mechanistic rationale for the unique ability of cGAS to produce 2′-5′ cGAMP. Three high-resolution crystal structures show that DncV and human cGAS generate CDNs in sequential reactions that proceed in opposing directions. We explain 2′ and 3′ linkage specificity, and test this model by reprogramming the human cGAS active site to produce 3′-5′ cGAMP, leading to selective stimulation of alternative STING adaptor alleles in cells. These results demonstrate mechanistic homology between bacterial signaling and mammalian innate immunity and explain how active site configuration controls linkage chemistry for pathway-specific signaling.
Introduction
Cyclic dinucleotides (CDNs) are key regulators of both bacterial physiology and mammalian innate immunity (Cai et al., 2014; Danilchanka and Mekalanos, 2013). Cyclic-di-guanosine monophosphate (cdG) was originally discovered as small molecule regulator of cellulose production in Acetobacter (Ross et al., 1987), but it is now appreciated that cdG and cyclic di-adenosine (cdA) regulate diverse processes including biofilm formation, antibiotic signaling and the bacterial stress response (Danilchanka and Mekalanos, 2013). In addition, cdG regulates developmental pathways in the eukaryote Dictyostelium (Chen and Schaap, 2012). More recently, a mixed base cyclic GMP–AMP molecule (cGAMP) was discovered in Vibrio cholerae 7th pandemic strains, and the responsible synthase, DncV, was identified as a virulence factor on pandemic island-1 (Davies et al., 2012). Remarkably, cyclic GMP–AMP was discovered shortly thereafter as the eukaryotic signal responsible for triggering innate immune responses downstream of cytosolic DNA recognition in mammalian cells (Sun et al., 2013), and the human enzyme cyclic GMP–AMP synthase (cGAS) is a major sensor for DNA-triggered innate immunity (Li et al., 2013b; Schoggins et al., 2014). Despite the prevalence of cyclic dinucleotide signaling in both prokaryotic and eukaryotic kingdoms of life, it remains to be proven if a direct connection exists between the signaling machinery that produces or responds to dinucleotides in either kingdom.
The bacterial cyclic dinucleotide synthases investigated to date function as obligate oligomers in which each active site in adjacent monomers generates one of the two phosphodiester bonds necessary to produce the final symmetric cyclized product (Chan et al., 2004; De et al., 2008; Witte et al., 2008). By contrast, the mammalian cGAS enzyme functions with a single active site, using a sequential series of reactions to produce a mixed dinucleotide species G[2′ –5′]pA[3′ –5′]p (cGAMP) containing a 2′ –5′ phosphodiester bond (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a; Zhang et al., 2013). The unique 2′ –5′ linkage in cGAMP has been shown to be responsible for enhanced immunity through increased stimulation of the adapter STING (Diner et al., 2013; Gao et al., 2013b; Zhang et al., 2013), but potential additional roles for the 2′ –5′ cGAMP linkage in stability or signal discrimination remain unknown.
The requirement for a mixed dinucleotide potentially drove evolution of human cGAS to rely on a single active site and sequential enzymatic reactions. However, the prokaryotic Vibrio DncV enzyme also produces a mixed base 3′ –5′ cGAMP molecule (Ablasser et al., 2013; Davies et al., 2012; Diner et al., 2013). Vibrio DncV has been hypothesized to function similarly to cGAS (Diner et al., 2013; Gao et al., 2013a), but the low sequence homology (<10%) between DncV and mammalian OAS/cGAS family enzymes precludes prediction about whether DncV’s mechanism of action is functionally similar to cGAS or to conventional oligomeric bacterial dinucleotide cyclases. We reasoned that investigation of the molecular mechanism of Vibrio DncV would reveal how enzymes that synthesize mixed cyclic dinucleotides evolved in the prokaryotic kingdom. Such insights could in turn provide a new framework to understand the distinct ability of human cGAS to catalyze the 2′ –5′ linkage specificity that is critical to innate immune signaling.
Here we present a series of structures of Vibrio cholerae DncV, capturing the enzyme in substrate bound and catalytically trapped linear-intermediate states. In spite of the minimal sequence identity, our results show that DncV is both a structural and functional homolog of mammalian cGAS, demonstrating for the first time a direct connection between the biosynthetic machinery for generating dinucleotide signals in multiple kingdoms of life. Although the active site shares a common architecture between DncV and cGAS, we establish that the sequential enzymatic reactions proceed in opposite directions, with DncV using a 5′ pppA[3′ –5′]pG intermediate and cGAS a 5′ pppG[2′ –5′]pA intermediate. By comparing the two enzymes, we derive a model that explains how single active site cyclases control phosphodiester linkage specificity, and how human cGAS rotates the substrate nucleotides to produce the 2′ –5′ linkage unique to mammalian innate immunity. This model allowed us to rationally redesign the human cGAS active site for production of enzymatic variants that now synthesize 3′ –5′ linked species. These results provide an evolutionary connection between prokaryotic and eukaryotic signaling systems, and elucidate enzymatic plasticity that allowed evolution of a unique metazoan innate immune molecule.
Results and Discussion
Structure of Vibrio cholerae DncV reveals an auto-activated cGAS-like enzyme
We isolated a near full-length construct of Vibrio cholerae DncV (M4–L414) that retains full enzymatic activity, and determined its 3.0 Å resolution crystal structure. Selenomethionine derivatized crystals were used for phase determination, and model refinement was additionally improved by a higher resolution catalytically trapped intermediate structure (discussed below). The core of DncV adopts a template-independent nucleotidyl-transferase fold defined by beta-strands β2–5, similar to the originally characterized CCA-adding enzyme (Figure 1) (Xiong and Steitz, 2004). In spite of minimal sequence identity (~10%), the overall structure of DncV is remarkably similar to that of human cGAS (Figure 1B, Figure S1) (Kato et al., 2013; Kranzusch et al., 2013; Li et al., 2013a; Zhang et al., 2014). In addition, the catalytic core of DncV is nearly as structurally conserved as the previously described relationship between cGAS and the dsRNA-sensor oligo-adenylate synthase (OAS) (RMSD ~3.8 Å vs. ~3.0 Å) (Civril et al., 2013; Donovan et al., 2013; Kranzusch et al., 2013).
Figure 1. Structure of Vibrio cholerae DncV reveals an auto-activated cGAS-like enzyme.
(A) Crystal structure of Vibrio cholerae DncV. Alpha helices are colored in green, beta strands forming the enzyme core are in blue, and a red circle marks the active site. (B) Superposition of the crystal structures of DncV (green) and human cGAS (purple) (PDB 4KM5). (C) Surface view representation of the DncV (green), apo human cGAS (purple, “inactive”) and dsDNA-bound mouse cGAS (purple, “active”) active site substrate channels (Mouse cGAS PDB 4K98). Nucleotide substrate binding pockets are colored in blue and the active site is marked by a red circle. (D) Superposition of an electrostatic surface view of DncV colored by charge (positive, blue and negative, red) and cGAS-bound dsDNA (green). The structures of DncV and dsDNA-bound mouse cGAS were superposed without manual fitting of the dsDNA ligand. See also Figure S1.
The structure of Vibrio DncV reveals an auto-activated state, explaining why DncV is constitutively active while cGAS requires stimulation by dsDNA binding. Although the overall fold of the core active site is shared, two unique features maintain constitutive activity of DncV. First, whereas dsDNA-binding is required to configure the cGAS active site for competent metal ion coordination (Civril et al., 2013; Gao et al., 2013a), in DncV alpha-helical lobes brace either side of the active site and buttress beta-strands β2–5 to automatically position D132, D134 and D194 for divalent ion binding (Figure 1A). In agreement with automatic positioning, the orientations of DncV D132, D134 and D194 do not change between our apo and substrate bound or catalytically trapped structures (discussed below). Second, regulatory loops that restrict the substrate channel of cGAS (Gao et al., 2013a) are missing in the DncV enzyme, resulting in an open active site permanently competent for substrate binding (Figure 1B, C, Figure S1). Recent structural studies have suggested that complete activation of cGAS requires protein dimerization (Li et al., 2013a; Zhang et al., 2014). We noted that purified DncV behaves as a monomeric species, and there is no indication of functional dimerization within or between neighboring asymmetric units of the crystals (Figure S2). In addition to remodeling that maintains an open and competent active site, these results demonstrate that DncV lacks regulatory dimerization contacts observed in eukaryotic cGAS and conventional bacterial dinucleotide cyclases (De et al., 2008; Li et al., 2013a; Witte et al., 2008; Xiong and Steitz, 2004; Zhang et al., 2014).
DncV does not require dsDNA binding to stimulate enzymatic activity, and we therefore expected the basic DNA recognition cleft found in cGAS to be absent. Strikingly, however, DncV retains a long basic cleft with conserved residues along the same face of the enzyme (Figure 1D, Figure S1). Superposition of the mouse cGAS–DNA bound structure immediately positions the dsDNA along this cleft without a need for manual docking (Figure 1D). We therefore tested the impact of DNA and RNA on DncV activity, but saw no change in enzymatic behavior or mobility of DncV in electrophoretic gel-shift assays (Figure S3). The possible role of this cleft in DncV biology and Vibrio pathogenicity remains an ongoing area of investigation.
DncV GTP recognition parallels cGAS substrate discrimination
We next compared the nucleotide substrate preferences of Vibrio DncV and human cGAS enzymes. As expected, enhanced product formation for each enzyme was observed in the presence of ATP and GTP (Figure 2A). We confirmed previous studies (Ablasser et al., 2013; Davies et al., 2012; Diner et al., 2013) that when provided only ATP or only GTP, DncV synthesizes cdA and cdG (Figure 2A). Promiscuity of the active site for nucleotide selection is shared with human cGAS, which also inefficiently synthesizes both cdA and cdG species when presented with ATP or GTP alone (Figure 2A). Previous results with purified mouse cGAS were unable to detect cdA formation (Gao et al., 2013a). Low levels of human cGAS cdA formation may therefore indicate further differences between the human and mouse homologs in addition to variation in dependency of DNA-stimulated activation requirements (Kranzusch et al., 2013; Sun et al., 2013).
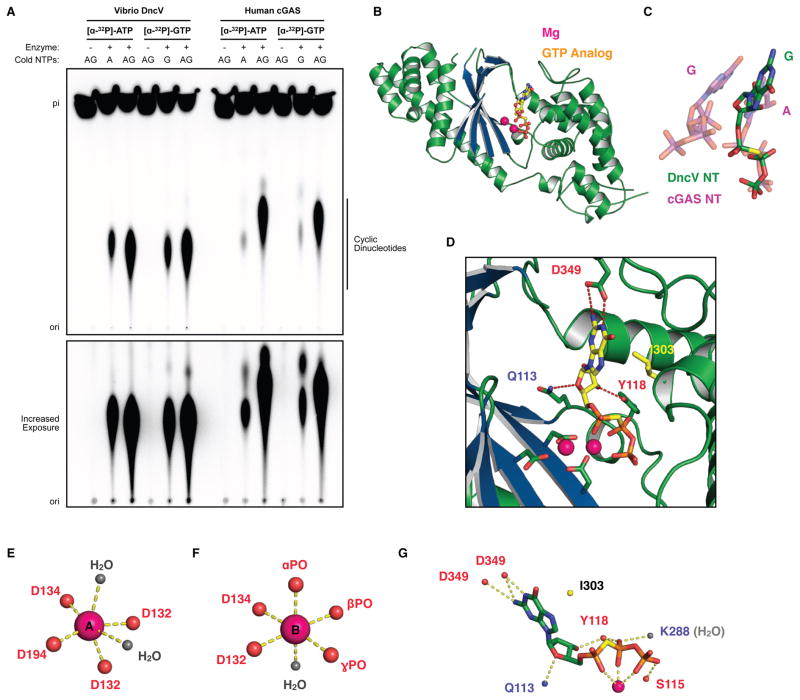
Figure 2. DncV GTP recognition parallels cGAS substrate discrimination.
(A) Analysis of DncV and human cGAS cyclic dinucleotide product formation. Reactions were labeled with indicated α-32P and unlabeled (“cold”) nucleotide substrate, and all cGAS reactions include dsDNA. Reactions were treated with alkaline phosphatase and separated by thin layer chromatography. The lower panel represents a longer exposure, and images are representative of multiple independent experiments. (B) Crystal structure of DncV bound to Mg ions (pink) and GTP analog (yellow/orange). (C) Alignment of DncV (green) and cGAS (purple) substrate nucleotides derived from superposition of the DncV–GTP and pig cGAS–ATP/GTP (PDB 4KB6) crystal structures. (D) Zoomed view of GTP analog bound in the DncV active site. (E,F) Cartoon schematic of DncV interactions with the catalytic Mg ions (metal A and metal B). Octahedral coordination was initially determined by building into the higher resolution 1.8 Å maps of the catalytically trapped structure. (G) Cartoon schematic of DncV interactions with the bound GTP analog as described in text. See also Figure S3.
To ascertain the molecular basis of initial substrate selection, we determined the crystal structure of DncV bound to a nonhydrolyzable nucleotide in which a methylene bridge replaces the α/β bridging oxygen (Apcpp or Gpcpp). The first step in cGAS product formation involves coordination of the adenosine triphosphate for 2′ hydroxyl attack by the guanosine ribose (Gao et al., 2013a), but our initial crystallization attempts using an ATP analog failed to reveal difference density compared to the apo DncV structure. We therefore used the GTP analog, and in this case obtained crystals with strong difference density for nucleotide, allowing us to determine the structure of DncV bound to Gpcpp analog at 2.8 Å resolution (Figure 2B). The bound GTP analog adopts an anti base configuration, and follows a similar path to the adenosine nucleotide residing in the phosphate-donor pocket observed in the catalytically inactive structure of the porcine cGAS (E200Q/D202N) enzyme (Figure 2C) (Civril et al., 2013). GTP is coordinated by base-specific hydrogen-bonding interactions between amino-acid D349 and the guanine N1 and N2 positions along the Watson-Crick edge, while bridging interactions between two bound divalent cations connect the triphosphate moiety and the conserved DncV catalytic triad D132, D134 and D194 (Figure 2D–G). DncV crystallization conditions require high magnesium concentrations, and the observed octahedral geometry of the bound metal ions (as seen in our higher resolution 1.8 Å catalytically trapped structure) is consistent with magnesium ion coordination (Figure 2E–F). In contrast to structures of cGAS, the base stacking interaction between human cGAS Y436 and bound nucleotides is notably absent in DncV. Instead, the tyrosine residue is replaced by I303 that retains active site pocket hydrophobicity but prevents stabilization of particular base orientation (Figure 2D, G).
Prokaryotic and metazoan dinucleotide cyclization reactions proceed in opposing directions
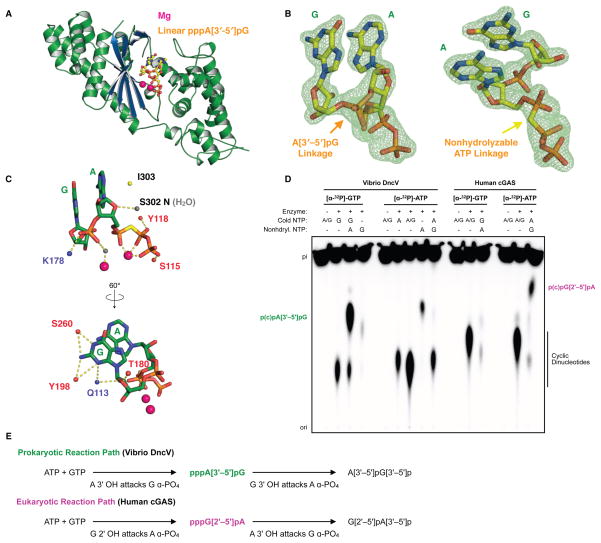
We next crystallized DncV in the presence of combinations of ATP/GTP and nonhydrolyzable Apcpp/Gpcpp analogs. Difference density was obtained for crystals of DncV with GTP and Apcpp, and we determined the 1.8 Å resolution structure of this complex (Figure 3A). Unexpectedly, the electron density can clearly be traced for unambiguous assignment of a bound linear dinucleotide intermediate pp(c)pA[3′ –5′]pG, implying that the first step in DncV nucleotide cyclization occurs through adenosine 3′ hydroxyl attack of the guanosine alpha-phosphate (Figure 3B). In this structure, the guanine and adenine bases adopt anti and syn base configurations, respectively. This observation suggests that the DncV reaction proceeds through a pppA(syn)[3′ –5′]pG(anti) state similar to the mouse cGAS pppG(syn)[2′ –5′]pG(anti) structure, where the second nucleotide to be hydrolyzed must pass through a syn conformation en route to product cyclization (Gao et al., 2013a).
Figure 3. Prokaryotic and metazoan dinucleotide cyclization reactions proceed in opposing directions.
(A) Crystal structure of DncV bound to Mg ions (pink) and a linear pppA[3′ –5′]pG intermediate (yellow/orange). (B) Fo-Fc omit map of the pppA[3′ –5′]pG ligand contoured to 4.5 σ. (C) Cartoon schematic of DncV interactions with the pppA[3′ –5′]pG as described in text. (D) Analysis of DncV and human cGAS cyclic dinucleotide product formation as in Figure 2A. Reactions include labeled/unlabeled NTPs and nonhydrolyzable, methylene-substituted ATP and GTP nucleotides as indicated. A[3′ –5′]pG (green) and G[2′ –5′]pA (purple) labels denote migration of trapped linear products from DncV and cGAS respectively, and image is representative of multiple independent experiments. (E) Reaction schematic of DncV and cGAS dinucleotide product formation.
Hydrogen bonding interactions occur between Q113 and the guanosine 2′ OH, and bridging water interactions between T180 and I303/S302 backbone amines coordinate the guanosine and adenosine riboses, respectively (Figure 3C). Interactions with S115 and Y118 as well as the bridging metal B interactions stabilize the adenosine triphosphate, and in the linear intermediate state, bridging interactions with metal A further stabilize the connecting A[3′ –5′]pG phosphate (Figure 3C). The only interactions with the nucleotide bases occur through the guanine position where S260 and Y198 make contacts with the Watson-Crick edge and Y198 and Q113 contact the Hoogsteen edge (Figure 3C).
We next used combinations of alpha-radiolabeled substrates and nonhydrolyzable nucleotides to test the order of DncV-catalyzed reactions leading to cGAMP formation. Robust DncV linear dinucleotide product formation occurred in reactions containing [α-32P]-GTP and nonhydrolyzable ATP, while no intermediate product was observed in DncV reactions with [α-32P]-ATP and nonhydrolyzable GTP (Figure 3D). Conversely, linear dinucleotide product formation was observed for human cGAS only in the presence of [α-32P]-ATP and nonhydrolyzable GTP (Figure 3D), confirming that the reaction with DncV proceeds primarily through the linear dinucleotide intermediate pppA[3′ –5′]pG, while reaction with cGAS proceeds through pppG[2′ –5′]pA (Figure 3D, E). Together with crystallographic identification of a linear pppA[3′ –5′]pG intermediate, these results show that in spite of shared structural homology, the active sites of DncV and cGAS produce cGAMP through sequential reactions that are chemically distinct and proceed in opposing directions.
Active site remodeling controls substrate rotation and phosphodiester linkage specificity
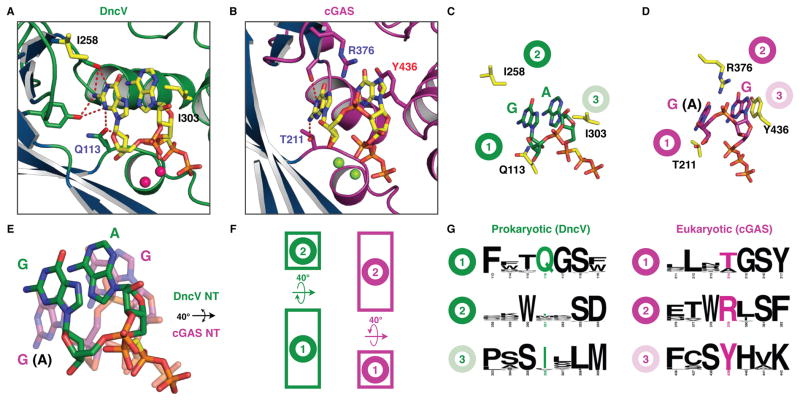
To understand mechanistic control of phosphodiester linkage specificity by cGAS-like enzymes, we compared the active site positioning of our DncV trapped linear intermediate structure (Figure 3) with that of the analogous mouse cGAS pppG[2′ –5′]pG structure (Gao et al., 2013a). Each dinucleotide is oriented by similar base and phosphate backbone interactions (Figure 4A–D), but superposition of the two bound ligands reveals significant rotation of the dinucleotide base and ribose geometries with respect to each other. In DncV the bases are rocked upwards ~40° away from the catalytic triad, consistent with repositioning of the 3′ or 2′ hydroxyl during the first phosphodiester bond formation (Figure 4E). Intriguingly, conserved residues in either enzyme would be predicted to clash with the alternative nucleotide orientations, indicating that each enzymatic pocket is specifically tailored to accept only one set of base rotation conformations.
Figure 4. Active site remodeling controls substrate rotation and phosphodiester linkage specificity.
(A,B) Zoomed view of the substrate binding pocket in the pppA[3′ –5′]pG bound DncV structure (green) and pppG[2′ –5′]pG bound mouse cGAS structure (purple). (C,D). Cartoon schematic illustrating site 1, 2 and 3 positions leading to altered substrate base rotation in DncV (green) and cGAS (purple). (E) Superposition of bound linear intermediate substrates from DncV (green) and cGAS (purple) illustrating an ~40° rotation in nucleotide base positioning. (F). Schematic illustrating substrate rotation model where elongating amino acids in site 1 or site 2 reposition prokaryotic (green) and eukaryotic (purple) bases respectively. (G) WebLogo illustration of site 1, 2 and 3 conservation in prokaryotic (green) and eukaryotic (purple) cGAS-like enzyme active sites. Text size correlates with conservation across alignment of all available DncV and cGAS sequences (Crooks et al., 2004). See also Figures S1, S2 and SI Table 2.
An extended side-chain (Q113) at the bottom of the pocket (site 1) and recessed side-chain (I258) at the top of the pocket (site 2) support the upward rotation of the bases in the DncV active site. Conversely a recessed side-chain T211 at site 1 below and an extended R376 side-chain at site 2 above the bases induce the downward rotation of the bases in the cGAS active site (Figure 4C, D, E). The amino acids at sites 1 and 2 within the active site pocket share conserved combinations of recessed or extended combinations of side-chains, such that in general prokaryotic DncV-like sequences encode an extended amino acid in the site 1 position while eukaryotic cGAS-like sequences encode an extended amino acid in site 2 (Figure 4F, G and SI Table 2). An additional discriminating characteristic is a conserved Y436 amino acid found in all metazoan cGAS enzymes, where Y436 stacks with the bases on the opposite side of the pocket (site 3); prokaryotic sequences favor nonpolar residues incapable of base-stacking interactions in site 3 (Figure 4C–G and SI Table 2). These differences indicate that the location of extended amino acids in alternate site 1 or site 2 positions, with a possible stabilizing role for base-stacking interactions in site 3, direct rotation of the nucleotide bases to favor 3′ –5′ or 2′ –5′ linkage specificity.
Rational reprogramming of human cGAS product formation
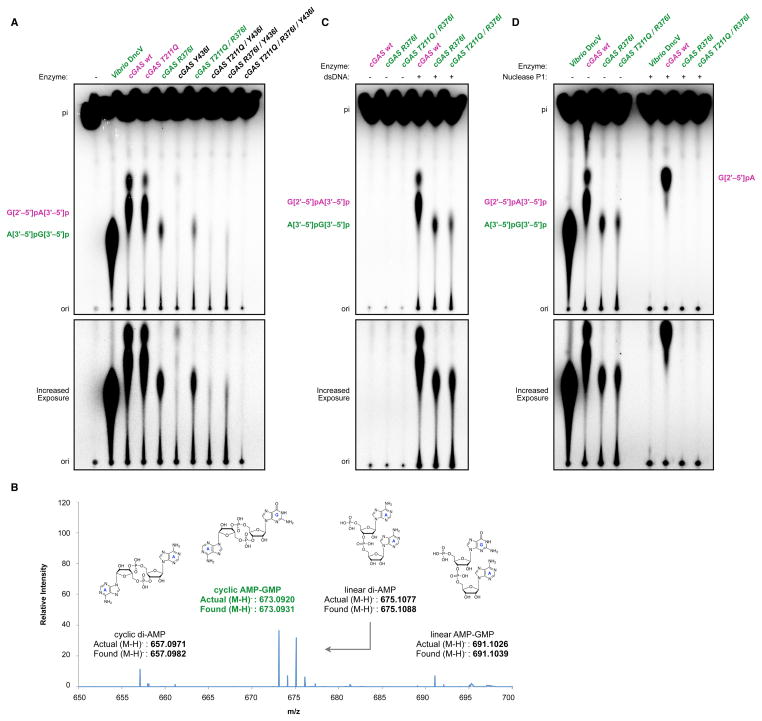
This proposed model of linkage specificity arising from our structure suggested that it may be possible to exchange recessed and extended amino-acid side-chains at sites 1–3 to re-engineer the catalytic activity of human cGAS. We therefore replaced human cGAS site 1, 2 and 3 residues with the corresponding prokaryotic DncV amino acids, and purified seven mutant enzymes bearing single, double or triple amino-acid replacements to test for potentially altered catalytic activity. We found that human cGAS R376I and T211Q/R376I variant enzymes exhibit a significant change in dinucleotide product migration as observed by thin layer chromatography, implying altered product linkage specificity compared to the wildtype G[2′ –5′]pA[3′ –5′]p enzyme product (Figure 5A). Moreover, the mutant enzymes produce a dinucleotide species that migrates similarly to the DncV 3′ –5′ cyclic dinucleotide species. The human cGAS site 2 mutation R376I is most critical for altered specificity, but the compensatory extended mutation of T211Q in site 1 appears to increase enzymatic signaling potential in a cellular context. Notably, alterations to this position are not tolerated in the prokaryotic DncV enzyme (discussed below). Conversely, the human cGAS site 3 Y436I mutation does not impact product migration, and addition of the site 3 mutation Y436I to create a triple mutant disrupts enzymatic activity, which may result from too large of an increase of the active site pocket or loss of base-stacking explicitly required for the altered activity of the cGAS mutant enzymes (Figure 5A).
Figure 5. Rational reprogramming of human cGAS product formation.
(A) Analysis of DncV and human cGAS cyclic dinucleotide product formation as in Figure 2A. Reactions include DncV, wildtype cGAS or the indicated cGAS mutant enzymes, and migration of A[3′ –5′]pG[3′ –5′]p (DncV, green label) and wildtype G[2′ –5′]pA[3′ –5′]p (cGAS, purple label) products are indicated. (B) High-resolution mass spectrometry analysis of cGAS R376I product formation confirms production of cyclic AMP-GMP, as well as cdA and linear intermediates, as discussed in text. (C) Dinucleotide analysis as in A, with wildtype cGAS or the indicated cGAS mutant enzymes in the presence of absence of dsDNA stimulation. (D) Dinucleotide analysis as in A, with alkaline phosphate-treated products additionally digested with Nuclease P1 to hydrolyze all 3′ –5′ linkages (right lanes). Wildtype cGAS G[2′ –5′]pA[3′ –5′]p product is digested by Nuclease P1 to the indicated G[2′ –5′]pA intermediate while DncV and reprogrammed cGAS products are digested to completion. Lower panels display increased exposures of the same experiments as in Figure 2, and images are representative of multiple independent experiments. See also SI Figures 3, 4 and 5.
In addition to binding pocket restrictions impacting rotation of the substrate nucleotides (Figure 4), the cGAS site 2 R376I mutation may also relax guanine-specific base interactions to allow altered product specificity. In each of our substrate-bound structures of DncV, nucleotide-specific interactions occur primarily with the guanine base (Figures 2 and 3). Contacts are made primarily through the guanine in the bound cGAS structures (Civril et al., 2013; Gao et al., 2013a), indicating that regardless of the opposing sequential order of reactions, DncV and cGAS are naturally tuned to use guanine-contacts for substrate positioning. Relaxing guanine-specific contacts while manipulating residues at sites 1–3 may afford increased flexibility in allowing altered rotation of substrate ligands.
To identify the altered products generated by human cGAS R376I and T211Q/R376I enzymes, we first isolated and analyzed the products with high-resolution mass spectrometry. Mass spectrometry results confirm that the reprogrammed cGAS enzymes produce cyclic AMP-GMP, demonstrating that altered product migration by thin layer chromatography is not due to changes in base or chemical composition (Figure 5B). In agreement with reprogramming mutations resulting in a relaxed requirement for guanine-specific contacts, we also detect cdA by high-resolution mass spectrometry (Figure 5B), and the requirement for selecting the order of guanine incorporation is now reduced (SI Figure 4). Importantly, the activity of reprogrammed cGAS enzymes retains the requirement for dsDNA stimulation (Figure 5C), indicating that reprogramming mutations that alter the active site properties do not change the overall enzyme function or ability to respond to DNA ligands.
Since mass spectrometry demonstrated that the engineered cGAS variants produce cyclic AMP-GMP, we reasoned that reduced product mobility is indeed caused by altered cyclic dinucleotide chemical linkage and reprogramming of the enzymatic activity away from a metazoan-specific 2′ –5′ linkage. To test this hypothesis, we conducted nuclease P1 digestions to determine overall linkage specificity. Nuclease P1 hydrolyzes the 3′ –5′ phosphodiester bonds in cyclic dinucleotides, but is unable to digest the unique cGAS 2′ –5′ linkage (Ablasser et al., 2013; Diner et al., 2013). As expected, nuclease P1 treatment completely degrades the DncV A[3′ –5′]pG[3′ –5′]p product, while the wildtype cGAS product is digested to a G[2′ –5′]pA linear intermediate (Figure 5D). In contrast to the wildtype enzyme, cGAS R376I and T211Q/R376I products gain sensitivity to nuclease P1, confirming they contain 3′ –5′ linkages (Figure 5D, right). Selective signaling of reprogrammed cGAS enzymes through only 3′-5′ permissive STING alleles in cells provides further confirmation of 3′ –5′ cyclic dinucleotide production (see below). Collectively, these data provide support for our structural model of DncV and cGAS phosphodiester linkage control, and demonstrate successful reprogramming of the human cGAS enzyme to produce 3′ –5′ linked dinucleotide products.
Active site plasticity is unique to eukaryotic cGAS enzymes
Since alterations to amino acids at sites 1 and 2 reprogrammed human cGAS to produce 3′ –5′ linked dinucleotide products, we tested the corresponding eukaryotic amino acids in Vibrio DncV to determine if cGAS-like active site residues are sufficient for conversion to 2′ –5′ product formation. We cloned seven Vibrio DncV constructs with single, double or triple human cGAS site 1, 2 and 3 substitutions, and tested potential cyclic dinucleotide synthesis activity in cells using a STING-dependent IFN-beta luciferase reporter assay. In contrast to human cGAS, DncV exhibited the opposite pattern of substitution tolerance, whereby mutation of site 1 to a recessed amino acid (Q113T) abolished all detectable cyclic dinucleotide production and mutation of site 2 to an extended amino acid (I258R) did not impact enzymatic activity (SI Figure 5). These results confirm a close relationship between “extended” and “recessed” amino acids at sites 1 and site 2 in both prokaryotic and eukaryotic cGAS-like enzymes where in each case the conserved extended amino acid plays a dominant role. The observation that site 1 mutations in DncV abolish all detectable activity suggests that enzyme active site plasticity leading to reprogramming is unique to eukaryotic cGAS enzymes (SI Figure 5). These results demonstrate that modulation of a prokaryotic cGAS-like enzyme requires further alteration to the enzyme active site in addition to a eukaryotic combination of recessed/extended mutations at sites 1 and 2. Increased tolerance in the human cGAS active site may be a prerequisite stage in the evolution of the ability to catalyze two distinct phosphodiester bonds. It is possible that the unique ability of human cGAS to allow reprogramming to 3′ –5′ linkage specificity enables recreation of a more ancestral state of the enzyme; further characterization of ancestral cGAS-like homologs will be required to evaluate this conclusion. The ability to reprogram human cGAS to synthesize 3′ –5′ specific products will allow screening for additional mutations that increase activity and further modulate active site chemistry.
Reprogrammed human cGAS enzymes signal discrete alleles of the adapter STING
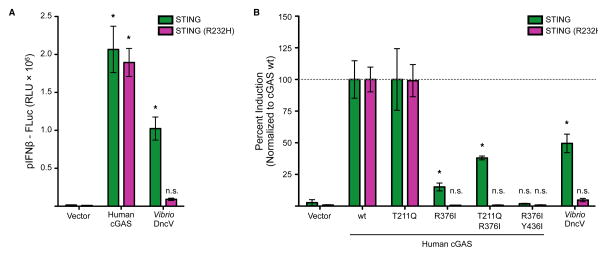
We next tested the reprogrammed cGAS enzymes to determine their ability to signal within cells and promote innate immune activation. Previous studies have demonstrated that distinct human STING alleles exist and respond to different cyclic dinucleotide repertoires (Ablasser et al., 2013; Diner et al., 2013; Jin et al., 2011). Mouse STING responds to both 2′ –5′ cGAMP and canonically linked 3′ –5′ cGAMP, cdA and cdG. In contrast, the human STING allele that encodes histidine at position 232 (R232H) responds only to 2′ –5′ cGAMP (Figure 6A) (Ablasser et al., 2013; Diner et al., 2013). In agreement with our in vitro results, both wildtype mSTING and hSTING R232H respond to the 2′ –5′ cGAMP product of wildtype cGAS enzyme, but only the mSTING wt allele responds to the product of reprogrammed cGAS enzymes, now producing 3′ –5′ linked cyclic dinucleotides (Figure 6B). Importantly, enzymatic reprogramming of the cGAS enzyme resulted in a signaling pattern similar to wildtype DncV, further verifying that the reprogrammed enzymes behave functionally as 3′ –5′ cyclic dinucleotide synthases, yet remain competent for cytosolic DNA recognition and triggering of the antiviral cascade. Since few mutations are required to alter cGAS product dinucleotide linkage specificity, it will be of interest to determine if naturally occurring cGAS enzymatic variants also synthesize 3′ –5′ linked products. Endogenous signaling by 3′ –5′ linked cGAMP could help account for the existence of STING alleles with increased promiscuity in the human population (Jin et al., 2011).
Figure 6. Reprogrammed human cGAS enzymes signal discrete alleles of the adapter STING.
(A) In-cell reconstitution of cyclic dinucleotide immune activation. Cells were co-transfected with empty vector, human cGAS or Vibrio DncV plasmids in the presence of wildtype STING or the R232H allele of STING incapable of responding to 3′ –5′ cGAMP. Activating dsDNA was co-transfected in all samples, and immune activation was monitored with a firefly luciferase reporter under control of the IFN-beta promoter (pIFNβ-FLuc). (B) Cells were transfected as in A, and included reprogrammed human cGAS enzymes as indicated. Data is normalized to the empty vector control and error bars represent the SD from the mean of at least three independent experiments (* denotes p < 0.002, and n.s. denotes not significant). See also SI Figure 5.
Conclusions
The results presented here show that DncV is both a structural and functional homolog of mammalian cGAS, demonstrating a direct connection between the biosynthetic machinery for generating dinucleotide signals in multiple kingdoms of life. Our results provide evidence for an evolutionary relationship between prokaryotic and eukaryotic signaling systems, and suggest an alternative to existing models hypothesizing that divergence within the ancestral metazoan immune repertoire led to creation of cGAS and OAS pathways separating cytosolic dsDNA and dsRNA recognition (Cai et al., 2014; Civril et al., 2013; Gao et al., 2013a; Kranzusch et al., 2013). It will be of great interest to identify and compare further DncV and cGAS homologs to determine if genetic exchange played a role in the development of cyclic dinucleotide signaling within mammalian innate immunity. Crystal structures of DncV in substrate bound and catalytically trapped linear-intermediate states explain why human cGAS produces the 2′ –5′ linked cGAMP that is unique to mammalian innate immunity. Rational redesign of the human cGAS active site to create enzymatic variants that synthesize 3′ –5′ linked species shows that relatively few mutations allow for specific signaling through alternative STING alleles. Our results reveal the potential existence of altered cGAS-signaling, and demonstrate the enzymatic plasticity that allowed evolution of a unique metazoan innate immune molecule.
Experimental Procedures
Protein purification
Vibrio cholerae DncV M4–L414 coding sequence (GenBank accession CP003069) was amplified by polymerase chain reaction (PCR) and cloned into a custom pET vector optimized for E. coli expression of an N-terminal 6×His (KSSHHHHHHGSS)–MBP–TEV fusion protein (Kranzusch and Whelan, 2011). Purification of DncV was adapted from a previously developed protocol optimized for full-length human cGAS, but for DncV an ion-exchange step to remove bound nucleic acid was not required. Briefly, BL21-RIL DE3 E. coli cells were co-transformed with DncV expression vector and pRARE2 tRNA (Agilent) and grown in 2×YT media at 37°C to an OD600 of ~0.5. Cells were cooled at 4°C for 15 min, induced with 0.5 mM IPTG and incubated with shaking at 16°C for ~18 h. Cell pellets were washed with PBS, re-suspended in lysis buffer (20 mM HEPES-KOH pH 7.5, 400 mM NaCl, 10% glycerol, 30 mM imidazole, 1 mM TCEP) supplemented with EDTA-free Complete Protease Inhibitor (Roche), and lysed by sonication. Clarified lysate was incubated with Ni-NTA agarose (Qiagen) for 1 h at 4°C, and recovered resin was washed with lysis buffer supplemented to 1 M NaCl by gravity-flow chromatography at 4°C. Bound protein was eluted using lysis buffer supplemented to 300 mM imidazole and MBP-tagged proteins were diluted to 50 mM imidazole, 5% glycerol and concentrated to ~40 mg ml−1 prior to digestion with Tobacco Etch Virus protease for ~15 h at 4°C. Digested protein was diluted in gel filtration buffer (20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM TCEP) and passed over a 5 mL Ni-NTA column (Qiagen) connected in line with a 5 mL MBP-Trap column (GE Life Sciences) to remove protease and digested MBP-tag. DncV protein was further purified by size-exclusion chromatography on a Superdex 75 16/60 column in gel-filtration buffer, and eluted protein was concentrated to 10–20 mg ml−1. Purified protein was used immediately for crystallography, or supplemented with 10% glycerol and flash-frozen in liquid nitrogen and stored at −80°C for biochemical experiments. For preparation of selenomethionine substituted DncV protein, cultures were grown in M9 minimal media supplemented with 1 mM MgSO4, 100 μM CaCl2, 1 μg ml−1 thiamine, 0.4% glucose and trace elements. After growth at 37°C to an OD600 of ~0.4, cultures were supplemented with leucine, isoleucine and valine at 50 mg l−1; phenylalanine, lysine and threonine at 100 mg l−1; and selenomethionine (Acros Organics) at 75 mg l−1. Cultures were grown at 37°C for an additional 30 min, then cooled at 4°C for 15 min, induced with 0.5 mM IPTG and incubated with shaking at 16°C for ~18 h. Selenomethionine-substituted protein was purified using identical conditions as native protein except buffers contained 2 mM TCEP.
Full-length human cGAS was expressed and purified as previously described (Kranzusch et al., 2013). cGAS mutants were generated by Pfu Ultra polymerase (Stratagene) and QuikChange PCR, and initially purified as MBP-tagged proteins prior to dialysis overnight at 4°C (20 mM HEPES-KOH 7.5, 150 mM KCl, 10% glycerol, and 1 mM TCEP. Dialyzed protein was concentrated, and stored for biochemistry similar to purified DncV. cGAS R376I and T211Q/R376I enzymes were subsequently purified with MBP-tag removal as previously described (Kranzusch et al., 2013).
Crystallization and structure determination
Crystals of DncV were grown at 18°C by hanging-drop vapor diffusion. Purified ~10 mg ml−1 DncV protein was incubated on ice for 30 min with 5 mM NTP combinations and 10.5 mM MgCl2. Following incubation, Easy-Xtal 15-well trays (Qiagen) were used to set 2 μL 1:1 or 0.8:1.2 (protein: reservoir) ratio drops over 350 μL of reservoir solution. Additionally, for some NTP-conditions, crystallization was optimized by pre-mixing protein–NTP and reservoir solutions and incubating at room temperature for 10 min prior to brief centrifugation and setting drops with mixture supernatant. Original crystallization conditions were determined by sparse-matrix screen using 400 nL drops set over 70 μL reservoir solutions in a 96-well format. Optimal crystallization conditions were 30 mM HEPES-KOH pH 7.5, 200–300 mM Mg(OAc)2 and 20–22% PEG-3350, but slight refinements to these conditions were required for growth with different substrates.
Crystals grew as clusters of rods and required incubation at 18°C for 2–4 days for complete growth. Individual crystals were harvested by gently tweezing the nucleation center with a nylon loop or a Kozak cat whisker to release single rods. Crystals were transferred with a nylon loop to a new drop containing reservoir solution supplemented with 15–20% ethylene glycol or glycerol as a cryoprotectant, and incubated for 15 s before flash freezing in liquid nitrogen. Selenomethionine crystals were grown and harvested using similar conditions, and optimized by micro or streak-seeding crushed native crystals with a Kozak cat whisker. Native and anomalous data were collected under cryogenic conditions at the Lawrence Berkeley National Laboratory Advanced Light Source (Beamline 8.3.1) and Stanford Synchotron Radiation Lightsource (Beamlines 11.1 and 12.2).
X-ray diffraction data were processed with XDS and SCALA (Kabsch, 2010) using the SSRL autoxds script (A. Gonzalez, SSRL). Indexed crystals belonged to the orthorhombic spacegroup P21 with two copies of DncV in the asymmetric unit. For phase determination, a series of independent datasets were collected from a single large selenomethionine crystal using a micro-focus beam at the peak selenium absorbance energy. These datasets were then merged to create a single high multiplicity dataset, allowing selenium sites to be identified with HySS within PHENIX (Adams et al., 2010). An initial map was calculated using SOLVE/RESOLVE (Terwilliger, 1999) providing interpretable electron density for de novo building of a mixed poly-alanine and native sequence model of one DncV monomer using Coot (Emsley and Cowtan, 2004). This partial structure was then used as a molecular replacement search model to extend phases into a 1.8 Å native pppA[3′ –5′]pG dataset. Iterative rounds of model building in Coot and refinement were conducted with PHENIX until all interpretable electron density was modeled. Data for refinement of the native pppA[3′ –5′]pG structure were extended to 1.8 Å, 0.2 Å beyond an I/σ resolution cutoff (~1.98 Å) as determined by CC* correlation and Rpim parameters (Karplus and Diederichs, 2012). The completed DncV model was then used as a search model and reference restraint to build the remaining apo and substrate bound structures.
In vitro reconstitution of cyclic dinucleotide synthesis and nucleic acid sensitivity
DncV and cGAS cyclic dinucleotide synthesis was performed and measured as previously described (Kranzusch et al., 2013). Briefly, ~1 μM of purified enzyme or equivalent volume of gel-filtration buffer was incubated with ~25 μM NTP substrates and ~10 μCi radiolabeled [α-32P]-ATP or [α-32P]-GTP (Perkin Elmer) as indicated. Reactions were performed in 10 μL of final reaction buffer (50 mM KCl, 5 mM Mg(OAc)2, 50 mM Tris pH 7.0, 1 mM TCEP, 0.1 mg ml−1 BSA (NEB)) and incubated at 37°C for 2 hours. All cGAS reactions included 2 μM of a chemically synthesized 45 bp double-stranded immunostimulatory DNA (Integrated DNA Technologies) (Stetson and Medzhitov, 2006), and DncV reactions include 2 μM of indicated nucleic acids as previously described (Kranzusch et al., 2013). Following incubation, 5 μL of the reaction volume were transferred to a new tube and treated with 5 units of alkaline phosphatase (NEB) at 37°C for 45 min to digest remaining unincorporated labeled NTPs. Reactions were terminated by spotting 1 μL onto a PEI-cellulose F thin-layer chromatography (TLC) plate (EMD Biosciences) and drying for 5 min at room-temperature. TLC plates were developed with 1.5 M KH2PO4 (pH 3.8) and run until ~2 cm from the top edge of a ~20 cm plate. Plates were dried at 80°C for 30 min and radiolabeled products were detected with a phosphorscreen and Storm phosphorimager (GE Life Sciences). Where indicated, reactions were additionally digested with 2 mU of Nuclease P1 from Penicillium citrinum (Sigma) in 1x P1 buffer (30 mM NaOAc pH 5.3, 0.4 mM ZnSO4) following alkaline phosphatase treatment by incubating at 45°C for 30 min as previously described (Rahmeh et al., 2009). DncV electrophoretic mobility shift assays were prepared with identical reaction conditions using nucleic acid substrates labeled at the 5′ end with T4PNK (NEB) and [γ-32P]-ATP as previously described, and separated on a 6% non-denaturing polyacrylamide gel with 0.5% TBE at 6 W for 1.5 h. Example images in figures are representative of multiple independent experiments.
R376I cGAS Product Purification and Structural Characterization
The enzymatic reaction products from R376I cGAS were purified as previously described using an Agilent HPLC with mixed components from the 1200 series and 1260 series (Diner et al., 2013). Briefly, purification was performed using an Agilent Polaris C18-A column (4.6 × 250 mm, 5 μm). Purification conditions included a 100% to 0% gradient of solvent A over 20 min at 50 C and a flow rate of 1 ml min−1, solvent A being 100 mM ammonium acetate in water at pH 7 and solvent B being acetonitrile. Purified elution fractions were evaporated multiple times in order to remove excess ammonia. Mass spectrometry analysis was performed in the University of California-Berkeley, QB3 Mass Spectrometry Facility.
HRMS (m/z): [M-H]− calculated for C20H23N10O12P2, 657.0971; found, 657.0982.
HRMS (m/z): [M-H]− calculated for C20H23N10O13P2, 673.0920; found, 673.0931.
Cell-based IFN-β luciferase assay
Full-length cDNA sequences for Vibrio DncV, human cGAS, human mutant STING (STING R232H) and wildtype mouse STING (STING wt) were cloned into pcDNA vectors (Invitrogen) for mammalian cell expression. 293T cells were seeded into opaque 96-well plates 20 hours prior to transfection at a density optimized to be 75–80% confluent at the time of transfection. Cells were transfected as indicated along with a control Renilla luciferase reporter plasmid and an IFN-β promoter Firefly luciferase reporter plasmid as previously described (Inn et al., 2011; Kranzusch et al., 2013). At 24 hours post transfection, Firefly and Renilla luciferase values were measured using the Dual-Luciferase Reporter Assay System (Promega) and a Veritas Microplate Luminometer (Turner Biosystems) according to the manufacturer’s protocols. IFN-β Firefly luciferase expression was normalized to transfection efficiency by calculating the relative expression of Firefly luciferase to Renilla luciferase activity. As indicated (Figure 6), data were combined from multiple independent experiments, and statistical significance was calculated with an unpaired, two-tailed t test in Prism.
Supplementary Material
Highlights.
Vibrio cholerae DncV is a structural and functional homolog of human cGAS
DncV and human cGAS cyclic AMP–GMP formation proceeds in opposing reaction orders
Structures of catalytically trapped DncV explain eukaryotic 2′ –5′ linkage specificity
Reprogrammed human cGAS produces 3′ –5′ cGAMP and pathway-specific signaling
Acknowledgments
X-ray data were collected at Beamline 8.3.1 of the Lawrence Berkeley National Lab Advanced Light Source (ALS) and Beamlines 11.1 and 12.2 of the Stanford Synchrotron Radiation Lightsource (SSRL). The authors are grateful to J. Holton and G. Meigs (ALS); T. Doukov, S. Russi and A. Gonzalez (SSRL) for technical assistance with data collection and processing; R. Nichiporuk (QB3 Mass Spectrometry Facility) for assistance with product determination; J. Cate, Y. Bai, S. Floor, R. Wilson and members of the Doudna and Berger labs for helpful comments and discussion. This work was funded by HHMI (R.E.V. and J.A.D.), the G. Harold and Leila Y. Mathers Foundation (J.M.B.), and the NIGMS Center for RNA Systems Biology (A.S.Y.L. and J.A.D.). P.J.K. is supported as an HHMI Fellow of the Life Sciences Research Foundation, and A.S.Y.L. is supported as an American Cancer Society Postdoctoral Fellow. R.E.V. and J.A.D. are HHMI Investigators.
Footnotes
Author Contributions
Experiments were designed by P.J.K. in consultation with R.E.V., J.M.B. and J.A.D. All structural and biochemical work was performed by P.J.K. A.S.Y.L. performed cell-signaling assays, S.C.W. conducted HPLC purification and mass spectrometry experiments, and M.S.S. assisted with initial protein purification and crystallization. The manuscript was written by P.J.K. and J.A.D., and all authors contributed to editing the manuscript and support the conclusions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS biology. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013a;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013b;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Molecular cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes and immunity. 2011;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O. Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS. PLoS One. 2013;8:e76983. doi: 10.1371/journal.pone.0076983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell reports. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Whelan SP. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19743–19748. doi: 10.1073/pnas.1112742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013a;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013b;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. Journal of virology. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Reciprocal-space solvent flattening. Acta crystallographica Section D, Biological crystallography. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Molecular cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, Zhang X, Chen ZJ. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell reports. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.