Abstract
mazEF is an Escherichia coli suicide module specific for a stable toxin and a labile antitoxin. Inhibiting mazEF expression appeared to activate the module to cause cell death. Here we show that several stressful conditions, including high temperatures, DNA damage, and oxidative stress, also induce mazEF-mediated cell death. We also show that this process takes place only during logarithmic growth and requires an intact relA gene.
Programmed cell death (PCD) systems are generally considered to be characteristic of multicellular organisms (12). However, PCD has also been found in unicellular eukaryotes (4) and even in prokaryotes (19, 23, 28, 42). One version of these genetic systems is found on extrachromosomal elements like plasmids and phages (8, 12, 25, 45). They consist of a pair of genes, of which the downstream gene encodes a stable toxin and the upstream gene encodes a labile antitoxin. When the extrachromosomal element is present in the cell, the antitoxin antagonizes the toxin. When the extrachromosomal element is lost, however, the labile antitoxin is degraded, allowing the toxin to kill the cell, a process called postsegregational killing. For this reason, it is believed that the addiction modules participate in maintaining the extrachromosomal elements in the cell (8, 12, 25, 45). The chromosomes of many bacteria have been found to carry toxin-antitoxin systems that are homologous to these extrachromosomal addiction modules (3, 10, 11, 20, 29, 30, 31, 33).
The first chromosomal addiction module found to be regulatable and to function as a PCD system was Escherichia coli mazEF (3), located in the relA operon (31). The product of mazF (MazF) is a stable toxin that cleaves mRNA at a specific site(s) (11, 47). The mechanism of this cleavage has not yet been clarified, as contradictory results have been previously reported (11, 47). The product of mazE (MazE) is an anti-MazF labile protein that is degraded by the protease ClpAP (3). Thus, continuous production of MazE is required to prevent MazF-mediated death. In contrast to the results seen with the addiction modules that are triggered by the loss of the element (8, 12, 25, 45), death mediated by the chromosomally borne mazEF is triggered by several conditions that prevent mazEF expression. Initially, we found that the mazEF module is under the control of ppGpp (3, 13), the amino acid starvation signal molecule produced by the RelA protein (7). Overproduction of ppGpp leads to the inhibition of the expression of mazEF and, thereby, to cell death (3, 13). This inhibition also takes place in the presence of general inhibitors of transcription and/or translation such as the antibiotics rifampin, chloramphenicol, and spectinomycin (40) and through the inhibition of translation by the Doc protein of prophage P1 (21). In each case, the inhibition of gene expression leads to a relatively low level of the labile MazE that allows the stable MazF to kill the cell. Recently, we found that mazEF-mediated death is also triggered by thymine starvation (41), which is known to cause a unique form of DNA damage (2, 34).
Here we asked the following question: do other agents that cause DNA damage and do other kinds of stressful conditions also induce mazEF-dependent death? We examined the following stress conditions: high temperature (50°C), DNA damage (caused by UV irradiation and nalidixic acid and mitomycin C exposure), and oxidative stress (H2O2). Under each of these stressful conditions, we observed mazEF-dependent cell death. In addition, we observed no mazEF-mediated cell death during stationary growth; we found mazEF-mediated cell death only during logarithmic growth, and it seems to require the production of ppGpp.
We used E. coli strains MC4100 relA1 and MC4100 relA+ and their ΔmazEF derivatives, all of which have been used by members of our group (3, 13, 21, 40, 41). We also used strain K38 (39), which we are currently studying in our laboratory. Its ΔmazEF derivative was constructed here by P1 transduction of the kanamycin resistance gene from MC4100mazEF::kanR. For the overproduction of MazF we used plasmid pQE30mazF, which is a derivative of the ampicillin resistance-encoding plasmid pQE30 (Qiagen) carrying the mazF gene under the control of the ptac promoter. This plasmid was cotransformed together with the plasmid pREP4, which harbors a lacIq gene (Qiagen) that enables the regulation of ptac-mazF. The bacteria were grown in liquid M9 minimal medium (40) and plated on rich Luria-Bertani (LB) agar medium (32) which we prepared with the following ingredients: 0.8% Bacto Tryptone, 0.5% yeast extract (both obtained from Difco, Sparks, Md.), 0.5% NaCl (Frutarom, Haifa, Israel), and 1.5% agar (Hispanagar, Burgos, Spain). Nalidixic acid, mitomycin C, rifampin, chloramphenicol, and ampicillin were obtained from Sigma (St. Louis, Mo.). H2O2 was obtained from Merck (Armstadt, Germany).
We studied the effects of various stressful conditions on cell viability by diluting (1/100) an overnight culture in M9 medium and growing the cells with shaking (150 rpm) in the same medium at 37°C until they reached logarithmic growth (optical density at 600 nm, 0.4 to 0.6) or, when indicated, for 18 h (optical density at 600 nm, about 2.0) to stationary phase. When the cells reached the stage of either logarithmic or stationary growth, they were incubated at 37°C for 10 min without shaking. The various stressful conditions were induced, and we then plated the cells on LB agar plates. We incubated the plates at 37°C overnight and then determined the ratio of the CFU of treated cells versus untreated cells.
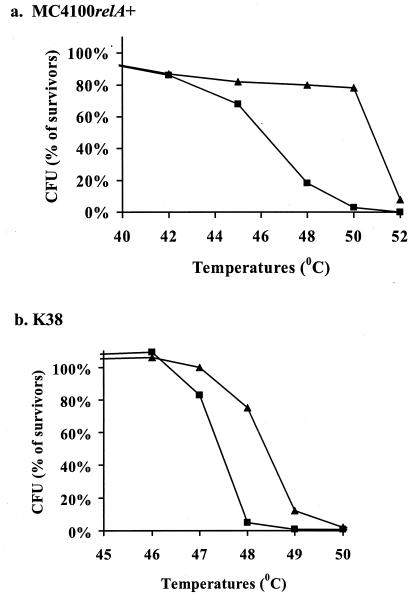
High temperatures induced mazEF-mediated cell death.
In previous work (37, 38), it was shown that E. coli cells are sensitive to short exposure (10 min) to temperatures higher than 48°C. Here we compared the sensitivities to high temperatures of the wild-type strains MC4100relA+ and K38 to those of their ΔmazEF derivatives. In contrast to the wild-type cells, most of the ΔmazEF cells survived exposure to high temperatures between 48 and 50°C (MC4100relA+) or at 49°C (K38). These ΔmazEF cells perished only at temperatures above 52°C (MC4100relA+) or 49°C (Fig. 1).
FIG. 1.
The effect of mazEF on bacterial viability after exposure to high temperatures. E. coli wild-type strains (▪) and their ΔmazEF derivatives (▴) were incubated for 10 min at various temperatures. Cell viability was determined by CFU counting on LB plates incubated at 37°C overnight. The survivor ratio was determined by comparing the number of CFU of treated cells to that of untreated cells. The results represent one experiment out of three independent similar experiments. (a) E. coli MC4100 relA+; (b) E. coli K38.
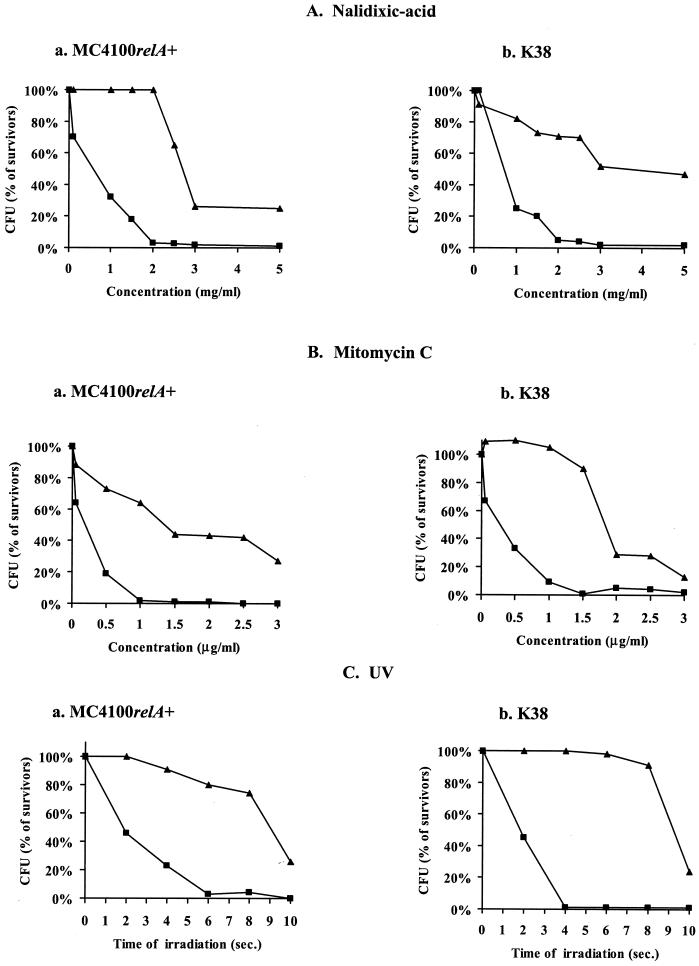
Damage to DNA-induced mazEF-mediated cell death.
In previous work (41), Sat et al. reported that the well-known phenomenon of thymineless death (2) is a result of the activation of the suicide module mazEF. As Sat et al. suggested then, thymine starvation might activate mazEF-mediated death by causing unique DNA damage (2, 34) that leads to the inhibition of the mazEF P2 promoter activity (41). Here we asked the following question: do other types of DNA damage also trigger mazEF-mediated cell death? We chose to examine three modes for generating DNA damage: those of UV irradiation and of the two chemical agents nalidixic acid and mitomycin C. UV irradiation mainly causes the generation of cyclobutane pyrimidine dimers in template DNA during replication (6, 16). Nalidixic acid inhibits the topoisomerase gyrase (9), and mitomycin C induces DNA damage by causing DNA cross-links (5, 24).
In our experiments, E. coli strains MC4100relA+ and K38 and their ΔmazEF derivatives were exposed to DNA-damaging conditions (nalidixic acid [Fig. 2A] or mitomycin C [Fig. 2B] treatment or UV irradiation [Fig. 2C] and the percentages of survivors were determined. The survival of the wild-type strains MC4100relA+ and K38 was affected by short exposures to nalidixic acid at concentrations of 1.0 mg/ml and higher (Fig. 2Aa) and to mitomycin C at concentrations of 0.05 μg/ml and higher (Fig. 2Ba). In contrast, the survival of the ΔmazEF derivative of MC4100relA+ was not affected by nalidixic acid at concentrations of 2.0 mg/ml and lower (Fig. 2Aa) or by mitomycin C at concentrations of 1.5 μg/ml and lower (Fig. 2Ba). A similar pattern was observed in strain K38 (Fig. 2Ab and Bb). Furthermore, its ΔmazEF derivative seems to be resistant to even higher concentrations of nalidixic acid and mitomycin C than MC4100relA+ (Fig. 2Aa versus 2Ab and Fig. 2Ba versus 2Bb). In addition, UV irradiation also affected the survival of the ΔmazEF derivatives less than it affected their corresponding wild-type strains. Only 2 s of irradiation (25 J/m2) was required to reduce the survival of both strains MC4100 relA+ and K38, while more than 8 s of irradiation (25 J/m2) was required to affect the survival of their ΔmazEF derivatives (Fig. 2C). Thus, we conclude that the mazEF system is involved in the process of death caused by DNA damage.
FIG. 2.
The effect of mazEF on bacterial viability after exposure to agents causing DNA damage. E. coli MC4100 relA+ (a) and K38 (b) and their ΔmazEF derivatives were treated for 10 min at 37°C under the following DNA-damaging conditions: nalidixic acid exposure (A), mitomycin C exposure (B), and UV irradiation (C). The cells were washed with saline and diluted in LB medium. For UV irradiation, a series of decimal dilutions were carried out and 10-μl drops were plated on LB plates, dried, and UV irradiated (25 J/m2). In all cases, the cells were plated on LB medium for CFU counting and the survivor ratio was determined by comparing the number of treated cells to the number of untreated cells as described in the legend to Fig. 1. The results represent one experiment out of three independent similar experiments. ▪, wild-type strains; ▴, ΔmazEF derivatives of the wild-type strains.
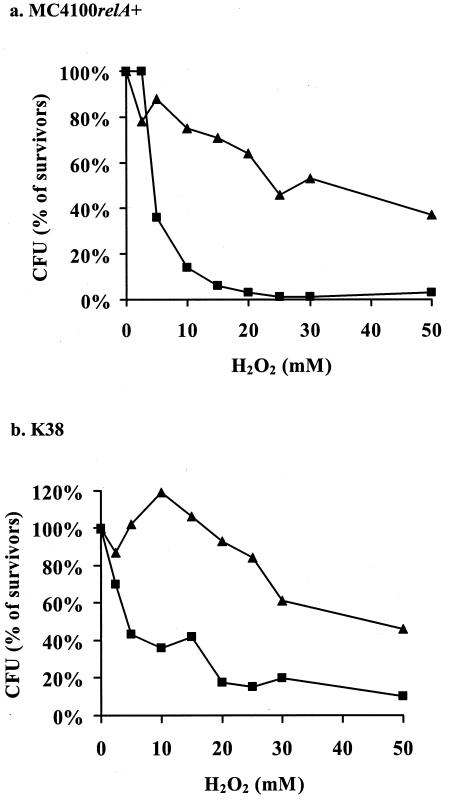
Oxidative stress induced mazEF-mediated cell death.
An increased level of reactive oxygen causes oxidative stress that leads to damage to all cellular components (43). To induce oxidative stress, we treated E. coli cells with hydrogen peroxide (H2O2) (Fig. 3). The wild-type strain MC4100 relA+ died after a short exposure to H2O2 at concentrations of 5 mM and higher; however, most of the cells of its ΔmazEF derivative survived at concentrations of H2O2 up to 50 mM (Fig. 3). A similar pattern was observed in strain K38, although this strain seems to be slightly less sensitive to H2O2 than MC4100relA+. Thus, mazEF-mediated cell death was also involved in death caused by oxidative stress in the presence of H2O2.
FIG. 3.
The effect of mazEF on bacterial viability after exposure to oxidative stress. E. coli strains MC4100relA+ (a) and K38 (b) and their ΔmazEF derivatives were incubated in the presence of various concentrations of H2O2 at 37°C for 10 min. Cell viability and survivor ratios were determined as described in the legend to Fig. 1. The results represent one experiment out of three independent similar experiments. ▪, wild-type strains; ▴, ΔmazEF derivatives of the wild-type strains.
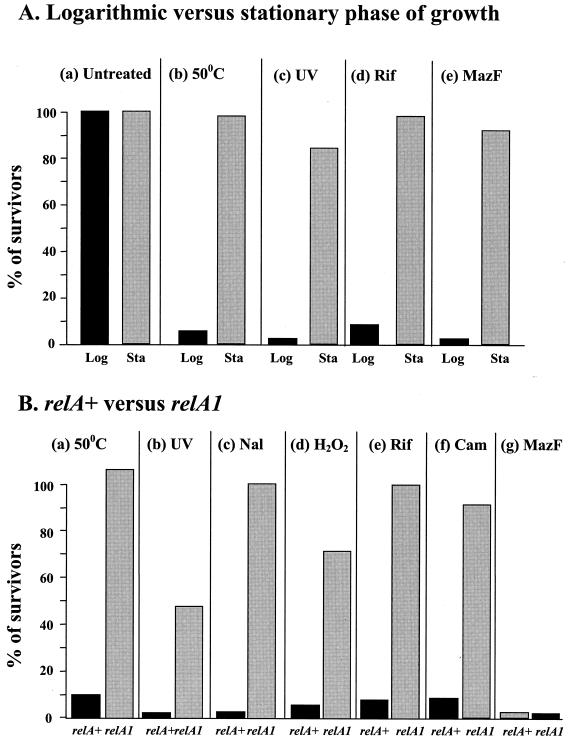
The induction of mazEF-mediated cell death by stress conditions is carried out at the logarithmic growth stage and requires an intact relA gene. All of the previous experiments of Aizenman et al. and Sat et al. (3, 40, 41) and all of the experiments described above showing that various stressful conditions induced mazEF-mediated death were carried out during logarithmic growth (Fig. 1 to 3). Here, we found that cells in the stationary growth phase survived these stressful conditions (Fig. 4A). These conditions included high temperature (50°C) (Fig. 4Ab), UV irradiation (Fig. 4Ac), and the addition of rifampin (Fig. 4Ad). During stationary phase, moreover, even the induction of the toxin MazF by IPTG (isopropyl-β-d-thiogalactopyranoside) was not lethal (Fig. 4Ae). Thus, we found that mazEF-mediated death was activated only during logarithmic growth. We do not yet understand why MazF appears to be inactive during stationary growth. However, this inactivity may contribute to the well-known resistance of cells to stressful conditions during stationary growth (22).
FIG. 4.
The effects of the growth stage (A) and ppGpp (B) on the induction of mazEF-mediated cell death. (A) Logarithmic (Log) and stationary (Sta) E. coli MC4100 relA+ cells were either left untreated (a) or submitted for 10 min to high temperature (50°C) (b), UV irradiation (c), rifampin exposure (20 μg/ml) (d), or the overproduction of MazF (e). (B) Logarithmic E. coli MC4100relA+ and MC4100relA1 cells were submitted for 10 min to high temperature (50°C) (a), UV irradiation (b), nalidixic acid (Nal) exposure (2 mg/ml) (c), H2O2 exposure (15 mM) (d), rifampin (Rif) exposure (20 μm/ml) (e), chloramphenicol (Cam) exposure (50 μg/ml) (f), or the overproduction of MazF (g). Cell viability and survivor ratios were determined as described in the legend to Fig. 1. The results represent one experiment out of three independent similar experiments.
All of the previous experiments in which Sat et al. used rifampin or chloramphenicol (40) and our experiments described above (Fig. 1 to 3) were done using bacterial strains that carried an intact relA gene. To test the involvement of the relA gene, we compared the effects of various stressful conditions which we found to induce mazEF-mediated death on MC4100relA+ and on MC4100relA1 strains during logarithmic growth (Fig. 4B). The relA1 mutation consists of an amino-terminal IS2 insertion between codons 85 and 86. This mutant allele produces only residual amounts of ppGpp (7). We treated each strain for 10 min under the following stressful conditions: 50°C (Fig. 4Ba), UV irradiation (Fig. 4Bb), nalidixic acid exposure (Fig. 4Bc), H2O2 exposure (Fig. 4Bd), rifampin exposure (Fig. 4Be), or chloramphenicol exposure (Fig. 4Bf). In contrast to the relA+ cells, which do not survive these conditions of treatment, the relA1 cells did survive (Fig. 4B). Interestingly, overproduction of MazF kills both relA+ and relA1 cells (Fig. 4Bg). Thus, it seems that ppGpp is involved in triggering the mazEF death rather than affecting the toxic action of MazF.
Loss of viability of E. coli cells due to high temperatures (37, 38), oxidative stress (43), and DNA damage by UV irradiation (6), nalidixic acid exposure (9), and mitomycin C exposure (24) was previously described. Here we have shown that mazEF is involved in cell death under stressful conditions (Fig. 1 to 3). In other words, it appears that under these conditions the cells die because of the activation of an internal death machinery. It should be noted that we observed mazEF-mediated death only in a window of mildly stressful conditions. Within this window of conditions, the ΔmazEF derivatives were relatively resistant to the stresses (Fig. 1 to 3). However, we found that under extreme stressful conditions the ΔmazEF cells also died (Fig. 1 to 3). It is still not clear whether the ΔmazEF cells died because of the induction of some other toxin-antitoxin systems or through the inactivation of some essential component.
As mentioned above, members of our group have previously shown that mazEF-mediated death can be triggered by various stressful conditions that inhibit the expression of mazEF. These conditions include the inhibition of transcription by the overproduction of ppGpp (3, 13) and by the presence of antibiotics that inhibit transcription and/or translation (rifampin, chloramphenicol, and spectinomycin) (40). The inhibition of mazEF expression may also be involved in the induction of mazEF-dependent death at high temperatures (Fig. 1). At such stressful temperatures, σ70, the normal transcription factor, becomes inactivated; σ70 is replaced by periplasmic σE (37, 38). Judging on the basis of investigations of the promoter recognition sites of σE (46), it should not initiate the transcription of the mazEF genes. Recently, Sat et al. also showed that DNA damage due to thymine starvation prevents mazEF transcription (41). Whether the herein-described mazEF-mediated death induced by UV, nalidixic acid, mitomycin C, and H2O2 also inhibits mazEF expression remains to be determined.
To the best of our knowledge, this report is the first to suggest that ppGpp has a role in E. coli cell death caused by various stressful conditions (Fig. 4B). These results are in addition to previous results showing that artificial overproduction of ppGpp can by itself cause mazEF-dependent death, which seems to be due to the inhibition of the transcription of mazEF (3, 13). Furthermore, we have shown here that the combined action of stress conditions together with ppGpp affects only the induction of mazEF and not the MazF toxicity (Fig. 4Bg). More particularly, since death by MazF action does not require the presence of ppGpp (Fig. 4Bg) we have concluded that the combined action of stressful conditions together with ppGpp only affects the induction of mazEF and not the toxicity of MazF itself.
Many gram-positive and gram-negative bacteria carry genes that are homologous to mazEF (14, 33). It is interesting that in several gram-positive bacteria, including Staphylococcus aureus, Bacillus subtilis (33), and Bacillus anthracis, the mazEF homologues are located immediately upstream of the sigB gene (27, 33). This gene encodes σB, which is a general stress-response component (26). Moreover, during heat stress the mazEF homologues are cotranscribed with sigB, at least in S. aureus (18). Thus, it seems that the link between stressful conditions and the mazEF system is not limited to E. coli. Moreover, even in eukaryotes various stressful conditions, including DNA damage, nitric oxide exposure, and heat shock, have been found to trigger apoptosis (35, 44).
Recently, it has been suggested that the chromosomal toxin-antitoxin system mazEF and its nonhomologous chromosomal module relBE (10) are not involved in cell death but rather induce a state of reversible bacteriostasis (36). This view is based on experiments showing that ectopic overexpression of the toxin MazF or RelE inhibits translation and cell growth, which can be resumed if the cognate antitoxin is expressed at a later time (36). However these experiments were carried out within a small window of time, during only the 5 h after MazF expression. Using the same system, we found that overexpressing MazE after a prolonged overexpression of MazF did not lead to the reversal of cell death. Thus, it seems that there is a point of no return (15; S. Amitai and H. Engelberg-Kulka, unpublished results). Even so, it should be emphasized that ectopic overexpression drastically affects bacterial pathways and networks, so that the conditions in the cell no longer reflect the actual physiological conditions under which toxin-antitoxin systems mediate cell death. The experiments described here and previously (3, 13, 21, 40, 41) were done when the mazE module was located on the E. coli chromosome as a single copy and in its natural context, which seems to be more appropriate than ectopic overexpression for studying bacterial pathways and physiological networks. These experiments clearly show that E. coli mazEF-mediated cell death is triggered by various stressful conditions. It may be that under stressful conditions, mazEF has some additional regulatory function(s), as suggested previously (10, 17); however, the present report and other previous reports (3, 13, 21, 40, 41) relate only to mazEF-mediated cell death, i.e., loss of viability.
Acknowledgments
We thank G. Glaser for kindly supplying us with plasmid pQE30mazF. We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
The research described here was supported by grant 215/99-2 from the Israel Science Foundation, administered by the Israel Academy of Science and Humanities.
REFERENCES
- 1.Ades, S. E., I. L. Grigorova, and C. A. Gross. 2003. Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 3.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameisen, J. C. 2002. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 9:367-393. [DOI] [PubMed] [Google Scholar]
- 5.Bizanek, R., B. F. McGuinness, K. Nakanishi, and M. Tomasz. 1992. Isolation and structure of an intra-strand cross-link adduct of mitomycin C and DNA. Biochemistry 31:3084-3091. [DOI] [PubMed] [Google Scholar]
- 6.Burger, A., J. Raymer, and R. Bockrath. 2002. DNA damage-processing in E. coli: on-going protein synthesis is required for fixation of UV-induced lethality and mutation. DNA Repair (Amsterdam) 1:821-831. [DOI] [PubMed] [Google Scholar]
- 7.Cashel, M., D. R. Gentry, V. Z. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 8.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, S. K., K. Pedersen, F. G. Hensen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response elements: ChpAK/MazF and ChpBK cleave translated mRNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 10.Couturier, M., E. M. Bahassi, and L. Van Melderen. 1998. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 11.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and anti-death in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 13.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 95:15481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., B. Sat, and R. Hazan. 2001. Bacterial programmed cell death and antibiotics. ASM News 67:617-625. [Google Scholar]
- 15.Engelberg-Kulka, H., B. Sat, M. Reches, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death as a target for antibiotics. Trends Microbiol. 12:66-71. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg, E. C., G. C. Walker, and W. Siega. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 17.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 19.Gonzallez-Pastor, E., E. C. Hobbs, and R. Losick. 2003. Cannibalism of sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 20.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 21.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 23.Hochman, A. 1997. Programmed cell death in prokaryotes. Crit. Rev. Microbiol. 23:207-214. [DOI] [PubMed] [Google Scholar]
- 24.Iyer, V. N., and W. A. Szybalski. 1963. A molecular mechanism of mitomycin action: linking of complementary DNA strand. Proc. Natl. Acad. Sci. USA 50:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 26.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kullik, I., R. Jenni, and B. Berger-Bachi. 1998. Sequence of the putative alanine racemase operon in Staphylococcus aureus: insertional interruption of this operon reduces D-alanine substitution of lipoteichoic acid and autolysis. Gene 219:9-17. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda, Y., and E. Ohtsubo. 1994. Mapping and disruption of the chpB locus in Escherichia coli. J. Bacteriol. 176:5861-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 263:15699-15704. [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Mittenhuber, G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1:295-302. [PubMed] [Google Scholar]
- 34.Nakayama, K., K. Kusano, N. Irino, and H. Nakayama. 1994. Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulsed field gel electrophoresis and evidence for involvement of homologous recombination. J. Mol. Biol. 243:611-620. [DOI] [PubMed] [Google Scholar]
- 35.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of bacteriostatic conditions by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 37.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russel, M., and P. Model. 1984. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J. Bacteriol. 159:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka.2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skulachev, V. P. 2002. Programmed death phenomena: from organelle to organism. Ann. N. Y. Acad. Sci. 959:214-237. [DOI] [PubMed] [Google Scholar]
- 43.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 44.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 45.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]
- 46.Yura, T., M. Kanemori, and T. M. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 47.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Quing, and M. Inouye. 2003. MazF cleaves cellular mRNA specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]