Abstract
In dogs, papillomaviruses are thought to cause oral and cutaneous papillomas and pigmented plaques. Eight canine papillomaviruses have been fully sequenced to date. Four of these canine papillomaviruses, including Canis familiaris papillomavirus (CPV)-3, CPV-4, CPV-5, and CPV-8, were amplified from pigmented plaques. Given this recent identification of several different canine papillomaviruses within pigmented plaques, it is likely that there are additional papillomavirus sequences that have not been previously identified. The aim of this study was to detect papillomavirus DNA sequences from pigmented plaques and identify potentially novel PV sequences through nucleotide sequence analysis. Polymerase chain reaction was used to amplify DNA sequences of the papillomavirus L1 gene from 27 pigmented plaques. Identification of novel papillomavirus sequences was based upon less than 90% shared DNA homology to any known papillomavirus. Ten different papillomaviruses were detected within the pigmented plaques, including 6 novel PV sequences. CPV-4 was detected within 41% (11/27) of the pigmented plaques, while CPV-5 was identified within 2 pigmented plaques and CPV-3 within a single pigmented plaque. A previously identified novel papillomavirus sequence was identified within 2 pigmented plaques in this study. The remaining 11 pigmented plaques contained 6 papillomavirus DNA sequences that have not been previously reported. These novel PV sequences were most similar to papillomaviruses that have been detected within canine pigmented plaques.
Keywords: Canine papillomavirus, pigmented plaque
Papillomaviruses (PV) are epitheliotropic, often species and tissue-specific viruses within the family Papillomaviridae. They are circular, double stranded DNA viruses approximately 8 kb in length1. PVs are classified into genus, species, and types based on the nucleotide sequence of the L1 open reading frame (ORF).1,2 The complete viral genome for 8 canine papillomaviruses has been identified. CPV-14 is believed to cause oral papillomas and CPV-216, CPV-66, and CPV-76 were amplified from cutaneous papillomas. The remaining four canine papillomaviruses, CPV-314, CPV-415, CPV-56, and CPV-87 were all amplified from cutaneous pigmented plaques. An additional novel PV sequence (GU220384) has also been detected within pigmented plaques from a dog10.
Given this recent identification of multiple different CPVs amplified from canine pigmented plaques, it is likely that there are additional PV sequences that have not been previously identified. This study aimed to detect and sequence PV DNA from cutaneous pigmented plaques and identify the associated PV or novel PV sequences through nucleotide sequence analysis.
The pathology archives from the Veterinary Medicine Teaching Hospital (VMTH) at the University of California, Davis (Davis, CA), Idexx Laboratories Inc. (West Sacramento, CA), Veterinary Diagnostics (VDx) Veterinary Pathology Services (Davis, CA), and An Independent Biopsy Service (Venice, CA) were searched for surgical biopsy specimens from dogs with the following diagnoses: viral plaque and pigmented plaque. Twenty-seven biopsies from 24 individual dogs were identified and included in this study. All biopsies had been routinely formalin fixed and paraffin embedded (FFPE), and a 4-μm section cut for hematoxylin and eosin (H&E) staining. The H&E stained sections were reviewed by two board-certified pathologists (JL and VA) to confirm the diagnosis.
Immunohistochemistry was performed using a horseradish-peroxidase staining method on 4-μm FFPE sections of tissue from each biopsy. A rabbit polyclonal anti-bovine PVa antibody against the L1 capsid protein was used as the primary antibody at a dilution of 1:600 for 1 hour. The secondary antibody, biotinylated goat anti-rabbit IgGb, was used at a dilution of 1:500 for 30 minutes. The positive control was an oral papilloma. Omission of the primary antibody served as the negative control.
Genomic DNA (gDNA) was extracted from two 25 μm thick sections of FFPE tissue from each case using a commercially available kit following manufacturer’s recommended protocolc. A 450 base pair region of the L1 ORF was amplified using a single set of degenerate consensus primers (MY09 and MY11).9 PV DNA was not detected within 4 pigmented plaques (4/27) using the degenerate primer set. For these cases, specific primers for CPV-4 were used in order to increase the sensitivity for detection of PV DNA in these remaining cases. Specific primers for CPV-4 were chosen since this was the most commonly detected CPV based upon initial sequencing results of the other 23/27 pigmented plaques. The specific primers for CPV-4 have been previously published and amplify a 194 base pair fragment11.
Omission of gDNA template served as a negative control for all PCR reactions. The PCR reaction conditions used in this study have been previously published9. All PCR products were electrophoresed through 1.5% agarose and stained with a commercially available nucleic acid staind. The PCR products obtained from every sample (including those generated with specific primers) were purified using a commercially available kit following the manufacturer’s recommended protocole and submitted to a commercial DNA sequencing laboratory following their specificationsf. The nucleotide sequences generated from the sequencing laboratory were routinely trimmed and edited using commercially available softwareg.
Identification of new PV types is based upon <90% shared DNA homology to any known PV type3. Therefore, identification of the PV type associated with each pigmented plaque was defined as having >90% shared nucleotide identity within the L1 gene. If a PV sequence had less than 90% similarity with other PV types, this was considered to be a novel PV sequence. Pairwise nucleotide alignments were performed on all novel PV sequences identified in this study and all known CPVs using commercially available softwareg. A phylogenetic tree was constructed using the neighbor-joining method with commercially available softwareh. The phylogenic tree was based on an alignment of the novel PV nucleotide sequences identified in this study and the most closely related CPVs.
Histologically, the pigmented plaques were characterized by locally extensive epithelial hyperplasia, marked orthokeratotic hyperkeratosis, prominent clumped keratohyalin granules, and hyperpigmentation within the stratum granulosum (Figure 1A). Koilocytosis can be a subtle feature in pigmented plaques5 and it was only identified within three biopsies in this study. Viral inclusions can be identified in pigmented plaques12, although this is not a consistent feature11,15, and it was not identified in any of the biopsies in this study. The clinical history, including breed of dog and location of the pigmented plaque, for all cases is listed in Table 1.
Figure 1.
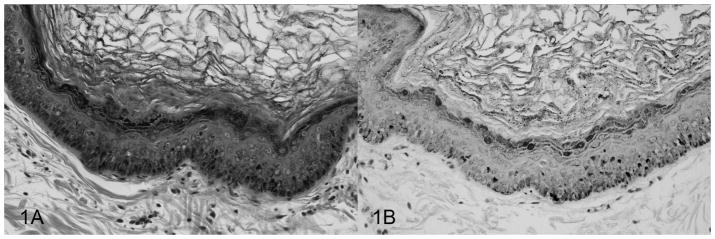
Photomicrographs of a canine pigmented plaque. A, pigmented plaque; locally extensive epithelial hyperplasia and orthokeratotic hyperkeratosis with prominent keratohyalin granules. H&E. B, pigmented plaque; demonstration of strong staining with the nuclei for papillomavirus antigen within keratinocytes in the proliferative epithelium. Anti-bovine papillomavirus antibody. Streptavidin-biotin staining method.
Table 1.
Breed of dog, location of pigmented plaque, and specific CPV* or novel PV† sequence (designated Novel CPV-A through F) detected for all biopsies included in this study.
| Breed | Location | CPV |
|---|---|---|
| Dachshund | Multiple: head, leg, abdomen | CPV-3 |
| Pointer | Left hind limb | CPV-4 |
| Pug | Right hind limb | CPV-4 |
| Chihuahua | Ventral thorax and right inguinal | CPV-4 |
| Bulldog | Multiple: abdomen | CPV-4 |
| Pug | Multiple: legs, chest, inguinal | CPV-4 |
| Wheaton terrier | Right and left hind limbs | CPV-4 |
| Australian terrier | Base of tail | CPV-4 |
| Unknown | Site unspecified | CPV-4 |
| Unknown | Site unspecified | CPV-4 |
| Pug | Site unspecified | CPV-4 |
| Pug | Site unspecified | CPV-4 |
| Mixed breed‡ | Limb | CPV-5 |
| Miniature pincher | Multiple: ventrum | CPV-5 |
| Pug | Ventral thorax | Novel CPV-A |
| Pug | Multiple sites: axillae, abdomen | Novel CPV-A |
| Pug | Site unspecified | Novel CPV-A |
| Unknown | Right medial thigh | Novel CPV-B |
| Unknown | Site unspecified | Novel CPV-B |
| Mixed breed‡ | Near ear | Novel CPV-C |
| Greyhound | Hock | Novel CPV-D |
| American hairless | Site unspecified | Novel CPV-E |
| Dalmatian | Abdomen | Novel CPV-E |
| Mixed breed‡ | Head | Novel CPV-F |
| Unknown | Site unspecified | Novel CPV-F |
| Mixed breed‡ | Limb | GU220384 |
| Mixed breed | Site unspecified | GU220384 |
Canine papillomavirus;
Papillomavirus;
Different biopsies from one dog
Immunohistochemistry revealed positive nuclear staining in all pigmented plaques. In some cases, the staining was evident within the keratinocytes throughout the stratum spinosum in the proliferative epithelium (Figure 1B) while in other cases, there were only one or two immunoreactive keratinocytes along the periphery of the acanthotic epithelium.
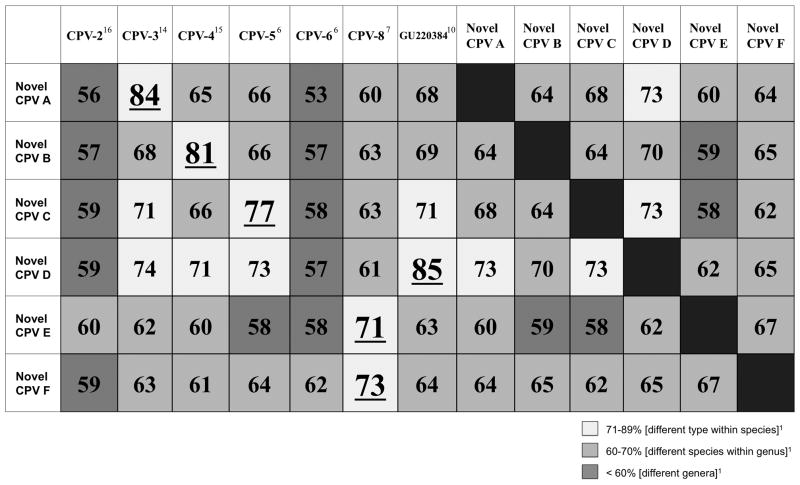
DNA from 10 different PVs was identified within the pigmented plaques (listed in Table 2). CPV-4 was detected within 11/27 pigmented plaques, CPV-5 within 2/27, and CPV-3 within 1/27. PV DNA sequences with 99% identity over 342 base pairs to the partial L1 PV sequence (GU220384) previously identified10 were detected within 2/27 of the pigmented plaques in this study (JQ040505). The 11 remaining pigmented plaques contained 6 previously unreported PV DNA sequences (designated novel CPV-A through F for the purpose of this study; listed in Table 2). The results of the percent pairwise nucleotide identities between the 6 novel PV sequences identified in this study and the most closely related CPVs are shown in Figure 2. Novel CPV-A (JQ040499) is most closely related to CPV-3 over 355 base pairs, novel CPV-B (JQ040500) with CPV-4 over 359 base pairs, novel CPV-C (JQ040501) with CPV-5 over 373 base pairs, and novel CPV-D (JQ040503) with GU220384 over 364 base pairs10. Both novel CPV-E (JQ040503) and CPV-F (JQ040504) were most closely related to CPV-8 over 342 and 372 base pairs, respectively. The nucleotide sequence for novel CPV-E was identical to a currently unpublished nucleotide sequence listed in the NCBI database (JF418155.1).
Figure 2.
Percent pairwise nucleotide identities comparing the 6 novel PV sequences identified in this study (designated novel CPV-A through F) to each other and the closest related CPVs. Nucleotide sequences were compared over a 342 to 373 base pair segment of the L1 gene. CPV-1 and CPV-7 shared <50% nucleotide identity to any novel PV sequence and were excluded from the table. The result with the highest percentage for each novel PV sequence is underlined. PV= papillomavirus; CPV= canine papillomavirus
The neighbor-joining phylogenetic tree generated from the alignment of an approximately 330 base pair segment of the L1 gene from the 6 novel PV sequences identified in this study and the most closely related CPVs is shown in Figure 3. Similar to the results of the pairwise nucleotide identities, the novel PV sequences are most closely related to CPV-3, CPV-4, CPV-5, and CPV-8.
Figure 3.
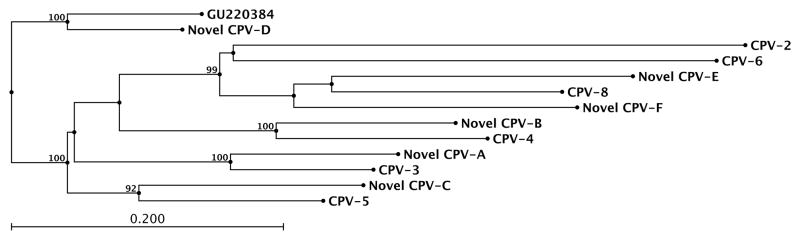
Neighbor-joining phylogenetic tree generated from an alignment of the nucleotide sequences (approximately 330 base pair segment) of the L1 gene from the 6 novel PV sequences identified in this study (designated novel CPV-A through F) and the most closely related CPVs. Bootstrap values were obtained from 100 resamplings of the data. Only bootstrap values >90% are shown. PV= papillomavirus; CPV= canine papillomavirus
CPV-8 DNA was not detected within any pigmented plaque in this study. However, the degenerate primers used were able to successfully amplify DNA from the most closely related CPVs, including the novel PV sequences, and it is likely that these primers would have been able to amplify DNA from CPV-8. Additionally, when the degenerate primers were aligned with the published nucleotide sequences for CPV-8, CPV-3, CPV-4, and CPV-5, there are similar numbers of mismatches at similar locations between the each of the target sequences and the primers. Given that CPV-3, CPV-4, and CPV-5 were successfully amplified with these mismatches, it is likely that these same primers should be able to amplify CPV-8. However, a co-infection with CPV-8 cannot be completely excluded.
A causal association between the PV detected and plaque formation cannot be definitively proven since PV DNA can be detected on non-lesional skin as well8. However, the presence of abundant PV L1 protein identified within the proliferative epithelium using immunohistochemistry is suggestive for causality. Additionally, PVs on non-lesional skin are maintained at low copy numbers (when compared to the large numbers of viral particles present in a productive PV infection in a papilloma or pigmented plaque), making is less likely that a rare PV associated with non-lesional skin would be amplified over the abundant PV DNA present within lesional skin from a single biopsy13. Furthermore, the study that detected PV DNA on normal skin extracted DNA from fresh samples, not FFPE biopsies, and the sensitivity for detection of a latent PV infection in formalin fixed skin is unknown.
Classification of PVs into different genera, species, and types is generally based upon the L1 nucleotide sequence identities, although interpretation is also based upon genome organization and biology of the virus1,2. The eight known CPVs identified to date are divided into three distinct genera1. CPV-1 and CPV-6 are classified into the Lambda papillomavirus genus, CPV-2 and CPV-7 into the Tau papillomavirus genus, and CPV-3, CPV-4, CPV-5, and CPV-8 into the Chi papillomavirus genus1. Definitive classification requires sequencing of the entire genome and analysis of the L1 gene1,2; however, the partial L1 nucleotide sequences generated in this study suggest the identification of 6 new CPV types. These 6 putatively novel CPVs are most closely related to CPV-3, CPV-4, CPV-5, and CPV-8 based upon the results of the pairwise nucleotide identities and phylogenetic analysis. Similar to their most closely related viruses, these putatively novel viruses are all associated with pigmented plaques. Based upon their similar sequence identity, shared tissue tropism, and clinical biology to CPV-3, CPV-4, CPV-5, and CPV-8, it is likely that these putatively novel viruses are within the Chi papillomavirus genus.
In conclusion, canine pigmented plaques were associated with a large diversity of viruses in this study, including CPV-3, CPV-4, CPV-5, six novel PV sequences not previously reported, and a seventh putatively novel CPV identified in a previous study10. These 6 novel PV sequences were most closely related to members of the Chi papillomavirus genus, all which have been identified in association with canine pigmented plaques.
Acknowledgments
The authors are grateful for the cases provided by Idexx Laboratories Inc. (West Sacramento, CA), Dr. John Peauroi at Veterinary Diagnostics (VDx) Veterinary Pathology Services (Davis, CA), Dr. Julie Yager at Yager-Best Histovet (Guelph, Ontario) and Dr. Emily Walder at An Independent Biopsy Service (Venice, CA).
Funding:
Funding for this study was provided by the Center for Companion Animal Health, School of Veterinary Medicine, University of California, Davis.
Footnotes
Dako Cytomation, Carpinteria, CA
Vector Laboratories, Burlingame, CA
DNeasy tissue kit, Qiagen, Valencia, CA
Cybr gold, Invitrogen, Carlsbad, CA
Promega Wizard SV Gel and PCR Clean-Up System, Promega, Madison, WI
DNA sequencing facility, University of California, Davis, CA
Vector NTI Software® sequence analysis software, Invitrogen, Carlsbad, CA
CLC Bio’s free CLC Work Bench, Germantown, MD
Declaration of conflicting interests:
The authors declare that they had no conflicts of interest.
References
- 1.Bernard HU, Burk RD, Chen Z, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–9. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Villiers EM, Fauquet C, Broker TR, et al. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 3.de Villiers EM, Whitley C, Gunst K. Identification of new papillomavirus types. Methods Mol Med. 2005;119:1–13. doi: 10.1385/1-59259-982-6:001. [DOI] [PubMed] [Google Scholar]
- 4.Delius H, Van Ranst MA, Jenson AB, et al. Canine oral papillomavirus genomic sequence: a unique 1.5-kb intervening sequence between the E2 and L2 open reading frames. Virology. 1994;204:447–52. doi: 10.1006/viro.1994.1552. [DOI] [PubMed] [Google Scholar]
- 5.Gross TL. Skin diseases of the dog and cat: clinical and histopathologic diagnosis. 2. Ames, Iowa: Blackwell Science; 2005. [Google Scholar]
- 6.Lange CE, Tobler K, Ackermann M, et al. Three novel canine papillomaviruses support taxonomic clade formation. J Gen Virol. 2009;90:2615–21. doi: 10.1099/vir.0.014498-0. [DOI] [PubMed] [Google Scholar]
- 7.Lange CE, Tobler K, Lehner A, et al. A case of a canine pigmented plaque associated with the presence of a Chi-papillomavirus. Vet Dermatol. 2011 doi: 10.1111/j.1365-3164.2011.01007.x. Epub. 2011/09/03. [DOI] [PubMed] [Google Scholar]
- 8.Lange CE, Zollinger S, Tobler K, et al. Clinically healthy skin of dogs is a potential reservoir for canine papillomaviruses. J Clin Microbiol. 2011;49:707–9. doi: 10.1128/JCM.02047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manos M, Ting Y, Wright D, et al. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. In: Furth M, Greaves M, editors. Molecular diagnostics of human cancer. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. pp. 209–14. [Google Scholar]
- 10.Munday JS, O’Connor KI, Smits B. Development of multiple pigmented viral plaques and squamous cell carcinomas in a dog infected by a novel papillomavirus. Vet Dermatol. 2011;22:104–10. doi: 10.1111/j.1365-3164.2010.00913.x. [DOI] [PubMed] [Google Scholar]
- 11.Narama I, Kobayashi Y, Yamagami T, et al. Pigmented cutaneous papillomatosis (pigmented epidermal nevus) in three pug dogs; histopathology, electron microscopy and analysis of viral DNA by the polymerase chain reaction. J Comp Pathol. 2005;132:132–8. doi: 10.1016/j.jcpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Stokking LB, Ehrhart EJ, Lichtensteiger CA, Campbell KL. Pigmented epidermal plaques in three dogs. J Am Anim Hosp Assoc. 2004;40:411–7. doi: 10.5326/0400411. [DOI] [PubMed] [Google Scholar]
- 13.Stubenrauch F, Laimins LA. Human papillomavirus life cycle: active and latent phases. Semin Cancer Biol. 1999;9:379–86. doi: 10.1006/scbi.1999.0141. [DOI] [PubMed] [Google Scholar]
- 14.Tobler K, Favrot C, Nespeca G, Ackermann M. Detection of the prototype of a potential novel genus in the family Papillomaviridae in association with canine epidermodysplasia verruciformis. J Gen Virol. 2006;87:3551–7. doi: 10.1099/vir.0.82305-0. [DOI] [PubMed] [Google Scholar]
- 15.Tobler K, Lange C, Carlotti DN, et al. Detection of a novel papillomavirus in pigmented plaques of four pugs. Vet Dermatol. 2008;19:21–5. doi: 10.1111/j.1365-3164.2007.00640.x. [DOI] [PubMed] [Google Scholar]
- 16.Yuan H, Ghim S, Newsome J, et al. An epidermotropic canine papillomavirus with malignant potential contains an E5 gene and establishes a unique genus. Virology. 2007;359:28–36. doi: 10.1016/j.virol.2006.08.029. [DOI] [PubMed] [Google Scholar]