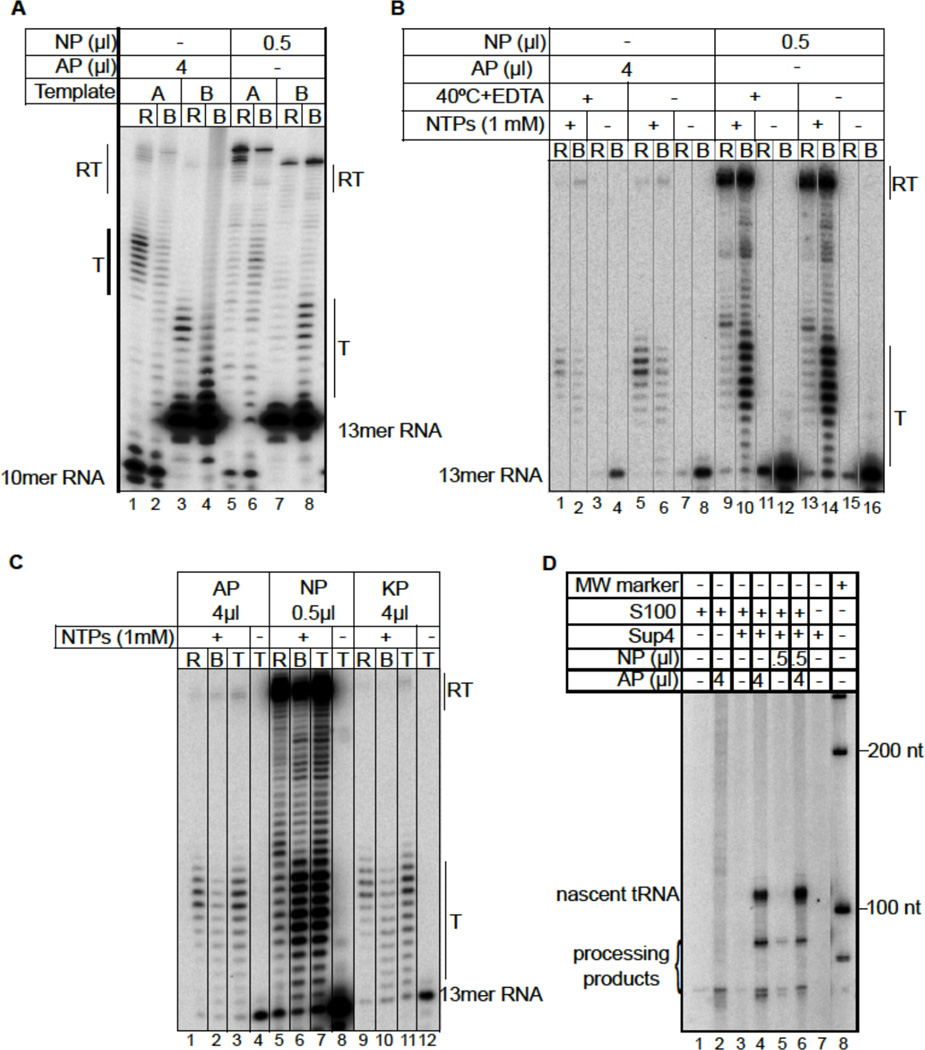
Figure 1. Direct comparisons of Nielsen (NP) polymerase and our (AP) pol III for termination and TFIIIB+TFIIC-dependent initiation.
A) Elongation complexes were assembled on templates A and B and immobilized, through the RNA polymerase His6 tag, on NiNTA agarose. Ribonucleoside triphosphates (NTPs) were added, incubated for 10 minutes and the released (R) and bound (B) transcripts separated, as indicated above each lane. B) Comparison of EC formation and transcription by AP and NP on immobilized template B. Quantities of AP and NP used are the same as in A; reactions were for 1 minute. Lanes 1–4, 9–12 were done as described (3) while lanes 5–8, 13–16 were done in the absence of EDTA and 40°C incubation. C) Comparison of transcript release by three different preparations of pol III. AP was purified on NiNTA agarose (3) (used in other figures) while untagged KP was highly purified by sequential ion exchange chromatography (6). Quantities of AP and NP used are the same as in A and B. Released, bound and total (T) transcripts are indicated. D) Pol III-specific transcription assay. Preinitiation complexes were formed by incubating a SUP4 tRNA gene plasmid with S. cerevisiae S100 extract deficient in endogenous pol III activity (see text) followed by addition of AP or NP as indicated above the lanes. Reactions were started by the addition of NTPs (with α-[32P] GTP) and MgCl2, and incubated for 20 minutes.