Abstract
In this account, we varied PEGylation density on the surface of hydrogel PRINT nanoparticles and systematically observed the effects on protein adsorption, macrophage uptake, and circulation time. Interestingly, the density of PEGylation necessary to promote a long-circulating particle was dramatically less than what has been previously reported. Overall, our methodology provides a rapid screening technique to predict particle behavior in vivo and our results deliver further insight to what PEG density is necessary to facilitate long-circulation.
Keywords: PRINT, nanoparticle, PEGylation, protein adsorption, macrophage association, pharmacokinetics
The full potential for nanotechnology has yet to surface even after decades of groundbreaking research. Nano-based drug delivery systems have produced advancements toward next generation cancer therapy (Doxil and Abraxane), but rapid elimination of nanoparticles (NPs) from the body continues to mitigate progress.1–4 Following intravenous (IV) administration, NPs are rapidly removed from circulation and accumulate mainly in the liver and spleen due to opsonization and recognition by the mononuclear phagocyte system (MPS).2,5–7 There have been many attempts to reduce uptake of nanoparticles by the MPS by controlling the physicochemical characteristics of the particles, such as size, surface charge, hydrophilicity, and surface functionality.2,5,8–10 Generally, nanoparticles that have a mean diameter of 200 nm or less with a neutral to negative ζ-potential and an extended hydrophilic polymer surface exhibit prolonged blood circulation.2,5,10,11
Poly(ethylene glycol) (PEG) has been extensively used on a variety of nanoparticle systems to increase surface hydrophilicity and improve circulation half-life by decreasing interactions with blood proteins and MPS cells.1,2,6,12–15 PEGylation can be conducted with several unique techniques using diblock PEG derivatives, or by covalently attaching, entrapping, or adsorbing PEG chains onto the surface of a nanoparticle.1,6,14 The effects of PEGylation are highly dependent on the PEG molecular weight (MW), polymer chain architecture, and surface density of the PEG coating, which leads to transitions in PEG conformations at the surface.13,14,16 There is a general consensus that stealth properties can be achieved by coating with a high density of PEG with MW ranging from 2K to 10K.14,17–21 Despite PEG extending particle circulation in vivo, there is no general standard as to what surface density is needed to accomplish this goal. This stems partially from a lack of easy and efficient PEG quantification strategies.
A number of techniques are currently used to measure degree of PEGylation on NP surfaces, yet few have reported precise techniques for PEG quantification on polymeric NPs.7 Most techniques described in literature are qualitative assessments of PEGylation such as NMR (PEG peak typically observed ~3.65 ppm), dynamic light scattering, and ζ-potential.22,23 In some instances, PEG surface density is determined by simply assuming complete saturation of PEG on the surface of the nanoparticle, and therefore surface PEG density is reported based upon PEG size and nanoparticle surface area. For other nanoparticle formulations, PEG is incorporated into the nanoparticle matrix and PEG surface coverage is then based upon the weight or mole percent of the PEG incorporated, assuming that all the PEG chains are on the surface and not embedded within the particle.14 Both of these methods typically offer an overestimation of PEG grafting. Few quantitative methods exist to accurately determine PEG surface density. Chromatography, such as size exclusion chromatography (SEC) and high-performance liquid chromatography (HPLC), are well-documented quantitation techniques for PEGylated liposomes.6,24 Thermogravimetric analysis (TGA) is used to calculate PEG weight, but is restricted to metallic NPs.25 Surface plasmon resonance (SPR) is another method to determine PEG loadings with the caveat that quantification is generally characterized on flat substrates.7 X-ray photoelectron spectroscopy (XPS) and Raman analyses can yield detailed information on conformation and quantification, however, these methods are beyond typical analytical practices.7,21 Fluorescent assays are commonly used for rapid PEG quantification and are amenable for a variety of nanoparticle core compositions.7,24,26
PEGylation density is commonly described in terms of the conformation that surface-bound PEG chains achieve, which is based upon the Flory radius (RF) of the PEG graft, the distance (D) between PEG grafts, or the length/thickness (L) of the grafted PEG layer.7,27,28 Upon the basis of these parameters, there are two main conformations that PEG chains can acquire, “mushroom” or “brush”.27,29,30 Mushroom conformation is dictated by having a low density PEG coverage, where D > RF, and therefore the PEG chains are not fully extended away from the nanoparticle surface, resulting in a thin PEG layer. As D decreases to that of RF, PEG chains arrange in a brush conformation with the PEG chains extending away from the nanoparticle surface, resulting in a thick layer. When L > 2RF, the brush conformation was further defined by Damodaran et al. to be a dense brush.31 Thus by knowing the three parameters, D, RF, and L, the conformation of the PEG chain can be distinguished. Unfortunately, there is a disconnect throughout literature as to what parameters are reported. For ease of comparing degree of PEGylation, we have compiled data (Table 1) from the literature consisting of these key parameters and how they affect protein binding, macrophage uptake and circulation half-life. Equations 1–3 and Supporting Information S1–S2 were used to calculate unreported values (see Supporting Information for details on calculations). Upon the basis of this compilation, it appears that the beneficial effects of PEGylation (protein and macrophage rejection and enhanced circulation half-life) occur when the PEG grafts are in the dense brush regime.
Table 1.
Comparison of Parameters Used to Determine Surface PEG Density on Nanoparticles and Their Biological Interactions
| PEG MW (g/mol) | RF (nm) | D (nm) | L (nm) | regime | protein resistance | macrophage resistance | t1/2 (h) | ref |
|---|---|---|---|---|---|---|---|---|
| 5K | 5.96 | 3.2 | 8.80 | brush | no | no | - | 14 |
| 5K | 5.96 | 2.8 | 9.80 | brush | - | no | - | 46 |
| 5K | 5.96 | 2.5 | 10.6 | brush | - | no | 0.3 | 39 |
| 3.4K | 4.76 | 1.8 | 8.70 | brush | no | - | - | 55 |
| 2K | 3.5 | 2.6 | 10.2 | dense brush | - | yes | - | 6 |
| 5K | 5.96 | 2.2 | 11.5 | dense brush | yes | yes | - | 14 |
| 5K | 5.96 | 2.0 | 12.4 | dense brush | - | yes | - | 46 |
| 5K | 5.96 | 1.8 | 13.1 | dense brush | - | yes | 34.3 | 39 |
| 5K | 5.96 | 1.7 | 13.5 | dense brush | - | yes | 6 | 19 |
| 3.4K | 4.76 | 1.7 | 9.50 | dense brush | yes | - | - | 55 |
| 5K | 5.96 | 1.3 | 16.6 | dense brush | yes | yes | 8.5 | 18 |
| 5K | 5.96 | 0.98 | 20.0 | dense brush | yes | yes | 11.3 | 18 |
The aim of this study was to evaluate PRINT hydrogel nanoparticles (80 nm × 80 nm × 320 nm) with varying PEG surface coverage. PEG conformation was determined through standard plate-reader analysis of fluorescein-labeled PEG grafts. Herein we report predictive screening methods that can rapidly asses the circulation fate of our PEGylated PRINT nanoparticles. We explored the use of in vitro assays (protein binding and macrophage uptake) to predict in vivo circulation behavior. Intravital microscopy was used to quickly screen the circulation profiles of our particles, and a biodistribution study was conducted to look at long time points and confirm the results of in vitro assays.
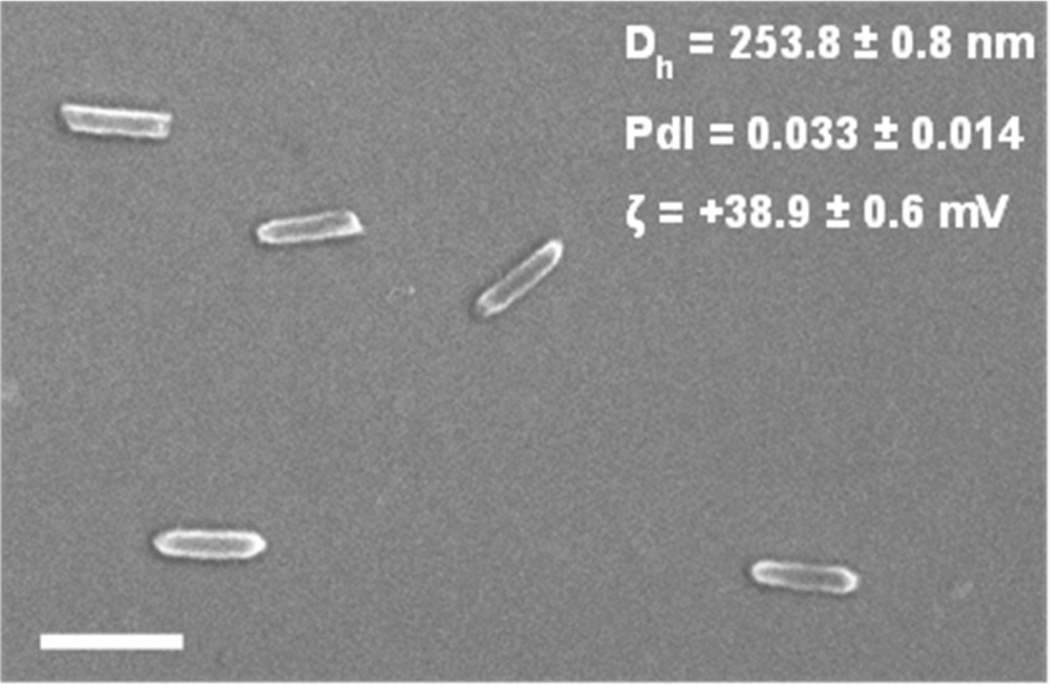
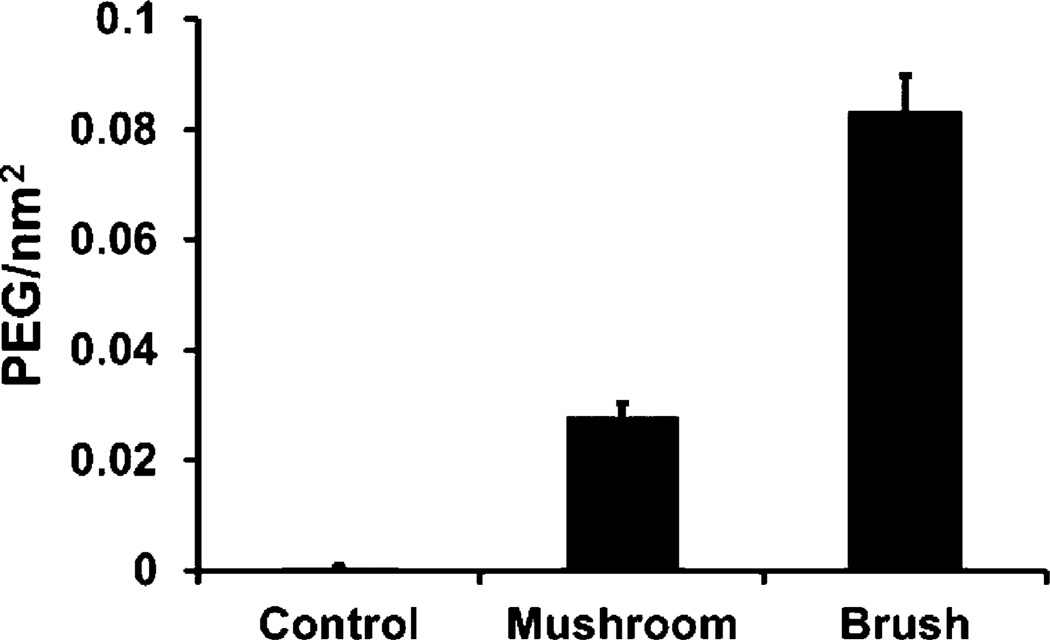
We synthesized calibration quality 80 nm × 80 nm × 320 nm hydrogel particles with a narrow polydispersity index (PdI) on a continuous roll-to-roll lab line using the PRINT process (Figure 1). Utilizing this roll-to-roll method, PRINT particles were successfully made continuously at a rate of 360 mg/hour, which also resulted in highly uniform populations of particles. Particles were fabricated with amine functional handles (from the amino ethyl methacrylate), which were reacted with either a methoxy- or fluorescein-terminated PEG5k-succinimidyl carboxy methyl ester (PEG5k-SCM). PEG grafting density was controlled by varying the PEG5k-to-nanoparticle ratio, and was quantified by fluorescence measurements of the fluorescein-PEG5k labeled particles. The fluorescence signal was correlated to the concentration of fluorescein-PEG5k with a standard curve. PEG density was calculated based upon the concentration of particles in solution and the surface area of hydrated particles (equations can be found in Supporting Information). PEG density was calculated to be 0.083 ± 0.006 and 0.028 ± 0.002 PEG/nm2 for the high and low PEG density NPs, respectively (Figure 2). In these calculations for PEG density, we assumed a uniform layer of PEG on the surface of the particle. Even though hydrogel particles swell, leading to possible penetration into the particle, it is entropically unfavorable for a large PEG molecule to permeate the hydrogel.
Figure 1.
Scanning electron image of 80 nm × 80 nm × 320 nm PRINT hydrogel nanoparticles, scale bar represents 500 nm. Dh, PdI, and ζ-potential were measured on a Malvern Zetasizer.
Figure 2.
PEG density quantification (n = 6) of 80 nm × 320 nm particles reacted with 14.0 and 2.0 mg of fluorescein-PEG5k-SCM resulting in brush (0.083 ± 0.006 PEG/nm2) and mushroom (0.028 ± 0.002 PEGs/nm2) conformations, respectively. A control was conducted with 14.0 mg of deactivated fluorescein-PEG5k-SCM leading to a negligible PEG density.
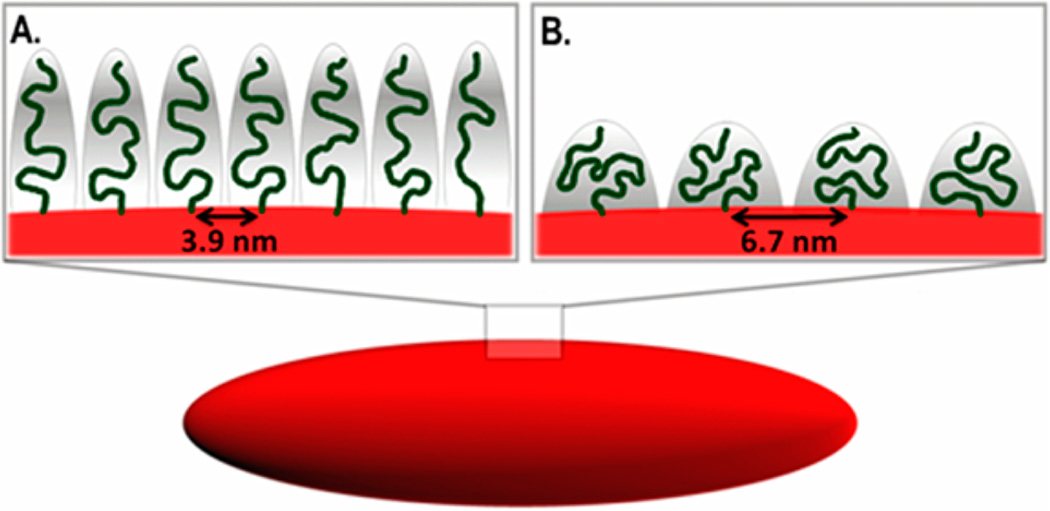
PEG density was converted to the area that one PEG chain occupies (A) and the distance between PEG graft sites (D) was calculated (eq 3) to be 3.9 and 6.7 nm for the high and low PEG density particles, respectively.32 As discussed in the introduction, the conformation of the PEG grafts is dictated by the relationship between D and RF. The Flory radius (eq 1) is determined by the number of monomers per polymer chain (N), and the length of one monomer (α).29 For PEG5k, α = 0.35 nm and N = 113, therefore RF = 5.96 nm.33,34 Upon the basis of RF and D, the PEG polymers were in the mushroom conformation for the low PEG density NPs and in the brush conformation for the high PEG density NPs (Figure 3). PEG brush thickness was calculated (eq 2) to be 7.9 ± 0.1 nm for high PEG density NPs. Upon the basis of Damodaran criterion, the high PEG density NPs were not in the dense brush regime.31 Hereafter, particles will be referred to as PEG mushroom and PEG brush NPs.
| (1) |
| (2) |
| (3) |
Figure 3.
Cartoon representation of 80 nm × 80 nm × 320 nm PRINT hydrogel nanoparticles with functionalized PEG in brush (A) and mushroom (B) conformations.
For all of the in vitro and in vivo studies, particles were PEGylated using the same method outlined above but with a methoxy-PEG5k-SCM. Following PEGylation, the NPs were acetylated to quench any remaining unreacted amines on the particle surface, resulting in particles with a slightly negatively ζ-potential (Table 2). The acetylation reaction was necessary because nanoparticles with positively charged surface groups typically exhibit higher protein adsorption and cell association than neutral or negative groups.35–37 After acetylation, particles were characterized by dynamic light scattering (DLS) and degree of PEGylation was determined by fluorescence (Table 2). DLS was used to determine particle hydrodynamic diameter (Dh), PdI, and ζ-potential (electrophoretic mobilities can be found in Table S1 in Supporting Information). Dh values listed in the table are not indicative of quantitative length scales and only act to compare relative trends between different particle types. Calculations for D and L were based upon measuring particle dimensions from SEM and AFM images (calculations can be found in Supporting Information).
Table 2.
Nanoparticle Characterization
| Sample | Dh (nm)a | PdIa | ζ (mV)a | D (nm)b | L (nm)b |
|---|---|---|---|---|---|
| non-PEGylated | 236.1 ± 4.4 | 0.063 ± 0.008 | −27.9 ± 0.9 | - | - |
| PEG mushroom | 341.2 ± 5.3 | 0.101 ± 0.030 | −8.15 ± 0.15 | 6.7 ± 0.3 | 5.5 ± 0.2 |
| PEG brush | 347.7 ± 7.7 | 0.042 ± 0.014 | −11.6 ± 0.5 | 3.9 ± 0.1 | 7.9 ± 0.1 |
Measured by dynamic light scattering.
Calculated from fluorescence analysis.
It is well established that phagocytosis is facilitated by the adsorption of plasma proteins to NP surfaces and that varying the surface chemistry of NPs can affect the identity and amount of proteins adsorbed.15,35,36,38–40 One metric commonly used to characterize “stealth” of PEGylated NPs is through in vitro classification including protein binding and/or macrophage uptake experiments.14,19,21,36,39–46 In an effort to typify our PEGylated PRINT nanoparticles, we conducted a comprehensive study of protein binding and macrophage association on NPs with varying surface coverage of PEG.
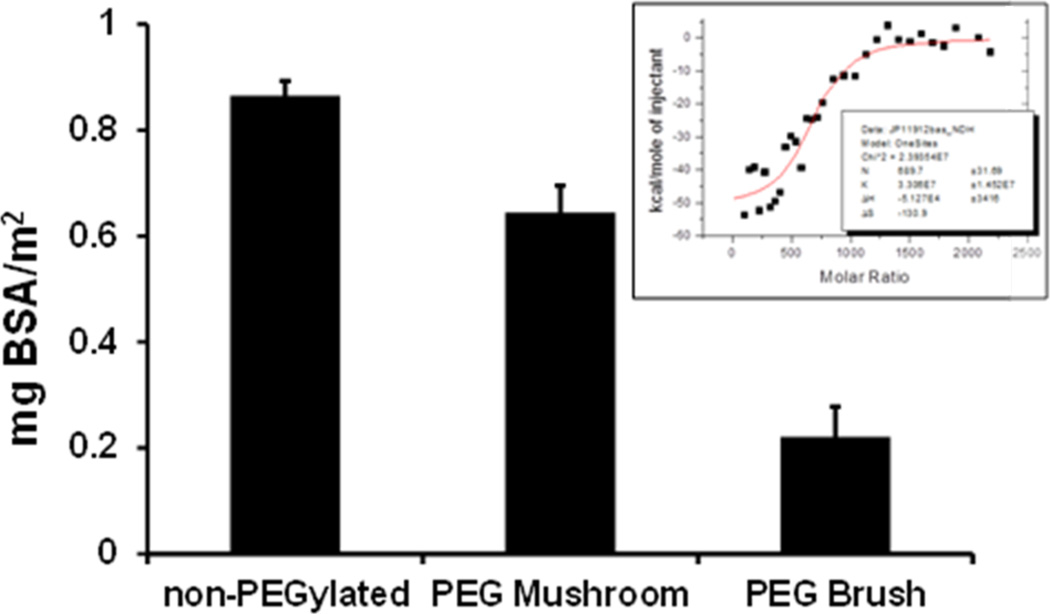
Previous reports indicated that varying the surface chemistry of particles can affect both the identity and amount of proteins adsorbed.35,36,46 Isothermal titration calorimetry (ITC) is the gold standard for measuring biomolecular interactions and has recently been utilized to determine nanoparticle–protein interactions.47–52 Following a protocol by Lindman and coworkers, we used ITC to screen the effect that PEG conformation (mushroom vs brush) had on protein adsorption.47 We investigated the interaction of bovine serum albumin (BSA) with non-PEGylated, PEG mushroom, and PEG brush NPs. BSA was chosen as a model protein because albumin is the most abundant protein in serum and is typically a major component in the protein corona surrounding nanoparticles.14,35,36,53,54 The stoichiometry of proteins bound per nanoparticle was derived from a simple fit to the data of a one-site binding model using ORIGIN software (Figure 4 inset). Figure 4 shows the amount of protein bound per particle significantly decreased with increasing surface PEG density. These results support previous findings where an increase in surface PEG density leads to decreased protein adsorption.14,42,43,46,55 However, in comparison to literature, we are reporting protein rejection properties at much lower surface PEG density.14
Figure 4.
Amounts of adsorbed BSA on non-PEGylated, PEG mushroom and PEG brush NPs. The protein amount is expressed as mg BSA/m2 of NP surface area. Error bars represent standard deviation for n = 3. The insert is a representative image raw data collected from the isothermal titration calorimetry experiments.
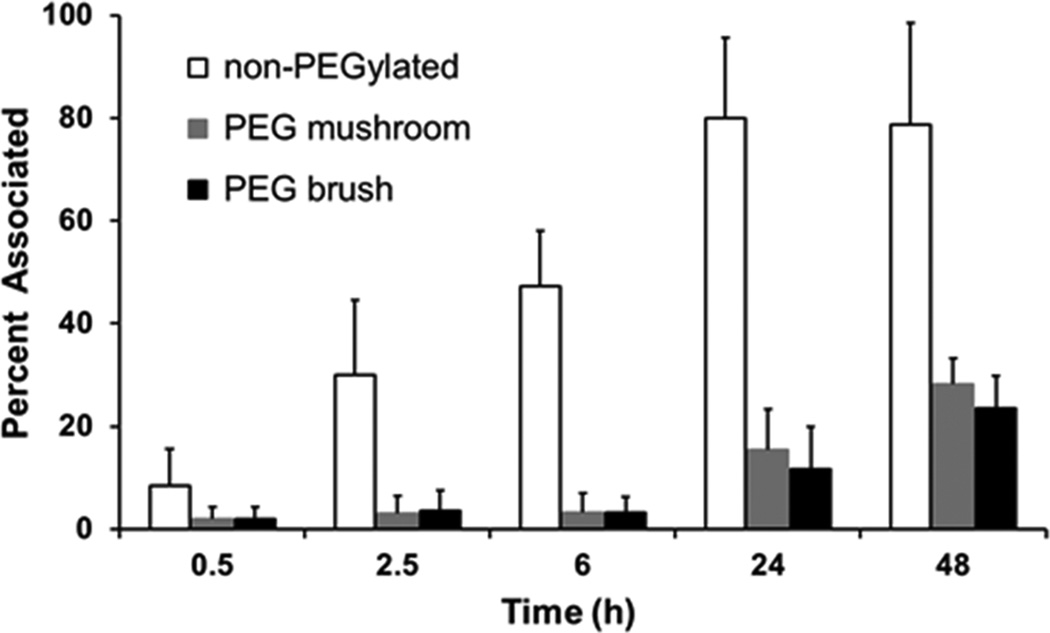
The ITC studies clearly indicate that surface PEG density regulated the quantity of adsorbed BSA, therefore we hypothesized that it would also regulate the efficiency of macrophage uptake. To address macrophage association, nanoparticles were incubated in cell culture medium with the MH-S cells. Under these conditions, protein adsorption (from the cell culture medium) onto the surface of the nanoparticles can lead to cellular recognition by the macrophages and thus initiate uptake. We measured nanoparticle association with macrophages as a function of PEG surface density and time (Figure 5). At early time points (0.5 to 6 h) the PEG mushroom and PEG brush particles behaved the same and were associated with MP-S cells 4–14 times less than non-PEGylated NPs. After 24 h, a slight difference between the PEG mushroom and PEG brush NPs became evident. These findings are in agreement with literature; as PEG surface coverage increases both protein adsorption and macrophage association decreases.31,35,43,46 However, as shown in Table 1, these protein/macrophage rejection properties are not typically observed until PEG grafts are in the dense brush regime, whereas we observed these properties for both PEG mushroom and PEG brush.
Figure 5.
Percent of MH-S cells associated with non-PEGylated, PEG mushroom, and PEG brush nanoparticles after various incubation times.
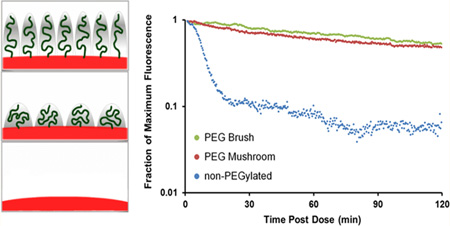
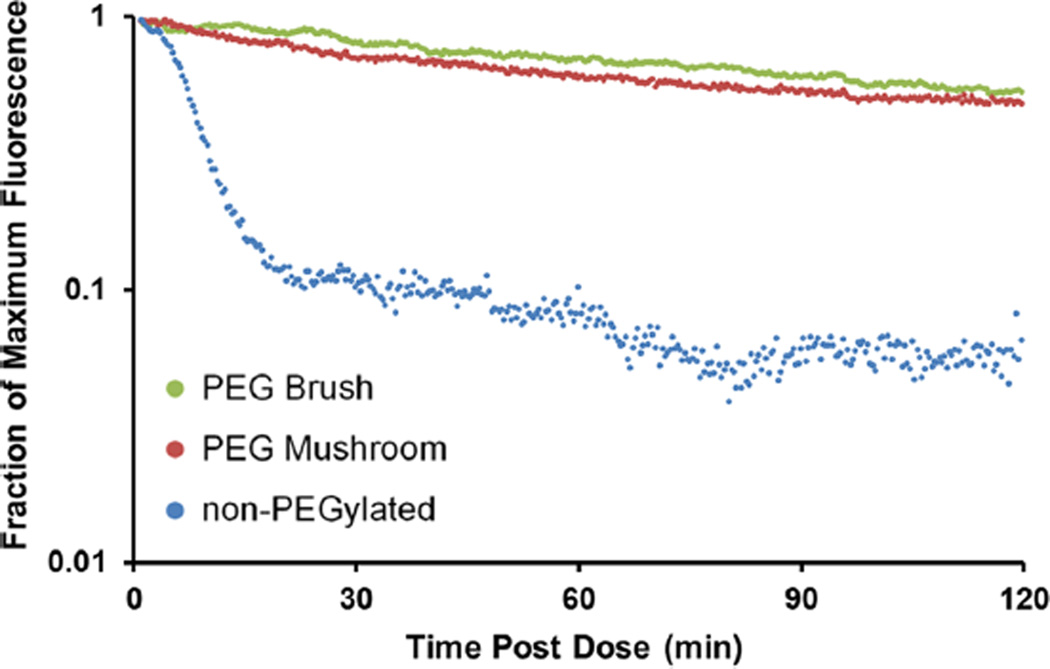
In an effort to translate in vitro data to circulation time in vivo, intravital microscopy (IVM) was utilized to track fluorescent nanoparticles in the ear vasculature of anesthetized mice over 2 h. Blood clearance curves of particles with varying surface PEG densities were generated from IVM fluorescence measurements and are displayed in Figure 6. The data plot clearly indicates that PEGylation is essential for extending circulation times. Both mushroom and brush PEG particles appeared to have similar long-circulation profiles, whereas the non-PEGylated particles were cleared rapidly. These findings are in line with those observed from the macrophage assay, where the PEG mushroom and PEG brush NPs exhibited nearly identical association.
Figure 6.
Intravital microscopy circulation profiles based upon fraction of maximum fluorescence remaining for non-PEGylated, PEG mushroom, and PEG brush nanoparticles.
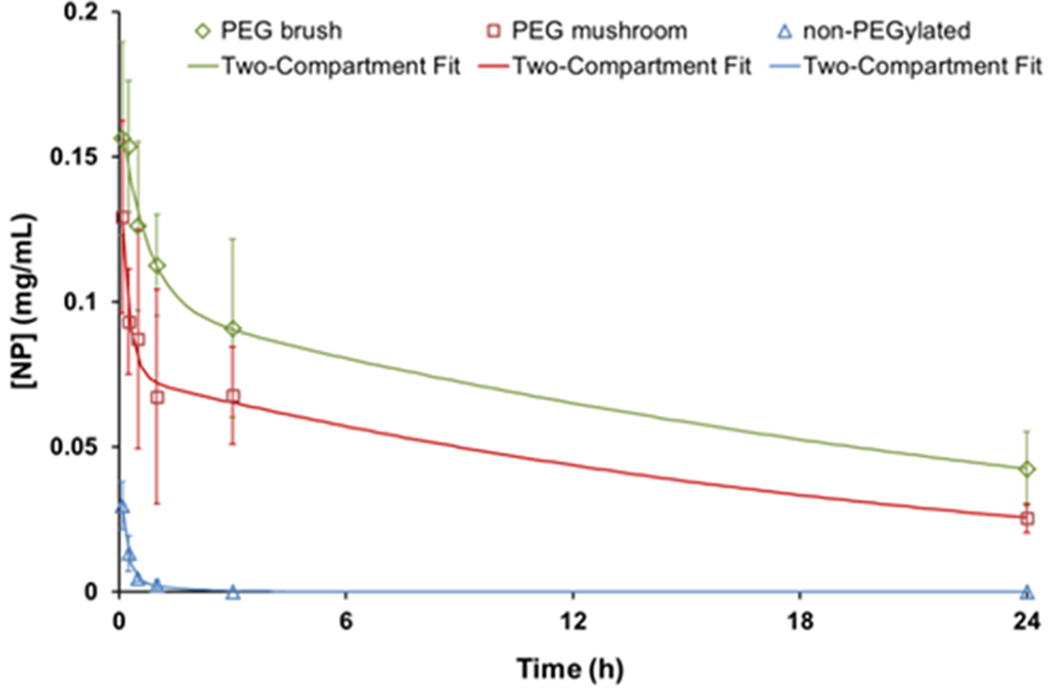
In order to distinguish the difference between a mushroom and brush conformation, the concentration of particles in blood at longer time points was investigated. Mice were dosed with particles and sacrificed at certain time points extending out to 24 h. Figure 7 depicts blood circulation curves with accompanying two-compartment PK model fits. The trend among the particle types closely follows that of the IVM profiles. The non-PEGylated particles exhibited the same rapid clearance. However, further delineation between a mushroom and brush was displayed during the initial time points (>3 h), with a consistent difference at 24 h.
Figure 7.
Concentration of nanoparticles in blood over time (open diamond, square, and triangle). Data is fit to a two-compartment PK model (solid lines). Error bars represent standard deviation for n = 4.
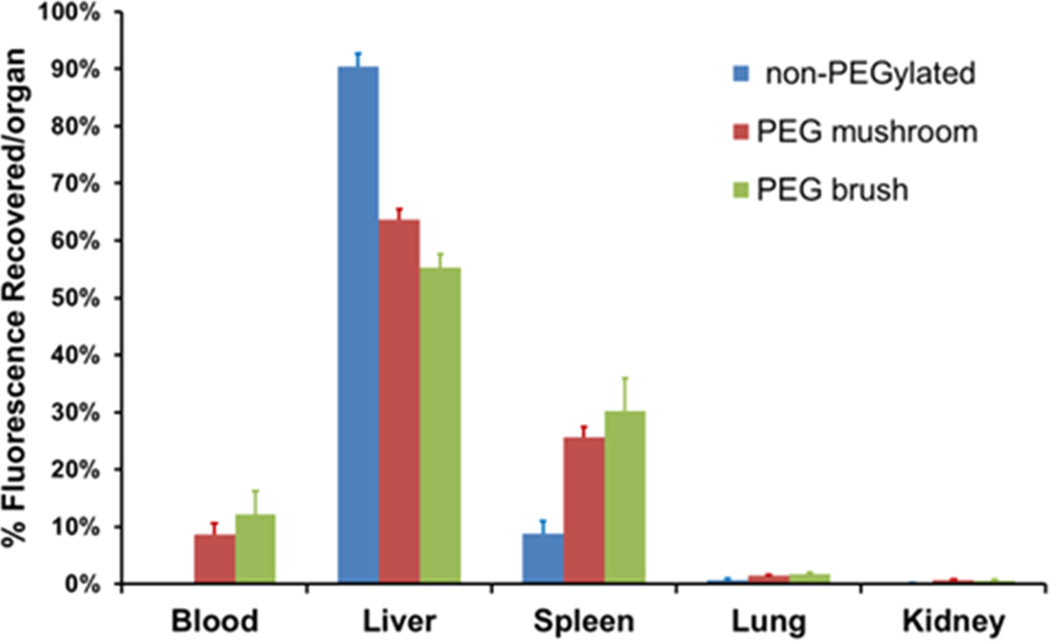
Additionally, the MPS organs were resected and analyzed for particle fluorescence to determine if organ accumulation was also dependent on surface PEG conformation. Figure 8 shows the organ accumulation as a function of PEG surface density at 24 h postinjection (organ accumulation at earlier time points can be found in Supporting Information). The blood concentration of non-PEGylated particles was undetectable at 24 h. Liver accumulation was high for all particle types but showed decreased uptake as PEG density increased. Furthermore, as PEG density increased, so did splenic uptake. This has been observed for other PEGylated polymeric particles compared to non-PEGylated particles, where a shift from liver to splenic accumulation was observed.19,56,57 It was hypothesized that longer circulation times allow more exposure of the particles to the spleen, resulting in higher splenic filtration and uptake by resident phagocytic cells.57 Our data fits this theory with a marked increase in splenic fluorescence between non-PEGylated and PEGylated particles, and a small increase as PEG density increased further. Thus, while PEGylation increases circulation time via delayed phagocytosis, a particle’s fate is ultimately the MPS organs.19 While the exact mechanism is still unclear, this trend has been observed for over a decade with many particle sizes, shapes, and compositions.1,5,14,30,58,59 Finally, lung and kidney accumulation was minimal and likely attributed to fluorescence in residual blood within the organ.
Figure 8.
Biodistribution of nanoparticles at 24 h postinjection, expressed as percent recovered fluorescence per organ. Error bars represent standard deviation from n = 4.
Pharmacokinetic (PK) parameters were determined from the blood concentration curves for each particle type. Data was subjected to both one- and two-compartment analysis, with elimination from the central compartment, using PKSolver. A two-compartment model was found to be the best for all three particle types upon comparison of Akaike Information Criterion values. Table 3 lists the secondary parameters calculated from the primary constants of integration from the two-compartment model (Supporting Information Table S2). As suggested by IVM data, PEGylation clearly extends particle circulation on longer time scales. The half-life (beta-phase) increased from 0.89 h for non-PEGylated to 15.5 h for PEG mushroom and 19.5 h for PEG Brush. Clearance and AUC (parameters important in drug delivery applications) also increased as PEG density increased. A PEG brush surface resulted in a 200-fold and 1.5-fold decrease in clearance versus a bare and PEG mushroom surface, respectively. Significant improvements in PK values upon PEGylation are widely cited in literature with similar results.14,57,59 Furthermore, longcirculation half-lives are generally only observed for NPs with a dense PEG brush surface. Again, we have shown these same improvements for NPs with PEG mushroom and brush surfaces.
Table 3.
Pharmacokinetic Parameters from Two-Compartment Model Fit of Blood Concentration Data
|
t1/2, α (h) |
t1/2, β (h) |
CL (mL/h) |
Vd (mL) |
AUC0-t (h*mg/ mL) |
|
|---|---|---|---|---|---|
| non-PEGylated | 0.116 | 0.892 | 17.5 | 4.75 | 0.013 |
| PEG mushroom | 0.144 | 15.5 | 0.128 | 1.40 | 1.12 |
| PEG brush | 0.478 | 19.5 | 0.087 | 1.48 | 1.66 |
We have developed a method for fabricating calibration quality hydrogel nanoparticles via a continuous process. This allowed us to conduct comprehensive in vitro and in vivo studies using highly uniform populations of particles and to fully characterize how NPs behave as a function of surface PEG density. PEG density was varied using a highly tunable method that could easily be quantified by fluorescence. Protein adsorption and macrophage association were significantly reduced upon PEGylation of the NPs. An increase in PEG density, resulting in a conformation change from mushroom to brush, showed slight improvements in these in vitro studies. Extended circulation due to PEGylation was confirmed using IVM with both PEGylated particles behaving similarly. Furthermore, a larger difference in behavior between the two regimes became evident through PK and biodistribution analysis. PEGylation resulted in at least a 17-fold increase in circulation half-life, a 136-fold decrease in clearance, and an 86-fold increase in AUC over non-PEGylated NPs. In conclusion, we show significant improvement in overall particle behavior with lower PEG densities than previously reported.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. William Zamboni for the helpful discussions regarding PK analysis, Dr. Ashish Pandya for the synthesis of HP4A, and the University of North Carolina Animal Studies Core for their assistance. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. This work was supported by Liquidia Technologies, the Carolina Center for Cancer Nanotechnology Excellence (U54CA151652) and by the University Cancer Research Fund.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Materials and methods, equations for calculating PEG density, and biodistribution data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): The authors declare competing financial interest. Joseph DeSimone is a founder, member of the board of directors, and maintains a financial interest in Liquidia Technologies. Liquidia was founded in 2004 to commercialize PRINT technology and other discoveries of Professor Joseph DeSimone and colleagues at the University of North Carolina, Chapel Hill.
REFERENCES
- 1.Owens DE, Peppas NA. Int. J. Pharm. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol. Pharmaceutics. 2008;5(4):505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muggia FM. J. Clin. Oncol. 1998;16(2):811–811. doi: 10.1200/JCO.1998.16.2.811. [DOI] [PubMed] [Google Scholar]
- 4.Gradishar WJ. Expert. Opin. Pharmacother. 2006;7(8):1041–1053. doi: 10.1517/14656566.7.8.1041. [DOI] [PubMed] [Google Scholar]
- 5.Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Matter. 2009;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 6.Li SD, Huang L. Biochim. Biophys. Acta. 2009;1788(10):2259–2266. doi: 10.1016/j.bbamem.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNeil SE. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2009;1(3):264–271. doi: 10.1002/wnan.6. [DOI] [PubMed] [Google Scholar]
- 9.Debbage P. Curr. Pharm. Des. 2009;15(2):153–172. doi: 10.2174/138161209787002870. [DOI] [PubMed] [Google Scholar]
- 10.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Mol. Pharmaceutics. 2008;5(4):487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SD, Huang L. Mol. Pharmaceutics. 2008;5(4):496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 12.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem., Int. Ed. 2010;49(36):6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 13.Mosqueira VCF, Legrand P, Morgat JL, Vert M, Mysiakine E, Gref R, Devissaguet JP, Barratt G. Pharm. Res. 2001;18(10):1411–1419. doi: 10.1023/a:1012248721523. [DOI] [PubMed] [Google Scholar]
- 14.Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH. Colloids Surf., B. 2000;18(3–4):301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 15.Moghimi SM, Szebeni J. Prog. Lipid Res. 2003;42(6):463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 16.Malmsten M, Emoto K, Van Alstine JM. J. Colloid Interface Sci. 1998;202(2):507–517. doi: 10.1006/jcis.1998.5727. [DOI] [PubMed] [Google Scholar]
- 17.Benhabbour SR, Sheardown H, Adronov A. Macromolecules. 2008;41(13):4817–4823. [Google Scholar]
- 18.Fang C, Shi B, Pei YY, Hong MH, Wu J, Chen HZ. Eur. J. Pharm. Sci. 2006;27(1):27–36. doi: 10.1016/j.ejps.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Bazile D, Prudhomme C, Bassoullet MT, Marlard M, Spenlehauer G, Veillard M. J. Pharm. Sci. 1995;84(4):493–498. doi: 10.1002/jps.2600840420. [DOI] [PubMed] [Google Scholar]
- 20.Stolnik S, Illum L, Davis SS. Adv. Drug Delivery Rev. 1995;16(2–3):195–214. [Google Scholar]
- 21.Zahr AS, Davis CA, Pishko MV. Langmuir. 2006;22(19):8178–8185. doi: 10.1021/la060951b. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Fuentes M, Torres D, Martin-Pastor M, Alonso MJ. Langmuir. 2004;20(20):8839–8845. doi: 10.1021/la049505j. [DOI] [PubMed] [Google Scholar]
- 23.Duncanson WJ, Figa MA, Hallock K, Zalipsky S, Hamilton JA, Wong JY. Biomaterials. 2007;28(33):4991–4999. doi: 10.1016/j.biomaterials.2007.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng T-L, Chuang K-H, Chen B-M, Roffler SR. Bioconjugate Chem. 2012;23(5):881–899. doi: 10.1021/bc200478w. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharya R, Patra CR, Earl A, Wang S, Katarya A, Lu L, Kizhakkedathu JN, Yaszemski MJ, Greipp PR, Mukhopadhyay D, Mukherjee P. Nanomed.: Nanotechnol. Biol. Med. 2007;3(3):224–238. [Google Scholar]
- 26.Xia XH, Yang MX, Wang YC, Zheng YQ, Li QG, Chen JY, Xia YN. ACS Nano. 2012;6(1):512–522. doi: 10.1021/nn2038516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degennes PG. Adv. Colloid Interface Sci. 1987;27(3–4):189–209. [Google Scholar]
- 28.Kenworthy AK, Hristova K, Needham D, McIntosh TJ. Biophys. J. 1995;68(5):1921–1936. doi: 10.1016/S0006-3495(95)80369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degennes PG. Macromolecules. 1980;13(5):1069–1075. [Google Scholar]
- 30.Storm G, Belliot SO, Daemen T, Lasic DD. Adv. Drug Delivery Rev. 1995;17(1):31–48. [Google Scholar]
- 31.Damodaran VB, Fee CJ, Ruckh T, Popat KC. Langmuir. 2010;26(10):7299–7306. doi: 10.1021/la9041502. [DOI] [PubMed] [Google Scholar]
- 32.Rex S, Zuckermann MJ, Lafleur M, Silvius JR. Biophys. J. 1998;75(6):2900–2914. doi: 10.1016/S0006-3495(98)77732-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hristova K, Needham D. Macromolecules. 1995;28(4):991–1002. [Google Scholar]
- 34.Soong R, Macdonald PM. Biochim. Biophys. Acta. 2007;1768(7):1805–1814. doi: 10.1016/j.bbamem.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 35.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U.S.A. 2008;105(38):14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdorster G, McGrath JL. Biomaterials. 2009;30(4):603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 37.Faure AC, Dufort S, Josserand V, Perriat P, Coll JL, Roux S, Tillement O. Small. 2009;5(22):2565–2575. doi: 10.1002/smll.200900563. [DOI] [PubMed] [Google Scholar]
- 38.Moghimi SM, Hunter AC, Murray JC. Pharmacol. Rev. 2001;53(2):283–318. [PubMed] [Google Scholar]
- 39.Sheng Y, Yuan Y, Liu CS, Tao XY, Shan XQ, Xu F. J. Mater. Sci.: Mater. Med. 2009;20(9):1881–1891. doi: 10.1007/s10856-009-3746-9. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv. Drug Delivery Rev. 2009;61(6):428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dos Santos N, Allen C, Doppen AM, Anantha M, Cox KAK, Gallagher RC, Karlsson G, Edwards K, Kenner G, Samuels L, Webb MS, Bally MB. Biochim. Biophys. Acta. 2007;1768(6):1367–1377. doi: 10.1016/j.bbamem.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Damodaran VB, Fee CJ, Popat KC. Appl. Surf. Sci. 2010;256(16):4894–4901. [Google Scholar]
- 43.Du H, Chandaroy P, Hui SW. Biochim. Biophys. Acta. 1997;1326(2):236–248. doi: 10.1016/s0005-2736(97)00027-8. [DOI] [PubMed] [Google Scholar]
- 44.Gessner A, Paulke BR, Muller RH, Goppert TM. Pharmazie. 2006;61(4):293–297. [PubMed] [Google Scholar]
- 45.Howard MD, Jay M, Dziublal TD, Lu XL. J. Biomed. Nanotechnol. 2008;4(2):133–148. [Google Scholar]
- 46.Walkey CD, Olsen JB, Guo HB, Emili A, Chan WCW. J. Am. Chem. Soc. 2012;134(4):2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 47.Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Nano Lett. 2007;7(4):914–920. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- 48.De M, You C-C, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129(35):10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 49.Baier G, Costa C, Zeller A, Baumann D, Sayer C, Araujo PHH, Mailander V, Musyanovych A, Landfester K. Macromol. Biosci. 2011;11(5):628–638. doi: 10.1002/mabi.201000395. [DOI] [PubMed] [Google Scholar]
- 50.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. U.S.A. 2007;104(7):2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierce MM, Raman CS, Nall BT. Methods. 1999;19(2):213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 52.Jelesarov I, Bosshard HR. J. Mol. Recognit. 1999;12(1):3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 53.Anderson NL, Anderson NG. Mol. Cell. Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 54.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. ACS Nano. 2010;4(7):3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 55.Meng FH, Engbers GHM, Feijen J. J. Biomed. Mater. Res., Part A. 2004;70A(1):49–58. doi: 10.1002/jbm.a.30056. [DOI] [PubMed] [Google Scholar]
- 56.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263(5153):1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 57.Peracchia MT, Fattal E, Desmaele D, Besnard M, Noel JP, Gomis JM, Appel M, d’Angelo J, Couvreur P. J. Controlled Release. 1999;60(1):121–128. doi: 10.1016/s0168-3659(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 58.Lankveld DPK, Rayavarapu RG, Krystek P, Oomen AG, Verharen HW, van Leeuwen TG, De Jong WH, Manohar S. Nanomedicine. 2011;6(2):339–349. doi: 10.2217/nnm.10.122. [DOI] [PubMed] [Google Scholar]
- 59.Sloat BR, Sandoval MA, Li D, Chung WG, Lansakara DSP, Proteau PJ, Kiguchi K, DiGiovanni J, Cui ZR. Int. J. Pharm. 2011;409(1–2):278–288. doi: 10.1016/j.ijpharm.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.