Abstract
The Bacillus subtilis rocG gene, encoding catabolic glutamate dehydrogenase, was found to be subject to direct CcpA-dependent glucose repression. The effect of CcpA required the presence of both the HPr and Crh proteins. The primary CcpA binding site was identified by mutational analysis and DNase I footprinting. In the absence of inducers of the Roc pathway, rocG was still expressed at a low level due to readthrough transcription. CcpA-dependent repression of rocG readthrough transcription proved to contribute to the slow growth rate of B. subtilis cells in glucose-glutamate medium. Increased readthrough expression of rocG was shown to be partially responsible for the growth defect of ccpA strains in glucose-ammonium medium.
Interconversions of α-ketoglutarate and glutamate are major links between carbon and nitrogen metabolism. These reactions are catalyzed by several enzymes: glutamate synthase [α-ketoglutarate + glutamine + NAD(P)H → 2× glutamate + NAD+(P)], glutamate dehydrogenase (GlutDH) [α-ketoglutarate + NH3 + NAD(P)H ⇄ glutamate + NAD+(P)], and glutamate-dependent aminotransferases [glutamate + keto acid (or aldehyde) ⇄ α-ketoglutarate + amino acid].
In Bacillus subtilis, two genes code for GlutDH proteins. The rocG gene product is the major catabolic GlutDH, while gudB encodes an intrinsically inactive GlutDH (7). Spontaneous gain-of-function mutations in gudB generate a second catabolic GlutDH, GudB1 (7). No anabolic glutamate dehydrogenase is present in B. subtilis, and all de novo synthesis of glutamate is catalyzed by glutamate synthase (3).
The B. subtilis rocG gene is a member of the RocR regulon (5). RocR also controls the rocABC and rocDEF operons, whose products catalyze degradation of arginine to glutamate (9, 16, 17). All three roc transcription units have SigL-dependent promoters, require RocR and AhrC proteins for expression, and are induced by arginine, ornithine, or proline. The RocG-catalyzed reaction (glutamate + NAD+ → α-ketoglutarate + NH3 + NADH) can be viewed as the final step in the utilization of arginine, ornithine, and proline, providing rapidly metabolizable carbon- or nitrogen-containing compounds for biosynthesis.
Many B. subtilis genes involved in utilization of alternative carbon sources are subject to carbon catabolite repression mediated by the CcpA protein (8, 11, 25, 34). We show here that rocG expression is also subject to CcpA-dependent carbon catabolite repression. In the course of this work, we discovered that rocG is transcribed not only from its own SigL-dependent promoter but also by readthrough from an upstream gene. In fact, the known growth defect of ccpA mutants in glucose-ammonium medium (13, 30, 33) can be partially relieved by blocking readthrough transcription of rocG.
MATERIALS AND METHODS
Bacterial strains and culture media.
The B. subtilis strains used in this study are listed in Table 1 or described in the table footnotes. The media and growth conditions for B. subtilis and Escherichia coli strains were described previously (7). TSS minimal medium was supplemented with 0.5% glucose, 0.4% succinate, or other carbon and nitrogen sources supplied at 0.2%.
TABLE 1.
B. subtilis strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| SMY | Wild type | P. Schaeffer |
| BR151MA ccpA::spc | ccpA::[(Tn917-lacZ)::spc] trpC2 lys-3 | T. Henkin |
| GM1222 | ptsH1 ykwC::pJH101 (′ykwC′ cat) trpC2 sacB′-lacZ | 12 |
| QB5618 | rocD::aphA3 trpC2 ΔamyE::Φ(rocD′-lacZ cat) | 17 |
| QB5626 | rocR(T120I) trpC2 ΔamyE::Φ(rocD′-lacZ cat) | 17 |
| QB7096 | crh::aphA3 trpC2 | 27 |
| SG82 | lacA::tet trpC2 | 10 |
| BB1179 | ccpA::[(Tn917-lacZ)::spc)] | SMY × DNA (BR151MA ccpA::spc) |
| BB1236 | rocD::aphA3 | SMY × DNA (QB5618) |
| BB1264 | rocG::pBB907 (′yweA rocG rocA′) neo | SMY × pBB907 |
| BB1271 | ΔrocG::ble ΔgudB::teta | 7 |
| BB1316 | rocR(T120I) | BB1236 × DNA (QB5626) |
| BB1434 | rocG::pBB923 [Φ(rocG′p+-lacZ erm)] | 5 |
| BB1512 | ΔamyE::Φ(DAS-rocG′p+-lacZ erm) | SMY × pBB996 (5) |
| BB1537 | ΔrocG::ble ΔgudB::tet ccpA::[(Tn917-lacZ)::spc] | BB1271 × DNA (BB1179) |
| BB1575 | ptsH1 ykwC::pJH101 (′ykwC′ cat) | SMY × DNA (GM1222) |
| BB1597 | rocR(T120I) ccpA::[(Tn917-lacZ)::spc] | BB1316 × DNA (BB1179) |
| BB1628 | yweA::pBB1013 (′yweA′ neo) | SMY × pBB1013 |
| BB1744 | rocGp1 ΔgudB::tet | BB1271 × pBB1048 |
| BB1770 | rocGp1 yweA::pBB1013 (′yweA′ neo) ΔgudB::tet | BB1744 × pBB1013 |
| BB1792 | rocGp1 yweA::pBB1013 (′yweA′ neo) rocR(T120I) | BB1316 × DNA (1770) |
| BB2062 | rocG::pBB1096 [Φ(rocG′p1-lacZ erm)] | SMY × pBB1096 |
| BB2063 | ΔamyE::Φ(DAS-rocG′p1-lacZ erm) | SMY × pBB1049 |
| BB2163 | ccpA::[(Tn917-lacZ)::spc] yweA::pBB1013 (′yweA′ neo) | BB1628 × DNA (BB1179) |
The ΔgudB::tet null mutation in the gene coding for the inactive GudB-GlutDH was present in some strains and served to prevent spontaneous gudB1 mutations from allowing expression of active GudB1-GlutDH (7).
Strain BB1316 [rocR(T120I)] was created by replacement of the rocD::aphA3 mutation in strain BB1236 by the wild-type rocD allele with chromosomal DNA from strain QB5626 carrying the rocR(T120I) mutation, which is very tightly linked to rocD. The rocD+ rocR(T120I) transformants were selected as small colonies on TSS-ornithine plates, which are unable to support growth of the parental strain. The presence of the rocR(T120I) mutation in strain BB1316 was confirmed by introducing a rocD-lacZ fusion and analyzing its expression pattern in different media (17).
Strain BB1575 (ptsH1) was constructed by transforming strain SMY with chromosomal DNA from strain GM1222 carrying a plasmid integrated into the chromosome close to the ptsH1 mutation. The PtsH− strains were identified among chloramphenicol-resistant transformants as slow-growing isolates on TSS-mannitol plates (12).
DNA manipulations and transformation.
Methods for plasmid isolation, agarose and polyacrylamide gel electrophoresis, use of restriction and DNA-modification enzymes, DNA ligation, PCR, and electroporation of E. coli JM107 cells were as described by Sambrook et al. (28). Isolation of chromosomal DNA and transformation of B. subtilis cells by chromosomal or plasmid DNA were done as described previously (7).
Construction of the rocGp1 mutant.
A double substitution mutation (rocGp1) within the putative cre site of the rocG gene (Fig. 1) was created by site-directed mutagenesis by the method of Kunkel (4, 20) with the mutagenic oligonucleotide oBB73 (5′-GATTTTTTAAAGGCCTTACATTAC [altered nucleotides are in bold]), and pBB907 (7) as the template. The presence of the mutation in the resulting plasmid, pBB1048, was confirmed by showing susceptibility to cleavage by StuI (the italics in oBB73) and DNA sequencing of the entire rocG regulatory region.
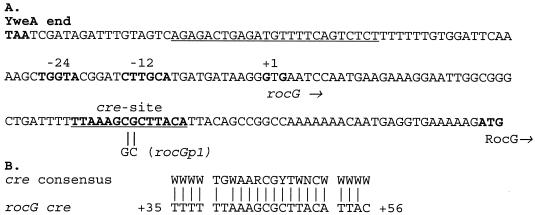
FIG. 1.
(A) Sequence of the rocG regulatory region. The termination codon of yweA, the −12 and −24 promoter regions, the transcription start points, a likely initiation codon, and the core of a putative cre site of the rocG gene are in bold. The directions of transcription and translation are indicated by horizontal arrows. The dyad-symmetry sequence of a putative yweA transcriptional terminator and the region protected by CcpA in DNase I experiments are underlined. The sequence changes in the rocGp1 mutant are indicated. (B) Comparison of rocG cre to the cre consensus sequence (36). Note that the second residue (G) of the core consensus sequence is not conserved in the rocG cre site.
pBB1048 was introduced into B. subtilis strain BB1271 (ΔrocG::ble) and neomycin-resistant transformants, arising from single-crossover homologous recombination events, were selected. Strain BB1744 (rocGp1) was isolated from one of the RocG+ transformants as a spontaneous neomycin- and phleomycin-sensitive derivative in which the plasmid carrying the neomycin resistance gene and the ΔrocG::ble allele, determining resistance to phleomycin, had been excised from the chromosome and lost. The replacement of the chromosomal rocG gene by the rocGp1 allele and excision of the plasmid in strain BB1744 were confirmed by detection of high GlutDH activity in glucose-ornithine minimal medium and by probing for the loss of vector sequences.
Construction of rocGp1-lacZ fusions.
pBB1096, containing the rocGp1-lacZ fusion, is identical to pBB923 but for the presence of the rocGp1 mutation from pBB1048 and was created in two steps as described earlier for pBB923 (5). To create pBB1049 containing a DAS-rocGp1-lacZ fusion with the downstream activating sequence (DAS) relocated upstream of the rocG promoter, the 0.71-kb BamHI-AgeI fragment of pBB996 containing the promoter part of the DAS-rocGp+-lacZ fusion (5) was replaced by a similar fragment of pBB1048. Both pBB1096 and pBB1049 were integrated into the chromosome either at the amyE locus by a double-crossover recombination event or at the rocG locus by a single-crossover recombination event as described previously (5).
Construction of pBB1013.
Plasmid pBB907 (7), containing the entire rocG gene and the promoterless coding region of the upstream gene yweA, was integrated into the chromosome of B. subtilis strain SMY, creating strain BB1264. To clone DNA adjacent to the site of integration of pBB907, the chromosomal DNA of strain BB1264 was digested with SacI, self-ligated, and introduced by electroporation into cells of E. coli strain JM107 pcnB80 zad::Tn10 (21). The isolated plasmid, pBB932, had a 4.04-kb insert of chromosomal DNA carrying most of the spsK gene and the entire spsL, yweA, and rocG genes. pBB932 was unstable in E. coli pcnB+ cells, apparently due to toxicity of the yweA product.
The 2.33-kb EcoNI-XhoI fragment was excised from pBB932 to create pBB960. Plasmid pBB1013 containing the internal, 0.25-kb-long part of the yweA gene was constructed by removing the 1.46-kb SacI-HpaI fragment from pBB960.
Gel shift and DNase I footprinting experiments.
His6-CcpA was purified as described previously (19). The 259-bp fragments (positions −181 to + 78 with respect to the rocG initiation codon) containing the rocGp+ or rocGp1 region were amplified by PCR from pBB907 and pBB1048, respectively, with oBB96 (5′-GTTAATCGATAGATTTGTAGTC) and 32P-labeled oBB56 (5′-CTTAATGATTGTTTGGGTAGAC) as primers. DNA-CcpA complexes were formed and analyzed as described before (19).
DNA sequencing.
Relevant parts of the rocG gene were sequenced by the dideoxy chain termination method of Sanger et al. (29) with vector- or rocG-specific oligonucleotides as primers and a Sequenase reagent kit (Amersham Life Science) as recommended by the manufacturer. Plasmid double-stranded DNA to be sequenced was purified with a QIAprep Spin miniprep kit (Qiagen). DNA and protein sequences were analyzed with the DNA Strider and Blast programs (1, 23) and the SubtiList web site (http://genolist.pasteur.fr/SubtiList/) (26).
Enzyme assays.
β-Galactosidase activity was determined as described previously (4) and expressed in Miller units (24). GlutDH activity was assayed as described previously (7). One unit of GlutDH activity converts 1 μmol of NADH to NAD+ per mg of protein per min. All numbers are averages of at least two experiments.
RESULTS
Glucose repression of rocG is dependent on CcpA.
Previously we reported that the activity of B. subtilis GlutDH, encoded by the rocG gene, is highly increased in the absence of glucose if an inducer of the RocR pathway, e.g., arginine or ornithine, is present in the growth medium (7). Since the CcpA protein is a major regulator of carbon catabolite repression in B. subtilis (11, 18), we measured GlutDH specific activity in a ccpA mutant. Inactivation of the ccpA gene caused a full induction of RocG activity in cells grown in glucose-ornithine medium (Table 2, strains SMY and BB1179).
TABLE 2.
GlutDH activity in minimal mediuma
| Carbon source | Nitrogen source | GlutDH activity (U)
|
||||
|---|---|---|---|---|---|---|
| SMY wild type | BB1179 ccpA | BB1597 ccpA rocR(T1201) | BB1744 rocGp1 | BB1792 rocGp1 rocR(T120I) | ||
| Glucose | Ammonium | ≤45 | ≤45 | NGb | ≤45 | 1,255 |
| Glutamate | ≤45 | ≤45 | 1,052 | ≤45 | 1,005 | |
| Ornithine | 74 | 681 | 791 | 677 | 666 | |
| Ornithine | Ornithine | 769 | 1,113 | NDc | 1,147 | 747 |
| Glucose | Proline | 76 | 62 | 883 | 63 | 967 |
| Proline | Proline | 246 | 177 | ND | 268 | ND |
Cells were grown in minimal medium with the indicated carbon and nitrogen sources and assayed for GlutDH activity (7).
NG, no growth under these conditions.
ND, not determined.
Expression of a transcriptional rocG-lacZ fusion, placed at the rocG locus, was also increased in a ccpA mutant in glucose-ornithine medium and was similar to the fusion activity seen in wild-type cells in the presence of ornithine as the sole carbon source (Table 3, strains BB1434 and BB1439). Such a fusion is expressed at a low level because its promoter is separated from the downstream activating sequence (DAS) required for rocG expression by the lacZ gene and plasmid sequences integrated at the rocG locus (5). A more active transcriptional fusion, in which the DAS region was relocated upstream of the rocG promoter (DAS-rocG-lacZ) (5), was regulated by CcpA in the same manner as RocG enzyme activity and the fusion at the rocG locus (Table 4, strains BB1512 and BB1521). Thus, CcpA regulates rocG expression at the transcriptional level, and this regulation is independent of the location of the DAS.
TABLE 3.
Activity of rocG-lacZ fusionsa
| Strain | Relevant genotype | Promoter region within the fusion | β-Galactosidase activity (U)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Medium with glucose plus:
|
Medium without glucose but with:
|
|||||||
| NH4Cl | Glutamate | Proline | Ornithine | Proline | Ornithine | |||
| BB1434 | Wild type | rocGp+ | 0.39 | 0.47 | 0.49 | 0.52 | 2.3 | 11.3 |
| BB1439 | ccpA | rocGp+ | 1.1 | 1.6 | 1.2 | 11.7 | 3.2 | 16.8 |
| BB1470 | ccpA rocR(T120I) | rocGp+ | NGb | 6.2 | 7.1 | 14.2 | ||
| BB2155 | lacA | rocGp+ | 0.15 | 0.10 | 0.54 | |||
| BB2157 | lacA ccpA | rocGp+ | 0.54 | 0.32 | 18.3 | |||
| BB2062 | Wild type | rocGp1 | 1.9 | 1.0 | 13.9 | 11.5 | ||
| BB2156 | lacA | rocGp1 | 2.8 | 1.2 | 19.9 | |||
| BB2158 | lacA ccpA | rocGp1 | 0.48 | 0.28 | 12.4 | |||
All strains are derivatives of BB1434 or BB2062 and contain a rocG-lacZ fusion integrated at the rocG locus. Cells were grown in minimal medium with the indicated carbon and nitrogen sources and assayed for β-galactosidase activity.
NG, no growth under these conditions.
TABLE 4.
Expression of DAS-rocG-lacZ fusionsa
| Strain | Relevant genotype | Promoter region within the fusion | β-Galactosidase activity (U)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Medium with glucose plus:
|
Medium without glucose but with:
|
|||||||
| NH4Cl | Glutamate | Proline | Ornithine | Proline | Ornithine | |||
| BB1512 | Wild type | rocGp+ | 0.12 | 0.76 | 11.4b | 11.9 | 35.1 | 233.9 |
| BB1521 | ccpA | rocGp+ | 1.6 | 1.4 | 19.6 | 187.8 | 50.1 | 160.8 |
| BB2131 | rocR(T120I) | rocGp+ | 3.6 | 13.5 | 14.6 | 9.6 | 138.7 | 228.8 |
| BB2139 | ccpA rocR(T120I) | rocGp+ | NGc | 247.7 | 268.6 | 250.0 | 252.1 | 279.2 |
| BB2063 | Wild type | rocGp1 | 0.16 | 1.1 | 275.8 | 251.0 | 93.9 | 336.4 |
| BB2064 | ccpA | rocGp1 | 1.2 | 1.0 | 11.2 | 242.0 | ||
| BB2132 | rocR(T120I) | rocGp1 | 56.5 | 166.0 | 245.1 | 200.0 | ||
Derivatives of strains BB1512 and BB2063 carrying DAS-rocG-lacZ fusions at the ectopic amyE locus (with the rocG DAS sequence positioned upstream of the fusion promoter) were grown in minimal medium with the indicated carbon and nitrogen sources and assayed for β-galactosidase activity.
Low activity of the rocG-lacZ fusion (1 U) was detected during early exponential growth.
NG, no growth under these conditions.
The rocR(T120I) allele, encoding a constitutively active version of the RocR protein, causes rocG expression to be largely independent of the presence of inducers (5, 17) but still subject to catabolite repression (Table 4, strain BB2131). In the ccpA rocR(T120I) double mutant, the activities of GlutDH and rocG-lacZ fusions were highly elevated not only in glucose-ornithine medium but also in glucose-glutamate medium. i.e., in the absence of inducers (Tables 2 to 4, strains BB1597, BB1470, and BB2139).
Role of auxiliary proteins in catabolite repression of rocG.
The activity of CcpA as a transcriptional regulator in most cases depends on the presence of one or two additional proteins, HPr and Crh, encoded by the ptsH and crh genes, respectively (11). rocG expression was still repressed by glucose even if only one of these two proteins was active in the cells (Table 5, strains BB2025 and BB2026). Inactivation of both proteins in strain BB2027 (ptsH1 crh) led to derepression of the DAS-rocG-lacZ fusion in glucose-ornithine medium (Table 5). Therefore, to repress rocG, CcpA must interact with either Hpr or Crh.
TABLE 5.
Role of the HPr and Crh proteins in catabolite repression of the rocG genea
| Strain | Relevant genotype | β-Galactosidase activity (U) in medium containing:
|
|
|---|---|---|---|
| Ornithine and glucose | Ornithine, no glucose | ||
| BB1521 | ccpA | 191.6 | 191.6 |
| BB2025 | ptsH1 | 7.0 | |
| BB2026 | crh | 13.1 | |
| BB2027 | ptsH1 crh | 242.1 | |
Derivatives of strain BB1512 carrying the DAS-rocGp+-lacZ fusion were grown in minimal medium with the indicated carbon and nitrogen sources and assayed for β-galactosidase activity.
Isolation and characterization of a rocG cre mutant.
A putative CcpA binding site (cre), corresponding closely to proposed cre consensus sequences, was found about 40 bp downstream of the rocG transcription start point and 30 bp upstream of the initiation codon (Fig. 1). The two conserved central nucleotides of this sequence were altered by site-directed mutagenesis (Fig. 1), and the mutant rocGp1 gene was substituted for the wild-type rocG allele in the B. subtilis chromosome as described in Materials and Methods. In contrast to wild-type cells, strain BB1744 (rocGp1) had high GlutDH activity in glucose-ornithine medium, similar to GlutDH activities found for the wild-type strain grown with ornithine as the sole carbon and nitrogen source and in the ccpA mutant grown in ornithine medium with or without glucose (Table 2). Similarly, rocGp1-lacZ fusions were highly expressed in ornithine medium even in the presence of glucose (Tables 3 and 4, strains BB2062 and BB2063). Thus, the rocGp1 mutation alters a site required for CcpA-dependent glucose repression of rocG. As expected, the rocGp1 promoter directed high GlutDH or lacZ fusion expression even in the absence of the RocR inducers if the constitutively active RocR(T120I) protein was present (Tables 2 and 4, strains BB1792 and BB2132).
CcpA binding to the rocG promoter.
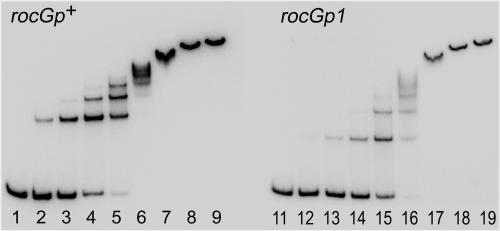
Purified CcpA bound efficiently to a 259-bp DNA fragment containing the putative wild-type cre site. CcpA formed several complexes with wild-type DNA, the number and relative abundance of which depended on the concentration of CcpA, indicating the presence of several binding sites or multimerization of the protein (Fig. 2, lanes 1 to 9). Unexpectedly, CcpA also bound to a similar fragment containing the rocGp1 mutation, although the binding was about fourfold less efficient. Some CcpA-DNA complexes formed with mutant DNA had different electrophoretic mobilities than those formed with wild-type DNA, indicating altered stoichiometry or geometry of the DNA-CcpA interaction and confirming the presence of sites which can interact with CcpA independently of cre (Fig. 2, lanes 11 to 19).
FIG. 2.
Gel mobility shift analysis of the interaction between the rocG promoter and CcpA. Radioactively labeled 259-bp rocGp+ (lanes 1 to 9) and rocGp1 (lanes 11 to 19) promoter fragments were incubated with increasing concentrations of purified CcpA. The CcpA concentrations were 0 (lanes 1 and 11), 0.15 nM (lanes 2 and 12), 0.6 nM (lanes 3 and 13), 2.4 nM (lanes 4 and 14), 9.8 nM (lanes 5 and 15), 39 nM (lanes 6 and 16), 156 nM (lanes 7 and 17), 625 nM (lanes 8 and 18), and 2,500 nM (lanes 9 and 19).
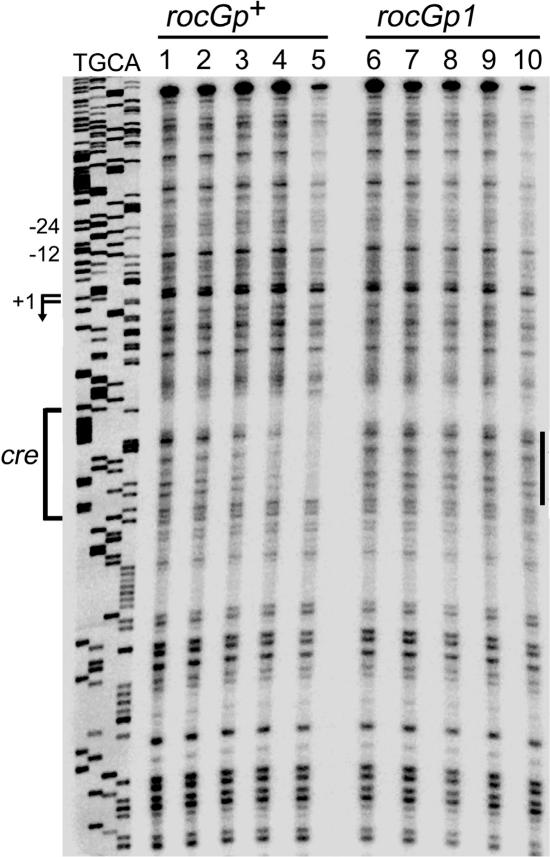
In DNase I footprinting experiments, CcpA, at a concentration of 39 nM or more, protected the wild-type DNA sequence corresponding to positions +39 to +52 with respect to the rocG transcription start point (Fig. 3, lanes 1 to 5). This sequence coincides with the core of the putative cre site. No detectable CcpA binding to the rocGp1 version of the same region was observed at any CcpA concentration used (Fig. 3, lanes 6 to 10), indicating that the rocGp1 mutation decreased CcpA affinity for the cre region at least 16-fold. An additional DNA region within both wild-type and rocGp1 fragments, which overlapped the rocG promoter and the upstream sequence, appeared to be partially protected at the highest CcpA concentration used (Fig. 3, lanes 5 and 10).
FIG. 3.
DNase I footprinting analysis of CcpA binding to the rocG promoter. The 259-bp rocGp+ (lanes 1 to 5) and rocGp1 (lanes 6 to 10) promoter fragments, labeled on the template strand, were incubated with purified CcpA and then with DNase I. The sequence of the template strand of pBB907 (7) was determined with oBB56 as a primer. The apparent transcription start sites and the direction of rocG transcription are shown by the bent arrows, and the cre site is indicated by a bracket. The −12 and −24 promoter regions are shown. The protected cre area is indicated by a vertical line to the right of the gel lanes. Lanes 1 and 6, no CcpA; other lanes contained increasing concentrations of purified CcpA: 9.8 nM (lanes 2 and 7), 39 nM (lanes 3 and 8), 156 nM (lanes 4 and 9), and 625 nM (lanes 5 and 10).
Contribution of RocG to the growth defect of the ccpA null mutant.
ccpA mutants are known to have a growth defect in glucose-ammonium medium that is relieved by addition of glutamate or by overexpression of glutamate synthase (13, 30, 33). We were therefore prompted to analyze the possible contribution of RocG, an enzyme of glutamate catabolism, to this growth defect. In contrast to other reports, in our hands ccpA mutants grew, albeit significantly more slowly than the wild-type strain, on glucose-ammonium plates and in liquid medium (generation time of 100 to 200 min versus 60 min for the wild type). In addition, the lag time and the generation time in liquid medium varied from experiment to experiment, although no accumulation of revertants was detectable in most of our experiments. The growth defect of the ccpA null mutant was noticeably alleviated by introduction of the rocG null mutation, although the resulting strain did not grow in glucose-ammonium medium as fast as the wild-type strain did (generation time of about 80 min versus 60 min). On the other hand, introduction of the rocR(T120I) mutation, which increases rocG expression in the absence of inducers (Table 4, strain BB2131) (5, 17), completely abolished the ability of ccpA strains to grow in glucose-ammonium medium (Tables 2 to 4, strains BB1597, BB1470, and BB2139).
Liquid cultures of the ccpA rocR(T120I) double mutant accumulated revertant strains that were able to resume growth in glucose-ammonium medium. The new mutation in at least some of the revertants had the phenotype of the rocG null mutation and mapped to the rocG locus (data not shown); as expected, introduction of a characterized rocG null mutation restored the ability of the ccpA rocR(T120I) strain to grow in glucose-ammonium liquid medium (data not shown). We conclude that rocG expression, even at the low level seen in the absence of inducers, is partially responsible for the growth defect of ccpA mutants in glucose-ammonium medium.
Strains BB1316 [rocR(T120I)], BB1744 (rocGp1), and especially BB1792 [rocGp1 rocR(T120I)] also had growth defects in glucose-ammonium medium, presumably due to higher expression of GlutDH in these mutants; the defects were more pronounced on agar plates than in liquid medium (data not shown). Because the growth defect of these strains was less severe than the defect of the ccpA mutant and because a rocG null mutation only partially alleviated the ccpA growth defect, other factors must contribute to the poor growth of ccpA mutants (22, 30).
Readthrough expression of rocG.
Though inactivation of the rocG gene greatly improved the ability of the ccpA mutant to grow in glucose-ammonium medium, inactivation of the rocR or sigL gene, both of which are required for expression of the rocG gene from its known promoter, did not affect the growth of the ccpA strain in this medium (data not shown). This result suggests that the low level of expression of rocG in glucose-ammonium medium initiates at a promoter that is independent of RocR and SigL.
The existence of such a promoter was demonstrated by integrating plasmid pBB1013, which carries an internal fragment of yweA, the gene immediately upstream of rocG, into the chromosome of a B. subtilis ccpA mutant (strain BB2163). Such a recombination event separates the rocG open reading frame and its SigL-dependent promoter from upstream chromosomal sequences. The presence of the intervening plasmid sequence relieved the growth defect of the ccpA mutant in glucose-ammonium medium to the same extent as did a rocG null mutation, indicating that rocG expression under these conditions initiated at an unidentified promoter located upstream of the integration point of pBB1013. Other, similar experiments showed that this promoter is located upstream of the yweA open reading frame. We suggest that transcription of rocG in glucose-ammonium medium is directed by the yweA promoter or the promoter of the upstream sps operon. Interestingly, this transcription would require readthrough of an apparent transcription terminator between yweA and rocG (Fig. 1).
The activity of RocG-GlutDH expressed from the upstream promoter was too low to detect by enzymatic assays. To quantitate the contribution of the upstream promoter to rocG expression, we tested the activity of rocG-lacZ fusions integrated at the rocG locus in strains carrying a mutation in lacA, which encodes an endogenous β-galactosidase (10). Expression of the lacA gene is subject to CcpA-dependent catabolite repression (25) and accounts for up to 1 Miller unit of β-galactosidase activity in minimal medium, obscuring expression of lacZ fusions with low activity. Under our growth conditions, the lacA strain with no lacZ fusion had 0.03 to 0.06 Miller units of β-galactosidase activity.
Very little activity of the rocGp+-lacZ fusion was detected in glucose medium in the absence of RocR activators (e.g., ornithine); in contrast, expression of the rocG-lacZ fusions in the ccpA mutants in glucose-ammonium or glucose-glutamate medium was increased about threefold (Table 3, strains BB2155, BB2157, and BB2158), consistent with our suggestion that an increased level of rocG expression is partially responsible for the growth defect of this strain in glucose-ammonium medium. This expression is apparently due to an increase in transcription readthrough which is normally blocked by binding of CcpA to the cre site. Unexpectedly, expression of the rocGp1-lacZ fusion with a mutation in the rocG cre site was severalfold higher in the ccpA+ strain than rocGp+-lacZ or rocGp1-lacZ expression in the ccpA mutant, although similar levels of expression were expected (Table 3, strains BB2156 and BB2158). Additional interactions between CcpA and the rocG regulatory region, other than with the cre site, could be responsible for this result.
Proline-mediated induction of rocG.
Proline was shown to be an inducer of RocG but less effective than ornithine or arginine if glucose was absent from the medium (7). Accordingly, no significant relief of glucose repression was observed in glucose-proline medium with the ccpA mutant or the rocGp1 strain containing a mutation in the CcpA binding site (Tables 2 to 4, strains BB1179, BB1744, BB1439, and BB1521). We suspect that this result reflects the fortuitous balancing of partial derepression of rocG and partial inactivation of TnrA, a global regulator of nitrogen metabolism genes (14, 15, 35), which may affect proline utilization. TnrA activity is abolished in cells grown in ammonium-containing medium (32). As expression of GlutDH in proline-containing medium is increased by inactivation of CcpA, by removal of glucose, or by the rocGp1 mutation, degradation of glutamate derived from proline will generate ammonium under such conditions. As a consequence, TnrA activity and proline utilization will be reduced (6), preventing high-level induction of rocG and other roc genes. Similarly, it was previously shown (and we confirmed) that enzymes of the Roc pathway can be induced by proline but not by the combination of proline and ammonium (which inactivates TnrA) (2).
In accord with this model, when a rocGp1-lacZ fusion was placed at either the rocG or amyE locus in strains that were otherwise wild type (in which GlutDH synthesis was still subject to catabolite repression and glutamate degradation to ammonium was not significantly enhanced), lacZ was as highly expressed in glucose-proline medium as it was in glucose-ornithine medium (Tables 3 and 4, strains BB2062 and BB2063). Additionally, in the ccpA rocR(T120I) double mutant strain, in which rocG expression does not depend on proline uptake, high rocG expression was observed in all media tested, including the proline medium (Table 2 and 4, strains BB1597 and BB2139).
Glutamate utilization.
The growth rate of B. subtilis cells in glucose-glutamate medium is severely limited by the low activity of RocG-GlutDH due to CcpA-mediated repression and inactivity of RocR under these conditions (7). The generation time in glutamate medium was decreased from 120 min for the wild-type strain to about 60 min if RocG-GlutDH activity was elevated by the presence of the constitutively active rocR(T120I) mutation or if total GlutDH activity was increased by the gudB1 mutation (7).
Interestingly, the generation time of strain BB1744 (rocGp1) in glucose-glutamate medium also decreased to 85 min. In contrast, the rocGp1 mutation had no effect on growth rate when the rocG gene was separated from the yweA promoter by insertion of plasmid DNA (strain BB1770). Therefore, even low-level readthrough transcription of rocG is sufficient for faster utilization of glutamate if catabolite repression of rocG is removed.
DISCUSSION
GlutDH, a ubiquitous metabolic enzyme, catalyzes a reversible reaction that can provide either the anabolic function of glutamate biosynthesis or the catabolic function of glutamate utilization. In B. subtilis, both RocG and the mutationally activated form of GudB (7) are exclusively catabolic enzymes. Neither RocG, even when fully derepressed by the rocGp1 and rocR(T120I) mutations, nor GudB1 is able to support the growth of a glutamate synthase mutant in the absence of glutamate (our unpublished results).
In our previous work, we showed that rocG, like the rocABC and rocDEF operons, requires SigL-containing RNA polymerase, RocR, AhrC, and an enhancer-like activating sequence for expression (5, 15). Unlike the rocABC and rocDEF operons, which are highly expressed in minimal glucose-arginine medium (2, 9, 16) and are not significantly affected by the presence or absence of CcpA (data not shown), the rocG gene is subject to carbon catabolite repression mediated by CcpA acting at the cre site located between the transcription and translation start points. This differential regulation of the genes of the roc regulon probably reflects the different functional roles of the corresponding products. While the first three reactions of the Roc pathway liberate nitrogen groups from arginine and ornithine (15), RocG has a dual role, providing both ammonium as a nitrogen source and α-ketoglutarate as a carbon source. High activity of GlutDH is presumably not needed when rapidly metabolizable carbon sources are available and could lead to depletion of the pool of glutamate, a major cellular anion (31), if unchecked. As shown in the accompanying paper (6), high activity of GlutDH could drastically alter the regulation of nitrogen metabolism genes due to production of ammonium and consequent inactivation of a global regulator TnrA (14, 15, 35). In addition, GlutDH interferes with the activity of GltC, the activator of glutamate synthase (6). Interestingly, expression of gudB is also reduced in the presence of glucose, although the mechanism of this regulation remains unknown (7).
Binding of CcpA to the rocG cre site not only reduces the level of RocR-dependent rocG expression but also serves as a roadblock to prevent readthrough transcription from an upstream promoter. The latter promoter only contributes significantly to rocG expression in the absence of RocR activators, because induced RocR-dependent expression from the rocG promoter is much stronger than readthrough. Although readthrough transcription is only a minor contributor to rocG expression in wild-type cells when RocR is active, it permits multiple levels of expression. Thus, the rocG gene has four physiologically relevant modes of expression in minimal medium: no detectable expression in glucose medium in the absence of RocR activators; a very low level of expression in the absence of both glucose and the activators; a low to moderate level of expression in glucose medium in the presence of the activators; and a high level of expression in the absence of glucose and with the activators present. Interestingly, we recently found that a cre site located between the ccpC gene and its upstream genes also regulates readthrough transcription of ccpC (19).
CcpA-dependent catabolite repression of GlutDH helps us to understand two previously unexplained phenomena. First, glutamate is known to be a poor nitrogen source for B. subtilis in glucose-containing medium. Since utilization of glutamate can be mediated by GlutDH activity, which is required to liberate ammonium for synthesis of glutamine, the negative effect of CcpA on rocG reduces the ability of cells to take full advantage of the potential nutritional value of glutamate. Second, ccpA mutants grow poorly in glucose-ammonium medium unless glutamate is added. The explanation in part seems to be that derepressed expression of rocG leads to degradation of endogenous glutamate. Additionally, higher RocG activity may interfere with glutamate synthesis due to inactivation of GltC (6). Reduced expression of glutamate synthase genes in ccpA mutants was documented previously (8, 13, 30).
The rocG, rocABC, and rocDEF genes have been shown in DNA array experiments to be regulated by glucose in a CcpA-dependent manner in cells growing in broth media (25, 34). In work to be published elsewhere, we found that this mode of regulation in broth media is unrelated to the cre site-dependent regulation of rocG described here for minimal media. Glucose catabolite repression of the roc genes in broth media is apparently due to poor uptake of arginine or proline or both.
Acknowledgments
We are grateful to M. Débarbouillé, S. Fisher, and N. Mani for helpful discussions and to R. Daniel, M. Débarbouillé, J. Errington, R. Gardan, T. Henkin, and J. Stülke for gifts of strains.
This work was supported by U.S. Public Health Service grant GM36718 to A.L.S. and a grant from the National Science Foundation (MCB-0110651) to B.R.B.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumberg, S., and C. R. Harwood. 1979. Carbon and nitrogen repression of arginine catabolic enzymes in Bacillus subtilis. J. Bacteriol. 137:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines, p. 203-231. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 4.Belitsky, B. R., P. J. Janssen, and A. L. Sonenshein. 1995. Sites required for GltC-dependent regulation of Bacillus subtilis glutamate synthase expression. J. Bacteriol. 177:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belitsky, B. R., and A. L. Sonenshein. 1999. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:10290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belitsky, B. R., and A. L. Sonenshein. 2004. Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J. Bacteriol. 186:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belitsky, B. R., and A. L. Sonenshein. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stulke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. [DOI] [PubMed] [Google Scholar]
- 9.Calogero, S., R. Gardan, P. Glaser, J. Schweizer, G. Rapoport, and M. Debarbouille. 1994. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R. A., J. Haiech, F. Denizot, and J. Errington. 1997. Isolation and characterization of the lacA gene encoding β-galactosidase in Bacillus subtilis and a regulator gene, lacR. J. Bacteriol. 179:5636-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 12.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faires, N., S. Tobisch, S. Bachem, I. Martin-Verstraete, M. Hecker, and J. Stulke. 1999. The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 1:141-148. [PubMed] [Google Scholar]
- 14.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, S. H., and M. Débarbouillé. 2002. Nitrogen source utilization and its regulation, p. 181-191. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 16.Gardan, R., G. Rapoport, and M. Debarbouille. 1995. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249:843-856. [DOI] [PubMed] [Google Scholar]
- 17.Gardan, R., G. Rapoport, and M. Debarbouille. 1997. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 24:825-837. [DOI] [PubMed] [Google Scholar]
- 18.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 19.Kim, H. J., C. Jourlin-Castelli, S. I. Kim, and A. L. Sonenshein. 2002. Regulation of the Bacillus subtilis ccpC gene by CcpA and CcpC. Mol. Microbiol. 43:399-410. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 21.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, H., C. Meinken, A. Matin, and J. Stulke. 2002. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J. Bacteriol. 184:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marck, C. 1988. DNA Strider: a C program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 26.Moszer, I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28-36. [DOI] [PubMed] [Google Scholar]
- 27.Presecan-Siedel, E., A. Galinier, R. Longin, J. Deutscher, A. Danchin, P. Glaser, and I. Martin-Verstraete. 1999. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol. 181:6889-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wacker, I., H. Ludwig, I. Reif, H. M. Blencke, C. Detsch, and J. Stulke. 2003. The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149:3001-3009. [DOI] [PubMed] [Google Scholar]
- 31.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 32.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:8841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray, L. V., Jr., F. K. Pettengill, and S. H. Fisher. 1994. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J. Bacteriol. 176:1894-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, K., H. Yamaguchi, M. Kinehara, Y. H. Ohki, Y. Nakaura, and Y. Fujita. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49:157-165. [DOI] [PubMed] [Google Scholar]
- 36.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 1998. Expression of the Bacillus subtilis acsA gene: position and sequence context affect cre-mediated carbon catabolite repression. J. Bacteriol. 180:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]