SUMMARY
We implement a joint model for mixed multivariate longitudinal measurements, applied to the prediction of time until lung transplant or death in idiopathic pulmonary fibrosis. Specifically, we formulate a unified Bayesian joint model for the mixed longitudinal responses and time-to-event outcomes. For the longitudinal model of continuous and binary responses, we investigate multivariate generalized linear mixed models using shared random effects. Longitudinal and time-to-event data are assumed to be independent conditional on available covariates and shared parameters. A Markov chain Monte Carlo (MCMC) algorithm, implemented in OpenBUGS, is used for parameter estimation. To illustrate practical considerations in choosing a final model, we fit 37 different candidate models using all possible combinations of random effects and employ a Deviance Information Criterion (DIC) to select a best fitting model. We demonstrate the prediction of future event probabilities within a fixed time interval for patients utilizing baseline data, post-baseline longitudinal responses, and the time-to-event outcome. The performance of our joint model is also evaluated in simulation studies.
Keywords: Idiopathic Pulmonary Fibrosis, Joint model, Mixed continuous and binary data, Multivariate longitudinal data, Prediction model, Shared parameter model, Survival analysis
1. Introduction
While longitudinal data from biomarkers or intermediate clinical assessments are often collected in Phase II and III clinical trials, the primary clinical trial outcome typically consists of time until the occurrence of a specified event, e.g., time to onset of illness or death. An example from the field of chronic lung disease research is the study of transplant-free survival in idiopathic pulmonary fibrosis (IPF). IPF is a chronic, progressive lung disease characterized by fibrosis of unknown etiology [24]. Here, our primary concern is the prediction of failure times by modeling the relationship between longitudinally measured indicators of pulmonary function and time to either lung transplant or death. Specifically, we utilize longitudinally assessed forced vital capacity (FVC), a continuous measure of pulmonary function, as a predictor of event times. We also utilize a second, binary, disease progression indicator defined as a decline of 5% or more in FVC from baseline FVC. Since these measures are potentially conjointly informative of time to lung transplant or death, it would be desirable to implement a modeling approach that allows prediction of event times using two or more longitudinal responses simultaneously.
Several approaches have been developed for handling multivariate longitudinal outcomes [4,20,16,23,29]. For example, some approaches factorize the joint distribution as the product of the marginal distribution of one variable times the conditional distribution of the second variable given the first [3,11]. Other approaches use random effects to model relationships between outcomes [8,13]. To date, however, most researchers have implemented joint models in which a univariate continuous longitudinal response is related to the time-to-event outcome [7,14,15,27]. In this paper, we develop a joint model for mixed continuous and binary longitudinal responses and a time-to-event outcome. For longitudinal responses we investigate mixed models with shared random effects. To build a joint model with a time-to-event outcome, we assume shared parameters exist between the longitudinal mixed models and the time-to-event outcome. The longitudinal responses and the time-to-event outcome are assumed to be independent conditional on the shared parameters and measured covariates.
We apply the proposed joint model to the dynamic prediction of mortality occurring within a fixed time for a subject still at risk just before time t [30]. A number of authors have proposed the prediction of future event probabilities for subjects based on the joint modeling of longitudinal measurements, time-to-event outcomes, and other covariates [9,25]. Fieuws et al. [9] investigated predicting renal graft failure using multiple longitudinal outcomes of biochemical and physiological markers utilizing a multivariate mixed model [22] and a pattern-mixture approach [19]. Rizopoulos [25] provided individualized prediction models of survival in AIDS patients who also had longitudinal CD4 cell count measurements. Our prediction approach is most closely related to that of Rizopoulos [25]. However, as is outlined in Section 2, we use a fully parametric model and a Bayesian approach similar to that of Guo and Carlin [14].
The remainder of the paper is organized as follows. In Section 2, we present our joint model assuming shared random effects for the longitudinal outcomes. In Section 3, we describe parameter estimation approach and model selection methods. In Section 4, we illustrate the dynamic prediction of event probabilities within a fixed time window. Section 5 presents simulation studies using the proposed model, whereas Section 6 applies the joint models to prediction of failure times in idiopathic pulmonary fibrosis data. Section 7 concludes with a brief discussion.
2. Joint modeling approach
2.1 Notation and definitions
For brevity, we focus the development of the model on the case of bivariate longitudinal responses, one continuous and normally distributed and the other binary. The generalization to higher dimensions and other members of the exponential family of distributions is conceptually straightforward (see Discussion). For subject i, i = 1,···, n, let y1ij and y2ij denote the jth outcome at time point tij consisting of continuous and binary components, respectively. Further, let denote the bivariate longitudinal outcome vector for subject i, where yhi = (yhi1,···, yhij)⊤, h = 1,2, j = 1,···, nhi is an nhi-dimensional column vector giving the hth longitudinal outcomes for subject i. For the longitudinal bivariate response vector, yi, with different data types, we assume a generalized linear mixed effects model
| (1) |
where gh(·) denotes known bijective link functions that differ across data types, Xhi and βh denote an nhi × ph design matrix of covariate values and a ph-dimensional vector of fixed effects, respectively, and Zhi and bhi denote nhi × q1 design matrix of covariates and a qh-dimensional vector of normally distributed random effects with zero mean and covariance matrix as described below. We assume that the elements of yhi are independent conditional on bhi [10,12].
Here, we choose the identity link for the continuous response and the logit link for the binary response. Thus the generalized linear mixed effects model (1) can be written in the form
| (2a) |
| (2b) |
We assume that b1i follows a normal distribution with a mean vector of zeros and variance-covariance matrix, Σ, and that b2i is proportional to b1i, i.e., b2i = A0b1i, where A0 is a diagonal matrix of unknown constants. A joint model with this assumption is termed a shared random effects joint model. We briefly outline another alternative, also implemented in our code, in the Discussion section.
2.2 Joint model of multivariate longitudinal outcomes and a time-to-event outcome
Let Ti denote the true event time for subject i, Ci be the censoring time, and δi = I(Ti ≤ Ci) be the event indicator. Let be the observed event time for subject i. We assume that censoring is non-informative [6]. A proportional hazards model is given by
| (3) |
where x3i is a p3-dimensional vector of covariates with regression coefficients β3, and λ0(t) is the baseline hazard function, which can be assumed to be of a parametric form or left unspecified. To express the effects of longitudinal outcomes on the time-to-event outcome, the shared parameters, U3i, are associated with the random effects of longitudinal outcomes b1i and b2i. The joint model connects the longitudinal response submodels (2a,b) and the event time outcome submodel (3)
| (4) |
where is a set of unknown constants and b3i is a normally distributed frailty term with mean zero and variance , independent of the . The hazard function of the time-to-event depends on the longitudinal outcomes through the shared b1i and b2i. Thus, the parameter a quantifies the degree of association explained by the random effects in (2a,b). In terms of a shared random effects joint model, Eq. (4) can be reduced in the form
The components of x1, x2, and x3 may or may not all be the same, allowing the longitudinal continuous and binary responses, and the time-to-event outcome to depend on different and/or overlapping covariate information.
3. Estimation and model selection
3.1 Parameter estimation
We assume that the longitudinal responses vectors yi and the time-to-event outcome T are independent conditional on covariates X, Z, and the random effects vector, b. The observed data for the ith subject, with ni repeated measurements, for the hth response are denoted by
Based on a full conditional independence assumption, we can express the joint distribution of the observed data for the ith subject as
where θ denotes the complete parameter vector and f(·) denotes a generic probability density function. Thus, the log-likelihood for the observed data is given by
where xhij and zhij are jth row of matrices xhi and zhi, respectively, and f(bi|Σ) is the multivariate normal density function of bi conditional on the covariance parameter matrix, Σ.
We implement a Bayesian approach for parameter inferences, using a Gibbs sampling algorithm. The algorithm was programmed using the R interface, rbugs, which accesses the software ‘OpenBUGS’ [21]. We implement the Gibbs sampler using weakly informative priors for all parameters. Fixed effects are normal with large variance components. Error variances and random effect covariances are distributed as inverse gamma and inverse Wishart distributions, respectively [2]. Code used in the analyses is available from the first author upon request.
3.2 Model selection criteria
Spiegelhalter et al. [28] proposed the Deviance Information Criterion (DIC) for model selection. The DIC is a generalization of the Akaike Information Criterion (AIC) [1] for hierarchical models based on the deviance of the posterior distribution. For parameter vector θ and observed data vector, y, let D(θ) be the deviance, D(θ) = −2 log f(y|θ) + 2 log h(y), where f(y|θ) is the likelihood function and h(y) is a standardizing function of the data alone [2]. We define D(θ) to be in our joint model, to denote the posterior expected deviance and θ̄ = Eθ|y[θ] to denote the mean of the posterior distribution of the parameters. The effective number of parameters, pD, which can capture the complexity of a model is . Then, the DIC is defined as . Smaller values of the DIC indicate better fitting models. As with the AIC, differences of the DIC between models are a tool used for model selection. Differences of 3 to 5 are considered meaningful. In this paper, we use the DIC for model selection.
4. Dynamic prediction of event probabilities
We compute dynamic probabilities of an event occurring within a fixed window as proposed by van Houwelingen and Putter [30]. Specifically, we considered the conditional survival function of time v ≥ t given survival to time t:
| (5) |
Equation (5) can also be expressed using a hazard function as
| (6) |
where Λ(·) denotes the cumulative hazard function and λ(·) denotes the instantaneous hazard function. Equation (6) implies that only the hazard on the interval [t, v] is necessary to predict the probability of event up to time v for a subject at risk just before time t. Let v = t + w be a fixed window of width w. We can relate the survival function to the cumulative distribution function as follows:
Fw(t) is evaluated at all-time points t where the estimates change value. The variance of this function is based on the Nelson-Aalen estimate of that cumulative hazard that is given as
where
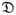 denotes the set of event times and R(t) denotes the size of the risk set, i.e., the number of subjects with no event and still being followed just before time t.
denotes the set of event times and R(t) denotes the size of the risk set, i.e., the number of subjects with no event and still being followed just before time t.
5. Simulation Studies
We conducted simulation studies to investigate the performance of the proposed joint models with the longitudinal continuous and binary outcomes and the time-to-event outcome. The simulated longitudinal data consisted of a quantitative outcome and a dichotomous outcome with seven repeated measurements at fixed times 0, 0.5, 1, 1.5, 2, 2.5, and 3 years. We considered three sample sizes, N=100, N = 200 and N = 500 subjects. Both longitudinal outcomes depended on the same fixed covariates: a continuous variable (x1) sampled from the normal distribution, x1 ~ N(0, 52), a dichotomous variable (x2) sampled from the Bernoulli distribution with probability 0.5, x2 ~ Bernoulli(0.5), and time (t), t = {0, 0.5, 1, 1.5, 2, 2.5, 3}, were included as fixed covariates. Subject-specific random intercepts and slopes were assumed. For the longitudinal continuous outcome, the measurement error term was normally distributed with mean zero and variance, . The continuous longitudinal outcome was generated from the model
The binary longitudinal outcome was generated from the model
and the random effects were assumed to have distribution
The time-to-event outcome was generated from an exponential distribution, T ~ exp(μ3), where μ3 = exp(β31 + β32x2 + α3b10 + α4b11 + b3) and with . A non-informative censoring time C was generated from a uniform distribution on [0.2, 2] which resulted in roughly 35% censoring on average. For each simulation study, 200 replications were performed. In each analysis, a total of 15,000 MCMC iterations were used, discarding the first 5,000 iterations as a burn-in.
Table 1 shows the results of the simulation studies, including true parameter values, bias (defined as the true parameter minus the mean estimated parameter), standard errors of the parameter estimates (SE), mean squared error (MSE), and the coverage probability of the estimated 95% credibility intervals (CP). With a few exceptions, most parameters in the joint models show acceptably low levels of bias and good coverage probabilities. For N=100, the biases of variance and covariance of random effects are relatively higher than for larger sample sizes, and it might create very low coverage probabilities. For N=200, CP averages close to 95% for all parameters. Most parameters have very small biases, with the exception of somewhat higher biases in the α parameters. However, biases in α are fairly small as a proportion of the size of the parameter, and decrease with larger sample sizes suggesting that a bias of the links between the longitudinal and time-to-event processes are strongly related to the number of events in the survival outcome. In general, as expected, larger sample sizes (N=500) show better results as indicated by the smaller bias, SE, MSE, and less variable CP that average very close to the nominal 95% level.
Table 1.
Simulation results
| Parameter | True | N = 100
|
N = 200
|
N = 500
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SE | MSE | CP | Bias | SE | MSE | CP | Bias | SE | MSE | CP | |||
|
|
|
|
|
|||||||||||
| β11 | 5.0 | −0.002 | 0.104 | 0.024 | 0.930 | −0.015 | 0.078 | 0.014 | 0.915 | 0.003 | 0.051 | 0.005 | 0.935 | |
| β12 | 0.8 | −0.005 | 0.138 | 0.047 | 0.920 | 0.008 | 0.107 | 0.029 | 0.880 | −0.010 | 0.071 | 0.010 | 0.955 | |
| β13 | −0.2 | 0.000 | 0.012 | 0.000 | 0.925 | 0.001 | 0.009 | 0.000 | 0.925 | 0.000 | 0.006 | 0.000 | 0.950 | |
| β14 | −0.2 | −0.004 | 0.056 | 0.007 | 0.930 | −0.005 | 0.044 | 0.004 | 0.910 | −0.004 | 0.028 | 0.002 | 0.945 | |
| β21 | 0.3 | 0.018 | 0.223 | 0.109 | 0.940 | −0.009 | 0.164 | 0.054 | 0.945 | −0.003 | 0.103 | 0.022 | 0.960 | |
| β22 | 0.3 | −0.016 | 0.266 | 0.163 | 0.920 | 0.013 | 0.198 | 0.084 | 0.930 | 0.001 | 0.125 | 0.029 | 0.965 | |
| β23 | −0.2 | −0.005 | 0.028 | 0.002 | 0.965 | −0.001 | 0.020 | 0.001 | 0.970 | −0.001 | 0.012 | 0.000 | 0.940 | |
| β24 | −0.25 | −0.013 | 0.114 | 0.028 | 0.930 | −0.015 | 0.083 | 0.015 | 0.920 | −0.004 | 0.052 | 0.006 | 0.925 | |
| β31 | 0.2 | −0.023 | 0.345 | 0.266 | 0.930 | −0.074 | 0.266 | 0.156 | 0.940 | −0.021 | 0.171 | 0.059 | 0.950 | |
| β32 | 1.0 | 0.003 | 0.419 | 0.398 | 0.945 | 0.031 | 0.307 | 0.216 | 0.915 | −0.002 | 0.194 | 0.072 | 0.985 | |
| α1 | 1.6 | 0.224 | 0.536 | 0.632 | 0.945 | 0.280 | 0.331 | 0.296 | 0.890 | 0.147 | 0.198 | 0.098 | 0.870 | |
| α2 | 1.2 | 0.154 | 0.244 | 0.147 | 0.935 | 0.040 | 0.161 | 0.055 | 0.935 | −0.012 | 0.100 | 0.022 | 0.930 | |
| α3 | 2.0 | 0.181 | 0.891 | 1.308 | 0.975 | 0.164 | 0.585 | 0.595 | 0.980 | 0.052 | 0.344 | 0.211 | 0.995 | |
| α4 | 3.5 | 0.369 | 0.647 | 0.836 | 0.975 | 0.153 | 0.457 | 0.379 | 0.985 | 0.087 | 0.291 | 0.158 | 0.960 | |
|
|
1.0 | 0.029 | 0.061 | 0.008 | 0.925 | 0.023 | 0.043 | 0.004 | 0.940 | 0.008 | 0.027 | 0.001 | 0.975 | |
|
|
0.25 | 0.005 | 0.058 | 0.003 | 1.000 | 0.001 | 0.055 | 0.003 | 1.000 | −0.011 | 0.049 | 0.003 | 1.000 | |
|
|
0.2 | −0.073 | 0.020 | 0.006 | 0.010 | −0.024 | 0.026 | 0.002 | 0.865 | 0.019 | 0.026 | 0.001 | 0.955 | |
|
|
0.25 | −0.081 | 0.022 | 0.007 | 0.080 | −0.016 | 0.025 | 0.001 | 0.895 | 0.013 | 0.021 | 0.001 | 0.950 | |
| ρ12 | 0.5 | −0.220 | 0.088 | 0.060 | 0.165 | −0.027 | 0.067 | 0.007 | 0.995 | 0.043 | 0.052 | 0.006 | 0.945 | |
6. Application to mortality in idiopathic pulmonary fibrosis outcomes study
6.1 Data description and Preliminary Analyses
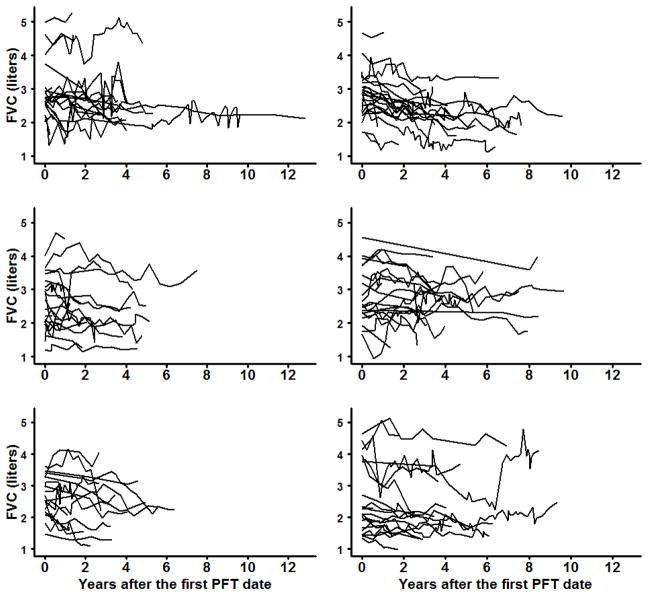
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive lung disease characterized by fibrosis of unknown etiology [24]. Our primary concern is prediction of failure times, defined as time to either lung transplant or death, utilizing longitudinally measured forced vital capacity (FVC), a continuous measure of pulmonary function. Individual profiles for the pulmonary function assessed by FVC are shown in Figure 1, with subjects randomly apportioned to six subplots in order to more clearly depict these trajectories. An additional disease progression indicator, defined as a decline of 5% or more in FVC from the baseline FVC measurement, is also used as a (binary) longitudinal response. Baseline covariates also considered for inclusion in the models were: age centered at time of diagnosis, baseline FVC, smoking, gender, and time in years.
Figure 1.
Subject-specific trajectories of FVC measurements. (PFT: pulmonary function tests)
Of the 125 patients considered in the study, 64 (51.2%) patients died or had lung transplants. Patient characteristics are summarized in Table 3. This dataset was comprised of irregular follow-up times across patients, but generally patients were followed every 3–4 months according to standard of care. The time origin for each patient was the baseline assessment of pulmonary function, performed upon evaluation at the Simmons Center for Interstitial Lung Disease (ILD). The FVC of each patient was repeatedly measured about 11 times on average, up to 43 times. Follow-up information for the study was reported through December 31, 2011. The average follow-up time was 3.7 years among all patients and 4.1 years among patients who did not die nor received a lung transplant. All IPF patients were evaluated at the University of Pittsburgh Medical Center and clinical data were obtained from the Simmons Center for ILD.
Table 3.
Bayesian Model Selection of shared random effects joint model
| Model | U1(t) | U2(t) | U3 | DICy1 | DICy2 | DICT | D̄ | pD | DICtotal |
|---|---|---|---|---|---|---|---|---|---|
| no random effects | |||||||||
| 1 | 0 | 0 | 0 | 1795 | 1715 | 356.2 | 3856 | 11.02 | 3867 |
| 2 | 0 | 0 | b3 | 1795 | 1715 | 354.3 | 3842 | 23.11 | 3865 |
| random intercepts | |||||||||
| 3 | b10 | 0 | 0 | 763.8 | 1715 | 356.1 | 2719 | 115.7 | 2835 |
| 4 | b10 | 0 | b3 | 763.9 | 1715 | 354.5 | 2706 | 127.9 | 2833 |
| 5 | b10 | 0 | α3b10 | 763.6 | 1715 | 357.3 | 2720 | 116.3 | 2836 |
| 6 | b10 | 0 | α3b10 + b3 | 763.6 | 1715 | 355.2 | 2706 | 128.2 | 2834 |
| 7 | b10 | α1b10 | 0 | 719.5 | 1189 | 356.2 | 2143 | 122.5 | 2265 |
| 8 | b10 | α1b10 | b3 | 718.8 | 1189 | 354.5 | 2128 | 134.3 | 2263 |
| 9 | b10 | α1b10 | α3b10 | 719.4 | 1189 | 357.8 | 2143 | 122.9 | 2266 |
| 10 | b10 | α1b10 | α3b10 + b3 | 718.8 | 1189 | 355.3 | 2128 | 134.7 | 2263 |
| random intercepts and random slopes | |||||||||
| 11 | b10 + b11t | 0 | 0 | 241.8 | 1715 | 356.2 | 2113 | 200.4 | 2313 |
| 12 | b10 + b11t | 0 | b3 | 241.9 | 1715 | 354.2 | 2099 | 212.5 | 2311 |
| 13 | b10 + b11t | 0 | α3b10 | 241.7 | 1715 | 355.2 | 2111 | 200.8 | 2312 |
| 14 | b10 + b11t | 0 | α4b11 | 241.9 | 1715 | 350.4 | 2104 | 203.1 | 2307 |
| 15 | b10 + b11t | 0 | α3b10 + b3 | 241.9 | 1715 | 353.0 | 2098 | 212.2 | 2310 |
| 16 | b10 + b11t | 0 | α4b11 + b3 | 241.0 | 1715 | 351.5 | 2095 | 212.1 | 2307 |
| 17 | b10 + b11t | 0 | α3b10 + α4b11 | 242.3 | 1715 | 351.1 | 2106 | 202.1 | 2308 |
| 18 | b10 + b11t | 0 | α3(b10 + b11) | 238.9 | 1715 | 352.7 | 2106 | 200.7 | 2307 |
| 19 | b10 + b11t | 0 | α3b10 + α4b11 + b3 | 238.9 | 1715 | 349.7 | 2092 | 211.4 | 2304 |
| 20 | b10 + b11t | α1b10 | 0 | 374.3 | 1227 | 355.9 | 1742 | 215.1 | 1957 |
| 21 | b10 + b11t | α1b10 | b3 | 373.7 | 1227 | 354.6 | 1728 | 227.3 | 1955 |
| 22 | b10 + b11t | α1b10 | α3b10 | 374.4 | 1226 | 357.5 | 1743 | 215.8 | 1958 |
| 23 | b10 + b11t | α1b10 | α4b11 | 372.6 | 1226 | 346.6 | 1727 | 217.7 | 1945 |
| 24 | b10 + b11t | α1b10 | α3b10 + b3 | 375.0 | 1228 | 355.1 | 1729 | 228.6 | 1958 |
| 25 | b10 + b11t | α1b10 | α4b11 + b3 | 372.6 | 1227 | 348.0 | 1719 | 228.6 | 1947 |
| 26 | b10 + b11t | α1b10 | α3b10 + α4b11 | 372.8 | 1225 | 347.0 | 1726 | 218.9 | 1945 |
| 27 | b10 + b11t | α1b10 | α3(b10 + b11) | 373.8 | 1227 | 356.2 | 1741 | 216.2 | 1957 |
| 28 | b10 + b11t | α1b10 | α3b10 + α4b11 + b3 | 373.6 | 1226 | 348.9 | 1719 | 229.4 | 1949 |
| 29 | b10 + b11t | α1b10 + α2b11t | 0 | 167.2 | 923.2 | 356.1 | 1235 | 211.6 | 1447 |
| 30 | b10 + b11t | α1b10 + α2b11t | b3 | 167.0 | 923.2 | 354.4 | 1221 | 223.9 | 1445 |
| 31 | b10 + b11t | α1b10 + α2b11t | α3b10 | 168.6 | 922.4 | 356.3 | 1235 | 211.9 | 1447 |
| 32 | b10 + b11t | α1b10 + α2b11t | α4b11 | 164.1 | 923.0 | 352.9 | 1229 | 210.6 | 1440 |
| 33 | b10 + b11t | α1b10 + α2b11t | α3b10 + b3 | 166.0 | 923.1 | 353.4 | 1218 | 224.0 | 1442 |
| 34 | b10 + b11t | α1b10 + α2b11t | α4b11 + b3 | 167.2 | 924.0 | 351.8 | 1219 | 224.3 | 1443 |
| 35 | b10 + b11t | α1b10 + α2b11t | α3b10 + α4b11 | 166.2 | 923.8 | 352.8 | 1230 | 212.7 | 1443 |
| 36 | b10 + b11t | α1b10 + α2b11t | α3(b10 + b11) | 165.3 | 922.5 | 354.8 | 1232 | 211.0 | 1443 |
| 37 | b10 + b11t | α1b10 + α2b11t | α3b10 + α4b11 + b3 | 167.6 | 924.3 | 351.0 | 1218 | 224.5 | 1443 |
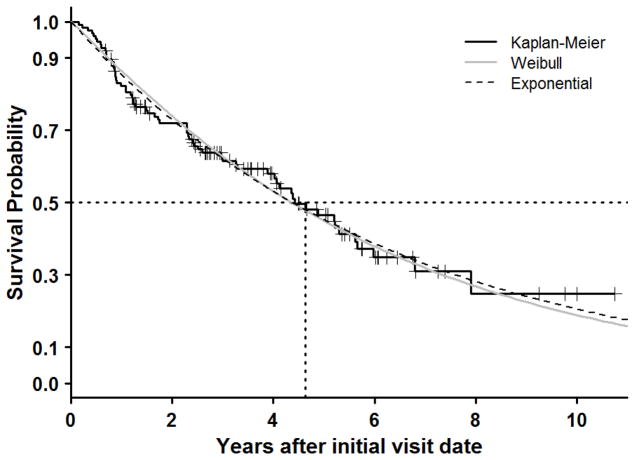
Fitted survival curves of the Weibull model and exponential model are shown in Figure 2. The survival curves of the Weibull model and exponential both show a good fit to the marginal survival function. Because the Weibull and exponential models are almost identical we used the more parsimonious exponential model. The estimated median transplant-free survival time from the initial visit date was 4.4 years.
Figure 2.
The Kaplan-Meier plot of transplant-free survival (black solid curve); Weibull survival curve (grey solid curve) and exponential survival curve (black broken curve).
6.2 Joint model
As before, let y1ij be the continuous longitudinal outcome indicating FVC measurements, y2ij be the binary longitudinal outcome indicating disease progression indicator, and Ti be the transplant-free survival time. More specifically, the model is given by
Under the shared random effects joint model, the random effects terms of the three submodels are defined as follows:
U1i(tij) = b10i + b11itij
U2i(tij) = α1b10i + α2b11itij
U3i = α3b10i + α4b11i + b3i
The association between longitudinal responses and time-to-event outcome is explained by the parameter α = (α1, α2, α3, α4). For the continuous longitudinal response, we included the covariates of baseline FVC and time in years. For the binary longitudinal response, age and time in years were found to be significant covariates. Gender, smoking, and baseline FVC were significant in the model for transplant-free survival.
Non-informative priors were used for all parameters. For the coefficients of the fixed effects β1, β2, and β3, we assume a multivariate normal distribution with mean zero and variance-covariance matrices 100I3, 100I3, and 100I4, respectively, where Ik indicates an identity matrix with dimension k. We assumed that the association coefficient, α, had a normal distribution with mean zero and variance equal to 100. A total of 15,000 MCMC iterations were used discarding the first 5,000 iterations for the burn-in period.
6.3 Results
We considered all possible combinations of random effects resulting in 37 assessed models. Table 3 summarizes DICs for each submodel, the posterior expected deviance, D̄, the effective number of parameters, pD, and total DIC scores (DICtotal) for each joint model. Note that DICy1, DICy2, and DICT denote DIC from the longitudinal continuous outcome, the longitudinal binary outcome, and the time-to-event outcome submodels, respectively. These scores show relative contributions of each component to the overall model DIC. DICtotal, was used to choose the “best” model.
Joint models with both subject-specific random intercepts and slopes in both the longitudinal continuous outcome submodel and binary outcome submodel (Models 29–37) have the smallest DICs, and hence the smallest DICtotal compared with other joint models with random effect structures. In the time-to-event submodel, adding a frailty term leads to only a very slight decrease in DICT. Model 32 with the smallest DICtotal was selected as our best model. Under Model 32, the longitudinal binary outcome shares the subject-specific random intercept and slope of the longitudinal continuous outcome and the time-to-event outcome is related to the subject-specific random slope of the longitudinal continuous outcome.
In Table 4, the posterior estimates for Model 32 parameters along with their standard errors and 95% credibility intervals are presented. For longitudinal continuous FVC, both the baseline FVC and time are statistically significant. Hence, patients with high values of baseline FVC have high FVC measurements and FVC measurements tend to decrease as time progresses. For the longitudinal binary outcome, there is a significant time effect. For the time-to-event outcome, male smokers with lower baseline FVC have a higher risk of death or transplant. The association parameter, α4 indicates that there is a nearly significant negative association between the subject-specific random slopes of FVC measurements and the hazard of the transplant-free survival outcome.
Table 4.
Posterior estimates for Model 32
| Parameter | Posterior Mean | Std. Error | 95% CI |
|---|---|---|---|
| Longitudinal continuous submodel | |||
| Intercept (β11) | 0.167 | 0.048 | (0.072, 0.263) |
| BaselineFVC (β12) | 0.933 | 0.014 | (0.906, 0.961) |
| Time (β13) | −0.094 | 0.016 | (−0.122, −0.061) |
| 0.081 | 0.008 | (0.066, 0.098) | |
| 0.068 | 0.007 | (0.056, 0.084) | |
| ρb1 | −0.068 | 0.069 | (−0.200, 0.067) |
| 0.060 | 0.002 | (0.055, 0.064) | |
| Longitudinal binary submodel | |||
| Intercept (β21) | −1.424 | 0.325 | (−2.043, −0.796) |
| Age (β22) | −0.015 | 0.014 | (−0.044, 0.012) |
| Time (β23) | 1.019 | 0.180 | (0.6513, 1.341) |
| α1 | −10.67 | 0.835 | (−12.38, −9.145) |
| α2 | −10.31 | 0.727 | (−11.82, −8.961) |
| Time-to-event submodel | |||
| Intercept (β31) | −1.967 | 0.530 | (−3.003, −0.939) |
| Male (β32) | 0.903 | 0.360 | (0.214, 1.648) |
| Smoking (β33) | 0.719 | 0.341 | (0.087, 1.422) |
| BaselineFVC (β34) | −0.405 | 0.203 | (−0.807, −0.013) |
| α4 | −1.613 | 0.810 | (−3.201, −0.025) |
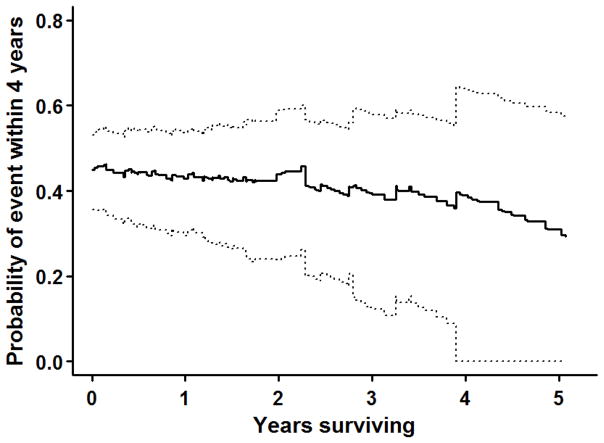
Figure 3 shows the predicted four-year-ahead failure probabilities for the IPF data set along with 95% confidence intervals, obtained from the best-fitting Model 32. In general, the probability of event within the next four years decreases slowly, with a deceleration apparent if the patient survives longer than two years, so that at baseline subjects have ~45% chance of failure in four years, whereas if a patient does not experience failure by year five, the probability of a failure in the next four years decreases to ~35%. As time progresses, confidence intervals become wider because of the sparsity of the data.
Figure 3.
Probability of transplant or death within the next four years using shared random effects joint model 32: Point estimates (solid curve) and 95 % confidence intervals (dotted curves).
7. Discussion
In this paper, we have developed a joint model for multivariate longitudinal continuous and binary responses and a time-to-event outcome, applying this model to the prediction of failure times from idiopathic pulmonary fibrosis. This general approach was introduced in Rizopoulos and Ghosh [26] who used natural splines to model three mixed longitudinal processes (two continuous and one binary) as predictors of a time-to-event outcome. In our case, we used parametric models for the longitudinal processes as the number of longitudinal observations per individual was relatively small and hence parametric random effect models were more tractable. Simulation studies indicate good performance for our models.
Our joint model was developed utilizing a unified Bayesian framework, including model fitting, parameter inferences, computation of dynamic prediction probabilities with posterior credible intervals, and a DIC model selection criterion that parses out the contributions of the joint sub-models to the overall fit. Results from the best fitting model indicate that the two longitudinal responses jointly contribute nearly significantly to prediction of failure times in IPF as indicated by the value of the shared parameter, α4, and its 95% credibility interval (Table 4). Our approach for choosing prior distributions followed that of Guo and Carlin [14], which is quite general and easier to implement than that used by Rizopoulos and Ghosh [26].
In our present investigation, we utilized shared random effects to characterize the relationship between the longitudinal continuous and binary outcomes. A disadvantage of the shared random effects joint model is that the structure of the random effects for both the longitudinal outcomes is limited. Nevertheless, shared parameter random effects models do gain efficiency by using a smaller number of random effects parameters compared to other joint model formulations.
A practical purpose our investigation was to illustrate the utility of joint models for computing dynamic predictions of the probabilities of events occurring within a fixed window of time. Given a subject is at risk just before time t, the probability of an event occurring within the next fixed window of time is predicted using the survival function obtained from our proposed joint models. By predicting the probability of event within a fixed window of time we can easily visualize conditional probabilities of events. Plots such as these can be an effective means for presenting dynamic risk of failure times to clinician researchers. Finally, we have implemented our shared random effects joint models in an R interface using OpenBugs, available from the first author upon request.
Finally, in related work, we have implemented a correlated random effects joint model [5] which we plan to investigate further. As part of this future investigation, we wish to extend models proposed by Lee and Nelder [17,18] that expand the correlated random effects models by introducing a general method called maximum h-likelihood estimation. In the correlated random effects model, the relationship between the continuous and binary outcomes can be investigated through correlations among the random effects b1i and b2i.
Table 2.
Patient characteristics of Idiopathic Pulmonary Fibrosis (IPF) outcome data
| N(=125) | % | |
|---|---|---|
| Gender | ||
| Male | 84 | 67.2 |
| Female | 41 | 32.8 |
| Race | ||
| White | 122 | 97.6 |
| Black | 1 | 0.8 |
| American Indian | 1 | 0.8 |
| Oriental | 1 | 0.8 |
| Smoking | ||
| Ever | 88 | 70.4 |
| Never | 37 | 29.6 |
| Diagnosis made | ||
| Clinically | 63 | 50.4 |
| Historically | 62 | 49.6 |
| Transplant | ||
| Yes | 23 | 18.4 |
| No | 102 | 81.6 |
| Mean | SD | |
| Age (year) | ||
| Overall | 65.2 | 9.34 |
| Male | 65.7 | 9.00 |
| Female | 64.2 | 10.04 |
| Follow-up (year) | ||
| All patients | 3.7 | 2.32 |
| Alive and not transplanted | 4.1 | 2.35 |
| Baseline PFTs | ||
| FVC | 2.7 | 0.85 |
| FEV1 | 2.2 | 0.65 |
| DLCO | 12.7 | 4.36 |
Definition of abbreviations: PFT = pulmonary function tests; FVC = forced vital capacity; FEV1 = forced expiratory volume in 1 second; DLCO = diffusing capacity of carbon monoxide.
Acknowledgments
The authors would like to thank Drs. Naftali Kaminski and Charles F. Reynolds III for providing data which helped motivate the methods demonstrated in this manuscript. This work was partially supported by NCI grant numbers U10-CA069974 and U10-CA069651 (Anderson) and NIMH grant number P30-MH-090333 (Anderson and Thompson).
References
- 1.Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csáki F, editors. Proc 2nd Int Symp Information Theory. Budapest: Akadémiai Kiadó; 1973. pp. 267–281. [Google Scholar]
- 2.Carlin BP, Louis TA. Bayesian Methods for Data Analysis. 3. Chapman and Hall/CRC Press; Boca Raton, FL: 2009. [Google Scholar]
- 3.Catalano PJ, Ryan LM. Bivariate latent variable models for clustered discrete and continuous outcomes. J Amer Statist Assoc. 1992;87:651–658. [Google Scholar]
- 4.Chi YY, Ibrahim JG. Joint models for multivariate longitudinal and multivariate survival data. Biometrics. 2006;62:432–445. doi: 10.1111/j.1541-0420.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- 5.Choi J-I. PhD dissertation. University of Pittsburgh; 2012. Prediction in the Joint Modeling of Mixed Types of Multivariate Longitudinal Outcomes and a Time-to-Event Outcome. [Google Scholar]
- 6.Collett D. Modelling Survival Data in Medical Research. Chapman & Hall; London: 1994. [Google Scholar]
- 7.Faucett CJ, Thomas DC. Simultaneously modeling censored survival data and repeated measured covariates: A Gibbs sampling approach. Stat Med. 1996;15:1663–1685. doi: 10.1002/(SICI)1097-0258(19960815)15:15<1663::AID-SIM294>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Fieuws S, Verbeke G. Joint modelling of multivariate longitudinal profiles: pitfalls of the random-effects approach. Stat Med. 2004;23:3093–3104. doi: 10.1002/sim.1885. [DOI] [PubMed] [Google Scholar]
- 9.Fieuws S, Verbeke G, Maes B, Vanrenterghem Y. Predicting renal graft failure using multivariate longitudinal profiles. Biostatistics. 2008;9:419–431. doi: 10.1093/biostatistics/kxm041. [DOI] [PubMed] [Google Scholar]
- 10.Fitzmaurice GM, Davidian M, Verbeke G, Molenberghs G. Longitudinal Data Analysis. Chapman and Hall/CRC Press; Boca Raton, FL: 2009. [Google Scholar]
- 11.Fitzmaurice GM, Laird NM. Regression models for a bivariate discrete and continuous outcome with clustering. J Amer Statist Assoc. 1995;90:845–852. [Google Scholar]
- 12.Fitzmaurice GM, Laird NM, Ware J. Applied Longitudinal Data. Wiley; Hoboken, New Jersey: 2004. [Google Scholar]
- 13.Gueorguieva RV, Sanacora G. Joint analysis of repeatedly observed continuous and ordinal measures of disease severity. Stat Med. 2006;25:1307–1322. doi: 10.1002/sim.2270. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Carlin BP. Separate and joint modeling of longitudinal and event time data using standard computer packages. Amer Statist. 2004;58:16–24. [Google Scholar]
- 15.Henderson R, Diggle P, Dobson A. Joint modelling of longitudinal measurements and event time data. Biostatistics. 2000;1:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- 16.Hogan JW, Laird NM. Mixture models for the joint distribution of repeated measures and event times. Stat Med. 1997;16:239–257. doi: 10.1002/(sici)1097-0258(19970215)16:3<239::aid-sim483>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Nelder JA. Hierarchical generalized linear models. Journal of the Royal Statistical Society Series B (Methodological) 1996:619–678. [Google Scholar]
- 18.Lee Y, Nelder JA, Pawitan Y. Generalized linear models with random effects: unified analysis via H-likelihood. CRC Press; 2006. [Google Scholar]
- 19.Little RJA. Pattern-mixture models for multivariate incomplete data. J Amer Statist Assoc. 1993;88:125–134. [Google Scholar]
- 20.Liu X, Daniels MJ, Marcus B. Joint models for the association of longitudinal binary and continuous processes with application to a smoking cessation trial. J Amer Statist Assoc. 2010;104:429–438. doi: 10.1198/016214508000000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lunn D, Spiegelhalter D, Thomas A, Best N. The BUGS project: Evolution, critique and future directions (with discussion) Stat Med. 2009;28:3049–3082. doi: 10.1002/sim.3680. [DOI] [PubMed] [Google Scholar]
- 22.Molenberghs G, Verbeke G. Models for Discrete Longitudinal Data. Springer; New York: 2005. [Google Scholar]
- 23.Molenberghs G, Verbeke G, Demetrio CGB, Vieira AM. A family of generalized linear models for repeated measures with normal and conjugate random effects. Statist Sci. 2010;25:325–347. [Google Scholar]
- 24.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, Klesen M, Zhang Y, Gibson KF. Peripheral Blood Proteins Predict Mortality in Idiopathic Pulmonary Fibrosis. American Journal of Respiratory and Critical Care Medicine. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizopoulos D. Dynamic predictions and prospective accuracy in joint models for longitudinal and time-to-event data. Biometrics. 2011;67:819–829. doi: 10.1111/j.1541-0420.2010.01546.x. [DOI] [PubMed] [Google Scholar]
- 26.Rizopoulos D, Ghosh P. A Bayesian semiparametric multivariate joint model for multiple longitudinal outcomes and a time-to-event. Stat Med. 2011;30:1366–1380. doi: 10.1002/sim.4205. [DOI] [PubMed] [Google Scholar]
- 27.Song X, Davidian M, Tsiatis AA. A semiparametric likelihood approach to joint modeling of longitudinal and time-to-event data. Biometrics. 2002;58:742–753. doi: 10.1111/j.0006-341x.2002.00742.x. [DOI] [PubMed] [Google Scholar]
- 28.Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian Measures of Model Complexity and Fit. J R Stat Soc Ser B Stat Methodol. 2002;64:583–639. [Google Scholar]
- 29.Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: An overview. Statist Sinica. 2004;14:809–834. [Google Scholar]
- 30.van Houwelingen HC, Putter H. Dynamic Prediction in Clinical Survival Analysis. Chapman and Hall/CRC Press; Boca Raton, FL: 2011. [Google Scholar]