Abstract
We utilized a full genome cDNA microarray to identify the genes that comprise the peroxide stimulon in the cyanobacterium Synechocystis sp. strain PCC 6803. We determined that a gene (slr1738) encoding a protein similar to PerR in Bacillus subtilis was induced by peroxide. We constructed a PerR knockout strain and used it to help identify components of the PerR regulon, and we found that the regulatory properties were consistent with the hypothesis that PerR functions as a repressor. This effort was guided by finding putative PerR boxes in positions upstream of specific genes and by careful statistical analysis. PerR and sll1621 (ahpC), which codes for a peroxiredoxin, share a divergent promoter that is regulated by PerR. We found that isiA, encoding a Chl protein that is induced under low-iron conditions, was strongly induced by a short-term peroxide stress. Other genes that were strongly induced by peroxide included sigD, sigB, and genes encoding peroxiredoxins and Dsb-like proteins that have not been studied yet in this strain. A gene (slr1894) that encoded a protein similar to MrgA in B. subtilis was upregulated by peroxide, and a strain containing an mrgA knockout mutation was highly sensitive to peroxide. A number of genes were downregulated, including key genes in the chlorophyll biosynthesis pathway and numerous regulatory genes, including those encoding histidine kinases. We used PerR mutants and a thioredoxin mutant (TrxA1) to study differential expression in response to peroxide and determined that neither PerR nor TrxA1 is essential for the peroxide protective response.
The appearance of oxygen on earth from oxygenic photosynthesis resulted in the production of potentially deleterious reactive oxygen species, such as superoxide and peroxide (19). Superoxide is not extremely reactive by itself, but the hydroxyl radicals formed can damage many biological macromolecules. All organisms have developed ways of protecting themselves against reactive oxygen species, including specific defenses and global responses that enable cells to survive oxidative stress. These defenses include superoxide dismutases, which convert superoxides to hydrogen peroxide, and the related catalases and peroxides, which degrade simple and organic peroxides. Bacteria display a classic adaptive response after treatment with low levels of oxidants, such as hydrogen peroxide; the treated cells have an enhanced ability to withstand subsequent treatment with a dose that normally would be lethal to the cells. This type of adaptive response is coordinated by transcriptional factors that sense the oxidative stress stimuli and control subsequent gene expression (60, 61). Escherichia coli and Bacillus subtilis are the microorganisms in which oxidative stress defense regulation has been studied most. In E. coli, significant progress has been made in elucidating the mechanisms by which the activities of the SoxR and OxyR transcription factors respond to O2·− and H2O2 stress, respectively. OxyR reacts with H2O2 to form an intramolecular disulfide bond, and the resulting conformational change activates the protein (8, 49, 61, 75). Similarly, in Streptomyces coelicolor oxidation of the RsrS anti-sigma factor by H2O2 or diamide leads to disulfide bond formation and results in release of σR and activation of target genes (28). However, not all bacteria have an OxyR homologue. Recently, a Fur-like peroxide-sensing repressor, PerR, was found in some organisms that lack OxyR. The PerR regulon in B. subtilis includes katA, ahpCF, mrgA, hemAXCDBL, fur, and perR itself. Moreover, PerR is a metalloprotein, and its regulation involves both oxidation and metal ions (17, 20, 21, 40).
PerR functions as the central regulator of the inducible peroxide stress response in certain gram-positive and gram-negative bacteria (17, 21, 40). PerR regulation was first described in B. subtilis, in which it regulates the peroxide stress response in regard to iron, manganese, and peroxide stress conditions (4). B. subtilis contains three Fur homologues that coordinate gene expression in response to iron (Fur), zinc (Zur), or H2O2 (PerR). All three proteins are dimeric, DNA-binding repressors that contain a single Zn(II) atom per monomer and a second regulatory metal ion, either Mn(II) or Fe(II), that acts as a cofactor that is necessary for binding to the target operator sites (17). When PerR binds to Mn(II), it is not readily dissociated from DNA by peroxide, whereas when PerR binds to iron, it can be readily dissociated from DNA by low levels of H2O2 (21).
The adaptive response to peroxide stress in B. subtilis is coordinated by at least two other transcription factors, σB and OhrR (20). The general stress response, coordinated by the σB protein, includes paralogs of many of the stress proteins, including two additional catalases (KatB and KatX). OhrR is representative of a recently identified class of peroxide sensors and acts as a repressor of the OhrA organic peroxide resistance gene (16, 62). Given the wide range of oxidative damage to the cell, it is conceivable that DNA repair, protein degradation, metabolic energy generation, cell division, and other cellular activities are all coordinately regulated. Together, these activities form an oxidative stress response network or stimulon.
Because cyanobacteria are the oldest known organisms that perform oxygenic photosynthesis, their responses to oxidative stresses have been studied extensively. However, until recently, even the identities of the protective enzymes were poorly characterized. Advances in this area were greatly aided by the complete genomic sequences of strains such as Synechocystis sp. strain PCC 6803 and Anabaena sp. strain PCC 7120, and the occurrence and biochemistry of cyanobacterial hydroperoxidases have been reviewed recently (31, 43, 48, 53). These studies have led to a better understanding of the role of catalase peroxidases (63), peroxiredoxins, glutathione peroxidases (53), and thioredoxin peroxidase (71) in these cyanobacteria. It is now recognized that there is a thioredoxin superfamily (proteins that have a thioredoxin-like fold and that interact with either thiol- or disulfide-containing substrates) that includes six subclasses: glutathione peroxidase, glutathione S-transferase, thioredoxin, glutaredoxin, Dsb, and peroxiredoxin (54, 55). In addition, insightful studies of the inhibition by oxidative stress of the repair of photodamage to photosystem II (PSII) have led to the conclusion that peroxide affects the translational machinery and not PSII directly (43).
To identify the genes that are involved with or regulate the anti-oxidative stress and repair system, we performed DNA microarray experiments with the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 (50). In this paper, we describe some of the key genes that are differently regulated by hydrogen peroxide. We immediately identified a gene encoding a putative PerR homologue in Synechocystis sp. strain PCC 6803 that responded strongly to peroxide stress, and below we outline our current view of the PerR regulon. We also constructed a perR knockout mutant and used microarray and physiological analyses to determine how this mutant responded to peroxide stress. We are also interested in redox regulation, and we performed a related analysis with a Synechocystis sp. strain PCC 6803 mutant that lacked one of the thioredoxins. We show here that some of the iron-stress-related genes were highly expressed under oxidative stress conditions, indicating that there is some coupling between the oxidative stress response and regulation of iron metabolism. Based on these experiments, we developed a general outline of the peroxide stimulon and the PerR regulon in this photosynthetic phototroph.
MATERIALS AND METHODS
Strains, growth conditions, and stress induction.
The bacterial strains and plasmids used in this work are listed in Table 1. Synechocystis sp. wild-type strain PCC 6803 and mutants were cultured at 30°C with 40 μmol of photons m−2 s −1 in a modified BG-11 medium as the basal medium. These growth conditions are referred to below as the control. The cell density of a culture was determined by absorption at 730 nm as previously described (10, 39).
TABLE 1.
Strains, plasmids, and primers used in this work
| Strain, plasmid, or primer | Relevant genotype, description, or sequence | Source or reference |
|---|---|---|
| Synechocystis strains | ||
| PCC 6803 (wild type) | Laboratory collection | |
| ΔperR mutant | PCC 6803 slr1738::Kanr | This study |
| ΔtrxA1 mutant | PCC 6803 slr0623::Spr | This study |
| Plasmids | ||
| P1738 | perR of Synechocystis sp. strain PCC 6803 in pGEM-T vector | This study |
| P0623 | trxA1 of Synechocystis sp. strain PCC 6803 in pGEM-T vector | This study |
| P1738- Kanr | Kanamycin gene cloned in P1738 plasmid | This study |
| P0623- Spr | Spectinomycin gene cloned in P0623 plasmid | This study |
| Primers | ||
| slr1738-forward | 5′-CCATTCTGAGGGAATAACACTGCC-3′ | |
| slr1738-reverse | 5′-ACAGGCAACAGGGATAATCAAGGC-3′ | |
| slr0623-forward | 5′-CAGATCGGTCAACTGGGCGTTTGG-3′ | |
| slr0623-reverse | 5′-CGCTATGGTCTTACCGACAAAGACC-3′ |
We used microtiter plates in order to analyze the impact of oxidative stress conditions on growth. Cultures of wild-type and mutant strains were grown to the exponential phase (∼7 × 107 cells ml−1) or the stationary phase (∼2.0 × 108 cells ml−1) and diluted with fresh BG-11 medium to obtain 4 × 107 cells ml−1. Aliquots (0.5 ml) were placed in a 48-well microtiter plate. Different concentrations of H2O2 (0, 0.6, 0.9, 1.2, 1.5, and 1.8 mM) or methyl viologen (0, 0.5, 1, 2, 3, and 4 μM; Sigma, St. Louis, Mo.) were added to the wells. The microtiter plates were incubated at 30°C with 40 μmol of photons m−2 s−1 and shaking for approximately 2 to 3 days. Cell growth was measured in 96-well UV-transparent microplates (BD Biosciences, San Jose, Calif.) by using a Spectramax Plus384 (Molecular Devices Corporation, Sunnyvale, Calif.) with 100 μl of cells in each well. The absorbance at 730 nm provided a way to monitor cell density and allowed us to calculate an approximate concentration required for 50% inhibition of the control for each oxidative stress treatment. The final results were recorded by digital photography. Growth experiments were repeated at least three times for each of the parameters measured.
To determine the effect of oxidative conditions on gene expression, cells were grown in BG-11 medium at 30°C until the late logarithmic phase (∼1.0 × 108 cells ml−1), 1.5 mM H2O2 was added to one-half of the culture, and the cells were harvested after 30 min.
Mutant construction.
PCR products that contained perR and trxA1 (Table 1) were cloned into the pGEM-T vector (Promega, Madison, Wis.). ΔtrxA1 was constructed by replacing a 252-bp fragment with the 2.0-kb spectinomycin resistance cassette (from plasmid pRL453), and trxM1 was inactivated by replacing a 590-bp fragment with the 1.1-kb kanamycin resistance cassette. Synechocystis sp. strain PCC 6803 was transformed with these plasmid constructs, and transformants were selected on plates containing antibiotics (40 μg ml−1). Segregation of the mutants was performed under low light (10 μmol of photons m−2 s−1) in the presence of 5 mM glucose. Colonies were streaked five times prior to analysis, and complete segregation was confirmed by Southern blot, PCR, and Western blot analyses (data not shown). It was also important to ensure that each resulting segregant was based on a single mutation in the gene of interest and that no second-site mutations were present. Therefore, complementation of the single and double mutants was performed, and the complemented strains responded like the wild type (data not shown). Overall, the results demonstrated that there were fully segregated knockout mutations in each of the genes. Although we obtained mutants with knockout mutations of trxA1 and trxM1, we were unable to obtain fully segregated trxM2 (sll1057), trxA2 (slr1139), and trxA3 (sll1980) mutants.
RNA isolation and Northern analysis.
Total RNA was extracted and purified by using phenol-chloroform extraction and CsCl2 gradient purification as previously described (52, 59). Five micrograms of total RNA was fractionated by electrophoresis on a 1.0% agarose gel with 0.6 M formaldehyde. RNA was transferred to a nylon membrane and fixed by baking the membrane at 80°C for 2 h in a vacuum oven. The blots were hybridized with α-32P-labeled DNA probes prepared by random primer labeling by using a Ready-To-Go kit (Pharmacia Biotech, Piscataway, N.J.). Hybridization was performed at 42°C with 50% formamide. Staining of the rRNA with ethidium bromide was used to standardize the loading of total RNA.
Microarrays and differential expression experiments.
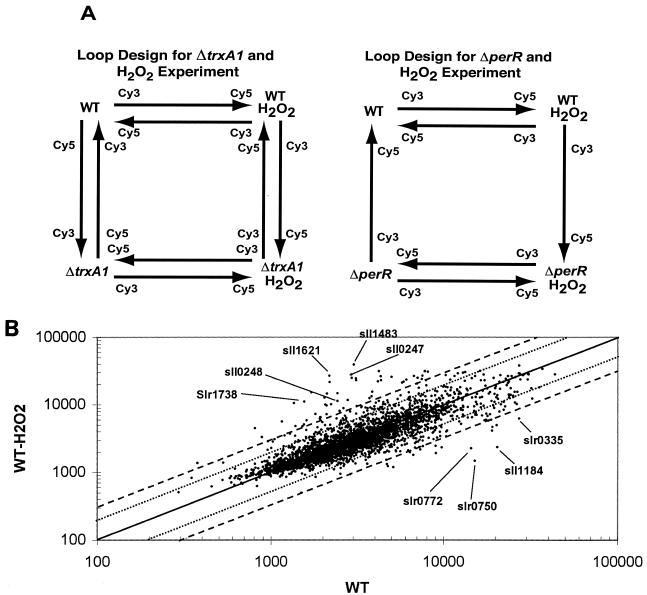
A complete description of array construction has been provided previously by Postier et al. (50). PCR products corresponding to the 3,165 genes identified in the Synechocystis genome on the Kazusa website (prior to May 2002) were amplified by using a two-stage process. The experimental loop design is shown in Fig. 1A. The cDNA labeling, glass treatment, prehybridization, and hybridization protocols have been described in detail previously by Singh et al. (58). Biological variation was sampled by extracting RNA from three separate experiments and pooling the RNA prior to hybridization. This strategy has been used in many microarray experiments (1, 47) and simplifies the final analysis.
FIG. 1.
(A) Diagrammatic representation of the loop designs utilized for identification of differentially expressed genes in response to peroxide stress. A total of eight slides were used with dye swaps for the experiment with and without peroxide with the wild type and the ΔtrxA1 mutant (left side), and six slides were used for the related experiment with the ΔperR mutant and the wild type (right side). (B) Scatter plot comparing the mean spot intensities of wild-type Synechocystis sp. strain PCC 6803 in the absence (x axis) and in the presence (y axis) of 1.5 mM H2O2. Data were normalized, and the mean signal intensities were plotted. The solid line indicates equal labeling for the two samples, whereas the dotted line (twofold) and the dashed line (threefold) identify genes that exhibited large labeling differences during the hybridization experiment. WT, wild type.
Data acquisition and statistical analysis.
Spot intensities of the images were quantified by using Quantarray 3.0 (Packard BioChip Technologies, Boston, Mass.). Data for the six slides in the ΔperR experiment and the eight slides in the ΔtrxA1 experiment were then collated into two data sets (one for each experiment) by using SAS (version 8.02; SAS Institute, Cary, N.C.). Previous testing has demonstrated that the results obtained with Quantarray are reliable and similar to the results obtained with Imagene 6.0 (41). The local background was subtracted from each spot.
For each replicate block on a slide, there were empty spots (422 or 326 spots depending on the specific slide batch), and there were three replicates per slide. We examined the distribution of spot intensities for these empty spots and declared data from a nonempty spot to be detected if the background-corrected intensity of the spot was greater than that for 95% of the empty spots. We examined the data for the ΔperR and ΔtrxA1 experiments separately. If all the spots for a given gene were not detected on all the slides in an experiment, then the gene was considered to be off and was not analyzed further (797 genes in ΔperR experiments and 864 genes in ΔtrxA1 experiments). We then calculated the log of the background-corrected signals that were normalized to the slide median (the median for all noncontrol spots detected).
Each experiment contained two genotypes (mutant and wild-type controls) and two stimuli (mutant and wild-type peroxide) for a total of four treatment combinations. The effects of the mutant and the peroxide stimulus were examined in an analysis of variance (ANOVA) modeling framework (9, 12, 29, 30, 44, 58, 68, 70). The model Yijklm = μ + ti + dj + ωk + ωk(ρl) + ɛijklm was fit, where Y is the intensity of the spot after correction for the local background signal, normalization, and log transformation, μ is the overall mean of the normalized values for the gene, and ωk and ωk(ρl) are the random effects of slide and of replicates within a slide, respectively. The fixed effects of treatment (ti), where i = (1,...4) for each of the four combinations of genotype and peroxide, and dye (dj), where j = (red, green), were also included in the model. To test the null hypothesis that a gene's expression level was not different across treatments, an F test of the effect of treatment for each gene was conducted, and a P value was calculated. We examined the model for conformation to the assumption of normality of the residuals by testing the null hypothesis that the residuals for each gene were normally distributed by using the Shapiro-Wilkes test. Additional tests comparing the effects of the genotype (mutant versus wild type) and peroxide stimulus (peroxide versus control) and the interaction between the genotype and peroxide stimulus were performed. All analyses were performed with SAS (SAS Institute).
We used a Bonferroni significance level as an initial criterion for rejecting the null hypothesis of a significant treatment effect (0.05/2,368 = 2.11 × 10−5 for ΔperR and 0.05/2,301 = 2.17 × 10−5 for ΔtrxA1). As type I and type II errors are inversely related, with decreases in false positives (type I) associated with increases in false negatives (type II), and as the Bonferroni correction is overly conservative as tests are correlated (11, 38, 69), we used a second nominal threshold of 0.001 to balance type I and type II results, and we bounded our considerations with an overly liberal threshold (0.05). In addition, we considered the test for dye effects and normality of the residuals. If the test of the null hypothesis of difference across times was rejected at the Bonferroni level and we had no evidence for dye effects or departure from normality of the residuals, we declared the gene differentially expressed across treatments and examined the contrasts. If the P value for the test of differences over time was less than or equal to 0.05 but larger than the Bonferroni level and we had no evidence for dye effects or departure from normality of the residuals, we considered the gene interesting. When dye effects were present or residuals showed evidence of departure from normality, we used caution in interpreting the results. Once the analysis was completed, we focused our attention on statistically significant and interesting genes (P < 0.001) that exhibited a change of at least 1.4-fold (68). In some cases, we included genes with a P value of <0.05 because they augmented or completed a functional category. Our objective was to identify genes that exhibited differential expression for further experimentation. Thus, we bracketed our interpretation of the results with a conservative (Bonferroni) threshold and a liberal 0.001 criterion, and we used a fold change filter to focus our efforts. The raw P values are shown in Table S1 in the supplemental material.
RESULTS
Array data and statistical analysis.
The effects of H2O2 on gene expression in the wild-type strain and two mutants of Synechocystis sp. strain PCC 6803 were monitored by using full genome microarrays printed in triplicate on each slide. Microarray experiments were performed as previously described (58) by using the loop design, as shown in Fig. 1A. For each RNA sample, either eight slides (ΔtrxA experiment) or six slides (ΔperR experiment) were used for hybridization; since each slide contained three replicates, 12 and 9 measurements per gene, respectively, could be used for the statistical analysis. The loop design allowed comparison of all conditions by using the ANOVA model (9, 58, 72). We could analyze the effect of peroxide on the wild type, the effect of peroxide on the mutant, the effect of peroxide on the specific genotype used in the loop, and the relationship between the wild type and each mutant. To obtain the results presented below, we used 1.5 mM H2O2 for 30 min to elicit stress, although we investigated the effects of peroxide concentrations ranging from 75 μM to 3 mM and times as short as 15 min. We found that 75 μM H2O2 for 15 min was sufficient to result in changes in expression of some of the genes but that other genes exhibited a temporal response to the stress and that it took some time for the amounts to increase or decrease (Li, Singh, and Sherman, data not shown).
The scatter plot in Fig. 1B shows the relationship between the average hybridization intensities of the wild-type control cells and cells that were treated with peroxide. This simple procedure provided an overview of the data and indicated that most of the spots fell along the diagonal and were equally labeled. The spots that were not on the diagonal were candidates for genes with expression changes, and lines indicating twofold and threefold changes are shown in Fig. 1B. In some of the previous microarray analyses the workers used an arbitrary cutoff of a twofold change to identify differentially expressed genes. However, it has been shown that changes in gene expression less than a twofold change can be reliably identified (1, 26, 35, 44, 72, 74), and this aspect has been discussed in detail previously (58). We selected genes for additional consideration using a significance level of 0.05 from the full ANOVA model (see Table S1 in the supplemental material) and at least a 1.4-fold change in transcript level intensity.
Genes differentially regulated by peroxide stress: the peroxide stimulon.
The effect of peroxide on gene expression based on the functional categories defined in Cyanobase is shown in Table 2. Table 2 also shows the differences in expression among the wild type and the ΔtrxA1 and ΔperR mutants. In all three experiments, approximately 18 to 20% of all genes showed some evidence of changes in levels of transcript accumulation that were at least 1.4-fold (P < 0.05). The use of a liberal filter, along with the requirement for similar significance in both experiments, provided a comprehensive overview of the impact of peroxide on gene expression for the different functional categories. We analyzed the statistically significant genes (P < 0.001) in detail, focusing particularly on the genes whose statistical significance was below the Bonferonni threshold. There were 158 and 198 genes in the ΔperR and ΔtrxA1 experiments, respectively, whose P values were under the Bonferonni threshold; for 103 of the 158 genes in the PerR experiment the difference also met the most stringent criterion in the TrxA experiment, and most of the other 55 genes had a P value of <0.001 (Table 3).
TABLE 2.
Functional categories of peroxide-responsive genes in Synechocystis sp. strain PCC 6803 and two mutantsa
| General pathway | No. of genes | No. of differentially regulated genes in:
|
||
|---|---|---|---|---|
| Wild type | ΔperR mutant | ΔtrxA1 mutant | ||
| Amino acid biosynthesis | 97 | 21 | 20 | 15 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 124 | 32 | 34 | 24 |
| Cell envelope | 67 | 9 | 14 | 12 |
| Cellular processes | 76 | 28 | 26 | 26 |
| Central intermediary metabolism | 31 | 7 | 8 | 6 |
| DNA replication, restriction, recombination, and repair | 60 | 8 | 7 | 10 |
| Energy metabolism | 132 | 29 | 31 | 15 |
| Hypothetical | 1,076 | 189 | 148 | 160 |
| Other categories | 306 | 74 | 46 | 85 |
| Photosynthesis and respiration | 141 | 63 | 50 | 47 |
| Purines, pyrimidines, nucleosides, and nucleotides | 41 | 3 | 5 | 7 |
| Regulatory functions | 146 | 28 | 24 | 39 |
| Transcription | 30 | 8 | 8 | 10 |
| Translation | 168 | 37 | 41 | 32 |
| Transport and binding proteins | 196 | 39 | 37 | 28 |
| Unknown | 474 | 79 | 75 | 67 |
| Total | 3,165b | 654 | 574 | 583 |
Genes were considered differentially regulated when the P value was <0.05 and the change was >1.4-fold.
Total number of genes based on Kazusa annotation prior to May 2002.
TABLE 3.
Selected differentially regulated genes of the peroxide stimulon in Synechocystis sp. strain PCC 6803 and ΔperR and ΔtrxA1 mutantsa
| Gene | Function | P(treatment) | Fold changeb
|
||
|---|---|---|---|---|---|
| Wild type | ΔperR mutant | ΔtrxA1 mutant | |||
| Biosynthesis of cofactors, prosthetic groups, and carriers | |||||
| Carotenoid | |||||
| slr1254 | Phytoene dehydrogenase (phytoene desaturase) | 2.0E-05 | 2.2 | 2.5 | 2.4 |
| Cobalamin, heme, phycobilin, and porphyrin | |||||
| sll1184 | Heme oxygenase (ho1) | 1.0E-10 | −7.7 | −5.4 | −5.0 |
| slr0772 | Light-independent protochlorophyllide reductase subunit (chlB) | 3.0E-10 | −6.3 | −6.5 | −4.0 |
| slr0750 | Light-independent protochlorophyllide reductase subunit (chlN) | 9.0E-10 | −9.2 | −6.8 | −3.8 |
| slr0749 | Light-independent protochlorophyllide reductase subunit (chlL) | 1.0E-09 | −4.5 | −3.8 | −1.8 |
| sll1091 | Geranylgeranyl hydrogenase (chlP) | 6.0E-07 | −3.2 | −2.2 | −1.6 |
| sll1185 | Coproporphyrinogen III oxidase, aerobic (hemF) | 3.0E-05 | −2.2 | −1.7 | −2.2 |
| sll1876 | Coproporphyrinogen III oxidase, anaerobic (hemN) | 1.0E-04 | −2.7 | −2.6 | −3.5 |
| slr0839 | Ferrochelatase (hemH, scpA) | 4.0E-04 | 1.3 | 1.6 | 1.9 |
| sll1685 | Light-induced Na+-dependent proton extrusion (pxcA, cotA) | 1.0E-03 | −1.4 | −1.4 | −1.3 |
| slr1055 | Mg protoporphyrin IX chelatase subunit H (chlH) | 3.0E-03 | −1.5 | −1.8 | −2.1 |
| slr0905 | Mg-protoporphyrin IX ester oxidative cyclase (bchE) | 9.0E-03 | −1.8 | −1.8 | −2.7 |
| Thioredoxin, glutaredoxin, and glutathione | |||||
| slr0623 | Thioredoxin (trxA1) | 2.0E-06 | −2.1 | −2.4 | −1.5 |
| ssr0330 | Ferredoxin-thioredoxin reductase, variable chain (ftrV) | 8.0E-06 | −3.1 | −2.1 | −2.0 |
| slr1846 | Hypothetical protein (ycf64) | 5.0E-05 | 1.6 | 1.6 | 1.4 |
| sll1057 | Thioredoxin M (trxM2) | 7.0E-04 | −1.8 | −1.8 | −2.9 |
| Cellular processes | |||||
| Cell division | |||||
| slr0228 | Cell division protein FtsH (ftsH) | 1.0E-08 | 4.5 | 4.6 | 2.4 |
| slr1604 | Cell division protein FtsH (ftsH) | 8.0E-06 | 2.8 | 2.3 | 2.8 |
| sll1633 | Cell division protein FtsZ (ftsZ) | 1.0E-03 | −2.0 | −1.7 | −2.0 |
| slr1267 | Cell division protein FtsW (ftsW) | 5.0E-03 | −1.3 | −1.6 | −2.0 |
| Cell killing | |||||
| slr1747 | Cell death suppressor protein Lls1 homolog | 1.0E-06 | 4.1 | 3.2 | 4.0 |
| Chaperones and chemotaxisc | |||||
| sll1533 | Twitching mobility protein (pilT2) | 4.0E-08 | −2.5 | −3.9 | −3.5 |
| sll0041 | Phytochrome-like photoreceptor (pixJ1, pisJ1, taxD1) | 2.0E-05 | 2.7 | 2.0 | 2.1 |
| slr1929 | Type 4 pilin-like protein (pilA6) | 6.0E-05 | −2.0 | −2.1 | −2.5 |
| slr1456 | gspG or pilA4 | 3.0E-06 | 2.0 | 2.0 | 3.1 |
| slr2015 | Type 4 pilin-like protein, essential for motility (pilA9) | 5.0E-03 | 1.6 | 1.1 | 1.5 |
| slr1930 | Type 4 pilin-like protein (pilA7) | 1.7E-02 | −1.7 | −1.3 | −2.0 |
| slr1928 | Type 4 pilin-like protein (pilA5) | 3.4E-02 | −1.4 | −1.2 | −2.6 |
| Detoxification | |||||
| slr1516 | Superoxide dismutase (sodB) | 3.0E-06 | 1.9 | 1.9 | 2.3 |
| sll1615 | Thiophen and furan oxidation protein | 7.0E-03 | 1.8 | 1.6 | 1.8 |
| Energy metabolism: fatty acid, phospholipid, and sterol metabolism | |||||
| sll0330 | Sepiapterine reductase | 6.0E-12 | 8.4 | 4.0 | 4.5 |
| sll1655 | Similar to biotin (acetyl coenzyme A carboxylase) ligase | 6.0E-11 | −1.6 | −2.4 | −1.4 |
| slr1020 | Sulfolipid biosynthesis protein SqdB (sqdB) | 3.0E-05 | −2.1 | −2.3 | −2.0 |
| slr1672 | Glycerol kinase (glpK) | 5.0E-05 | −2.5 | −2.4 | −2.2 |
| Hypothetical | |||||
| slr1687 | Hypothetical protein | 9.0E-12 | 11.0 | 4.4 | 2.7 |
| sll1483 | Periplasmic protein | 2.0E-11 | 15.3 | 14.4 | 6.5 |
| sll2013 | Hypothetical protein | 4.0E-11 | 6.4 | 5.7 | 4.1 |
| slr0270 | Hypothetical protein | 3.0E-10 | 8.6 | 5.1 | 4.2 |
| sll1620 | Hypothetical protein | 4.0E-10 | 5.8 | −1.3 | 4.7 |
| slr1215 | Hypothetical protein | 7.0E-10 | 4.4 | 4.5 | 2.5 |
| sll0185 | Hypothetical protein | 1.0E-09 | 4.9 | 4.4 | 2.4 |
| slr0967 | Hypothetical protein | 2.0E-09 | 5.4 | 5.6 | 2.1 |
| slr1235 | Hypothetical protein | 3.0E-09 | 2.6 | 2.5 | 1.9 |
| slr0888 | Hypothetical protein | 4.0E-09 | −2.6 | −5.5 | −2.4 |
| sll0939 | Hypothetical protein | 5.0E-09 | 6.4 | 4.8 | 3.6 |
| Other categories | |||||
| Adaptations and atypical conditions | |||||
| sll0947 | Light-repressed protein A homolog (lrtA) | 6.0E-06 | −2.6 | −3.6 | −1.6 |
| ssl2542 | HliA, CAB/ELIP/HLIP superfamily (hliA, scpC) | 2.0E-05 | 2.8 | 1.6 | 2.0 |
| ssr1789 | CAB/ELIP/HLIP-related protein HliD (hliD, scpE) | 1.3E-02 | 1.6 | 1.4 | 2.0 |
| Drug and analog sensitivity | |||||
| sll0086 | Putative arsenical pump-driving ATPase | 3.0E-08 | 2.4 | 2.2 | 2.0 |
| slr0946 | Arsenate reductase (arsC) | 7.0E-08 | 3.5 | 3.2 | 2.7 |
| sll1159 | Probable bacterioferritin comigratory protein | 1.0E-07 | 2.8 | 5.2 | 5.9 |
| slr1198 | Antioxidant protein | 9.0E-05 | −2.0 | −2.0 | −1.6 |
| sll1154 | Putative antibiotic efflux protein | 6.0E-04 | 2.1 | 1.5 | 1.4 |
| Other | |||||
| sll1621 | AhpC-peroxiredoxin | 5.0E-14 | 13.1 | −1.0 | 2.3 |
| slr0381 | Lactoylglutathione lyase | 1.0E-10 | 8.3 | 4.5 | 2.7 |
| slr0298 | FraH protein homolog | 7.0E-08 | 9.4 | 5.1 | 3.1 |
| sll1534 | Probable glycosyltransferase | 3.0E-07 | −3.6 | −4.6 | −2.7 |
| sll1245 | Cytochrome cM (cytM) | 2.0E-06 | −2.5 | −2.0 | −1.6 |
| slr2094 | Fructose-1,6-sedoheptulose-1,7-bisphosphatase (fbpl) | 4.0E-06 | −1.9 | −2.5 | 1.1 |
| slr1942 | Circadian clock protein KaiC homolog (kaiC3) | 7.0E-05 | −2.4 | −2.2 | −2.1 |
| Photosynthesis and respiration | |||||
| NADH dehydrogenase | |||||
| slr1291 | NADH dehydrogenase subunit 4 (ndhD2) | 3.0E-08 | 4.2 | 2.9 | 6.4 |
| slr0331 | NADH dehydrogenase subunit 4 (ndhD1) | 7.0E-05 | −1.7 | −2.2 | −1.6 |
| PSI | |||||
| sll0247 | Iron stress chlorophyll-binding protein (isiA) | 1.0E-12 | 11.3 | 9.9 | 5.8 |
| sll0248 | Flavodoxin (isiB) | 4.0E-08 | 4.7 | 3.8 | 4.9 |
| sll0249 | Hypothetical | 2.0E-05 | 2.2 | 3.9 | 3.3 |
| ssr2831 | PSI subunit IV (psaE) | 1.0E-05 | −1.9 | −1.8 | −1.3 |
| ssl0563 | PSI subunit VII (psaC) | 7.0E-05 | −1.9 | −2.5 | −2.3 |
| PSII | |||||
| slr1181 | PSII D1 protein (psbA1) | 1.0E-05 | 1.6 | 1.6 | 1.2 |
| ssl2598 | PSII PsbH protein (psbH) | 7.0E-05 | −1.7 | −1.7 | 1.1 |
| sml0003 | PSII reaction center M protein (psbM) | 1.0E-04 | 2.2 | 1.6 | 2.1 |
| slr1739 | PSII 13-kDa protein homolog (psb28-2) | 2.0E-02 | 1.7 | 1.1 | 1.7 |
| sml0005 | PSII PsbK protein (psbK) | 2.1E-02 | −1.6 | −1.5 | −1.9 |
| Phycobilisome | |||||
| slr0335 | Phycobilisome core membrane linker protein (apcE) | 4.0E-10 | −4.5 | −4.6 | −2.1 |
| slr2067 | Allophycocyanin alpha subunit (apcA) | 1.0E-09 | −4.4 | −3.1 | −2.0 |
| sll0928 | Allophycocyanin B (apcD) | 2.0E-07 | −1.8 | −1.6 | −1.7 |
| slr1986 | Allophycocyanin beta subunit (apcB) | 3.0E-07 | −2.9 | −3.0 | −1.5 |
| ssl0453 | Phycobilisome degradation protein NblA (nblA2) | 4.0E-07 | 4.3 | 3.3 | 4.0 |
| sll1579 | Phycobilisome rod linker polypeptide (cpcC2) | 1.0E-05 | −2.0 | −2.0 | −2.5 |
| sll1577 | Phycocyanin beta subunit (cpcB) | 2.0E-05 | −3.0 | −2.3 | −1.4 |
| sll1471 | Phycobilisome rod-core linker polypeptide (cpcG2) | 5.0E-05 | −2.3 | −1.4 | −2.0 |
| sll1580 | Phycobilisome rod linker polypeptide (cpcC1) | 9.0E-05 | −2.0 | −2.3 | −1.8 |
| slr1459 | Phycobilisome core component (apcF) | 2.0E-04 | −1.3 | −2.0 | −2.7 |
| ssr3383 | Phycobilisome small core linker polypeptide (apcC) | 3.0E-04 | −1.5 | −1.6 | −1.2 |
| ssl0452 | Phycobilisome degradation protein NblA (nblA1) | 8.0E-03 | 1.4 | 1.3 | −1.7 |
| slr1878 | Phycocyanin alpha-subunit phycocyanobilin lyase | 2.8E-02 | −1.5 | −1.3 | −1.9 |
| Soluble electron carriers | |||||
| ssl3044 | Probable ferredoxin | 1.0E-04 | 2.7 | 1.5 | 4.4 |
| slr1828 | Ferredoxin, PetF-like protein (petF, fdx) | 2.0E-04 | −1.5 | −1.6 | −2.0 |
| slr1643 | Ferredoxin-NADP oxidoreductase (petH) | 3.0E-04 | −1.9 | −2.2 | −1.8 |
| ssr3184 | 4Fe-4S-type iron-sulfur protein | 5.0E-04 | −1.6 | −1.5 | −1.6 |
| slr0150 | Ferredoxin, PetF-like protein (petF, fdx) | 8.0E-04 | −1.5 | −1.7 | −2.3 |
| ssl0020 | Ferredoxin I, essential for growth (petF) | 3.0E-03 | −1.9 | −1.7 | −1.5 |
| sll0199 | Plastocyanin (petE) | 5.0E-03 | −1.6 | −1.4 | −1.5 |
| sll1382 | Ferredoxin, PetF-like protein (petF, fdx) | 2.0E-02 | −1.4 | −1.6 | −2.0 |
| Regulatory functions | |||||
| slr1738 | Transcription regulator, Fur family | 2.0E-08 | 7.0 | 1.3 | 4.0 |
| sll1161 | Probable adenylate cyclase | 2.0E-06 | 3.4 | 3.7 | 2.9 |
| sll1742 | Transcription antitermination protein (nusG) | 3.0E-06 | 1.3 | 1.9 | 2.0 |
| slr1214 | Two-component response regulator, PatA subfamily | 1.0E-05 | 2.2 | 2.3 | 1.5 |
| sll1626 | LexA repressor | 2.0E-05 | −2.3 | −2.3 | −1.5 |
| slr0474 | Regulator for phytochrome 1 (Cph1) (rcp1) | 2.0E-05 | −1.8 | −2.8 | −1.9 |
| sll1005 | MazG protein homolog | 7.0E-05 | −1.9 | −1.8 | −1.8 |
| slr0152 | Serine/threonine protein kinase | 9.0E-05 | −2.0 | −2.0 | −1.3 |
| sll0998 | LysR family transcriptional regulator (ycf30) | 9.0E-05 | −1.3 | −2.2 | −2.0 |
| sll0789 | Two-component response regulator, OmpR subfamily | 1.0E-04 | 1.8 | 1.8 | −1.1 |
| sll1672 | Two-component hybrid sensor and regulator (hik12) | 1.0E-04 | −1.7 | −1.6 | −1.6 |
| sll2014 | Sugar fermentation stimulation protein | 2.0E-04 | 1.8 | 2.5 | 3.0 |
| sll0790 | Two-component sensor histidine kinase (hik31) | 6.0E-04 | −2.1 | 1.2 | −1.6 |
| slr1760 | Two-component response regulator | 1.0E-03 | −1.3 | −1.3 | −3.5 |
| sll1544 | Two-component response regulator, NarL subfamily | 2.0E-03 | 2.0 | 1.8 | 3.2 |
| sll1329 | Inositol monophosphate family protein | 4.0E-03 | 1.3 | 1.3 | 2.2 |
| sll1353 | Two-component sensor histidine kinase (hik15) | 5.0E-03 | −1.4 | −1.5 | −2.6 |
| slr0081 | Two-component response regulator, OmpR subfamily | 5.0E-03 | −1.3 | −1.4 | −1.4 |
| slr1584 | Two-component transcription regulator, OmpR subfamily | 7.0E-03 | −1.6 | −1.4 | −1.8 |
| sll0750 | KaiC-interacting protein (hik8, sasA) | 1.1E-02 | 1.8 | 1.7 | −2.1 |
| sll1871 | Two-component system sensory histidine kinase (hik6) | 1.3E-02 | −1.7 | −1.2 | −2.1 |
| slr1414 | Two-component sensor histidine kinase (hik11) | 1.7E-02 | 1.6 | 1.4 | 2.2 |
| slr2099 | Two-component hybrid sensor and regulator (hik40) | 2.1E-02 | −1.5 | −1.2 | −1.5 |
| sll1292 | Two-component response regulator, CheY subfamily | 2.8E-02 | −1.3 | −1.4 | −2.0 |
| Transcription: RNA synthesis, modification, and DNA transcription | |||||
| sll2012 | Group2 RNA polymerase sigma factor (sigD) | 9.0E-11 | 7.9 | 5.5 | 5.7 |
| sll0306 | RNA polymerase group 2 sigma factor (sigB) | 3.0E-08 | 2.7 | 3.0 | 1.7 |
| sll1818 | RNA polymerase alpha subunit (rpoA) | 6.0E-06 | −1.7 | 1.8 | 1.4 |
| sll1689 | Group2 RNA polymerase sigma factor (sigE) | 2.0E-04 | 1.2 | −1.1 | 1.6 |
| sll1787 | RNA polymerase beta subunit (rpoB) | 3.0E-03 | −1.6 | −1.3 | 1.3 |
| sll0184 | Group2 RNA polymerase sigma factor (sigC) | 4.0E-03 | 1.7 | 1.7 | 2.4 |
| Translation: degradation of proteins, peptides, and glycopeptides | |||||
| slr0008 | Carboxyl-terminal processing protease (ctpA) | 5.0E-06 | 2.0 | 2.3 | 2.1 |
| slr1204 | Protease | 1.0E-04 | 2.6 | 1.2 | 3.0 |
| slr0156 | ClpB protein (clpB2) | 2.0E-03 | 1.5 | 1.7 | 1.5 |
| slr1641 | ClpB protein (clpB1) | 4.4E-02 | 1.4 | 1.5 | 4.8 |
| Transport and binding proteins | |||||
| slr0513 | Iron transport system substrate-binding protein | 6.0E-10 | 2.4 | 5.7 | 4.2 |
| slr0447 | ABC-type urea transport system (urtA) | 2.0E-09 | −2.6 | −2.2 | −1.9 |
| sll0771 | Glucose transport protein (glcP,gtr) | 1.0E-07 | −3.8 | −3.7 | −1.4 |
| slr1740 | Oligopeptide binding protein of ABC transporter | 4.0E-07 | 3.2 | 2.6 | 3.9 |
| Unknown | |||||
| slr1544 | Unknown protein | 1.0E-09 | 9.1 | 6.9 | 6.1 |
| sll1135 | Unknown protein | 7.0E-09 | 7.7 | 3.3 | 5.6 |
| sll1830 | Unknown protein | 2.0E-08 | −3.6 | −4.9 | −2.5 |
| slr0108 | Unknown protein | 3.0E-08 | 3.7 | 4.2 | 2.8 |
| slr1484 | Unknown protein | 6.0E-08 | 3.3 | 4.9 | 4.7 |
| slr1855 | Unknown protein | 9.0E-08 | −1.9 | −2.6 | −1.6 |
| sll1515 | Glutamine synthetase-inactivating factor IF17 (gifB) | 3.0E-07 | 4.5 | 2.6 | 3.1 |
| sll1009 | Unknown protein | 6.0E-07 | −2.5 | −2.7 | −1.8 |
| slr1854 | Unknown protein | 2.0E-06 | −2.4 | −1.9 | −2.7 |
| slr0572 | Unknown protein | 3.0E-06 | −3.0 | −2.5 | −2.2 |
| sll1851 | Unknown protein | 7.0E-06 | 3.0 | 1.4 | 1.3 |
| sll0923 | Unknown protein | 1.0E-05 | 3.9 | 2.7 | 6.6 |
Genes were considered differentially regulated when the P value was <0.05 and the change was >1.4-fold. The genes are listed in descending order of P(treatment) within each category.
The changes for the wild type and the ΔperR and ΔtrxA1 mutants were calculated by dividing the normalized mean intensities of peroxide-treated strains by the normalized mean intensities of nontreated strains.
See Table 5.
When the effect of peroxide were examined, in some categories, such as central intermediary metabolism and DNA replication and related processes, few genes were affected, whereas more than one-third of the genes encoding photosynthesis and respiration proteins and cellular processes were affected. In the case of categories such as photosynthesis or biosynthesis of cofactors, most of the differentially expressed genes were in specific functional categories involved in pigment-protein complexes (Table 3). In most cases, there were few differences between the levels in the mutants and the levels in the wild type. However, there were a few significant exceptions. Many fewer genes involved in energy metabolism were affected in the ΔtrxA1 mutant than in the wild type and the ΔperR mutant. Conversely, the number of regulatory genes affected by peroxide was greater in the ΔtrxA1 mutant than in the other two strains. Finally, many fewer genes categorized as “other” showed differential expression in the ΔperR mutant compared to the expression in the wild type and the ΔtrxA1 mutant. It should be noted that the total numbers of differentially expressed genes in these experiments were quite similar; these findings indicate the reproducibility and correspondence of the data.
One objective of this study was to determine the genes that were most responsive to peroxide stress in our wild-type strain, and we identified some of these key genes. A gene with one of the greatest increases in transcript level in every experiment was isiA, which encoded a Chl protein that was also highly induced by low-iron conditions (58). In the wild type, this gene was induced 11.3-fold (P = 1 × 10−12). The adjacent genes isiB (4.7-fold; P = 4 × 10−8) and sll0249 (2.2-fold; P = 2 × 10−5), were also upregulated (Table 3), which was also observed under low-iron conditions (58). For convenience, we placed all three of these genes in the PSI group in Table 3, although the functions of all three genes remain to be clarified. When cells were treated with 75 μM H2O2, the isiAB operon was still the most peroxide-responsive operon, although the transcript levels did not increase as much when cells were stressed with a lower peroxide concentration (data not shown). Another highly responsive gene, sigD (sll2012), encoding a group 2 sigma factor, was induced eightfold (P = 9 × 10−11). In addition, the slr1738 gene was upregulated sevenfold (P = 2 × 10−8). Analysis of the protein sequence encoded by this gene indicated that the protein is very closely related to the PerR protein that was first identified in B. subtilis as a repressor inactivated by peroxide (reference 20 and references therein). Therefore, we designated this gene perR in Synechocystis sp. strain PCC 6803. Some of the other highly upregulated genes are shown in Table 3. The effects of peroxide stress on ΔperR and ΔtrxA1 are also shown in Table 3. Most of the key genes were induced to different degrees in the absence of TrxA1, and the differences are discussed below. In addition, several gene clusters in which all genes were either upregulated or downregulated in the wild type were identified from the microarray results, as described by Singh et al. (57).
A selected subset of the genes that were either up- or downregulated by peroxide are listed by functional category in Table 3. The entire data set is shown in Table S1 in the supplemental material, which includes the fold changes and the P values from the ANOVA analysis. Some of the more important categories are discussed below.
Pigment and photosynthesis genes.
Peroxide stress led to large changes in the transcript levels of genes encoding components of the photosynthetic apparatus. This was especially noticeable for genes involved in pigment biosynthesis and for genes associated with phycobilisome synthesis and assembly; the transcript levels of these genes were decreased substantially by peroxide stress. The largest changes were in the genes associated with the conversion of divinyl protochlorophyllide a to monovinyl protochlorophyllide a, and the genes encoding protochlorophyllide reductase (slr0749, slr0750, and slr0772) were downregulated approximately four- to ninefold (see Table S2 in the supplemental material for a comparison of changes due to growth under low-iron conditions with changes due to peroxide stress in the heme and pigment biosynthesis pathways). The transcript levels of the sll1214 and sll1874 genes, both of which exhibit strong homology to Arabidopsis CHL27, also decreased approximately two- to threefold (see Table S2 in the supplemental material). CHL27 is required for synthesis of protochlorophyllide and is a prime candidate for the cyclase in chlorophyll biosynthesis (64). The transcript levels of the genes encoding phycobilisome components decreased approximately two- to threefold. The one gene in this category whose transcript level increased was the gene encoding the phycobilisome degradation protein, NblA (ssl0453); in this case the transcript level increased approximately fourfold. The transcript levels of the structural genes of the photosynthetic and respiratory apparatus did not change greatly during the short peroxide stress. The obvious exception was isiA, which was the most peroxide sensitive of all of the genes. The only other gene in this category whose transcript level increased significantly was slr1291 (ndhD2), which encodes a component of the NADH dehydrogenase complex; the transcript level of this gene increased approximately fivefold. The other major changes in transcription involved genes encoding the soluble electron transfer components, such as ferredoxins and plastocyanin, all of whose transcript levels decreased approximately twofold.
Detoxification genes.
Peroxide stress induced expression of a number of genes thought to be involved in detoxification. The main gene in this category was the peroxiredoxin gene, aphC (sll1621; ∼13-fold induction; P = 5 × 10−14). Expression of another putative peroxiredoxin gene, sll1159, was also induced (approximately threefold; P = 1 × 10−7). This was part of a four-gene cluster whose members were all strongly expressed in the presence of peroxide and that included a gene encoding a putative adenyl cyclase (sll1161) (57). Two other detoxification genes were upregulated in both experiments; expression of both slr1516 (sodB) and sll1615 was induced approximately twofold. Other genes that may belong in this category that were upregulated by peroxide included arsC (slr0946; arsenate reductase), whose expression was induced approximately threefold, and a series of genes encoding flavoproteins and putative oxidoreductases (Table 3). Interestingly, the two genes normally associated with peroxide scavenging, katG (sll1987) and tpx (sll0755), were not significantly upregulated under these oxidative stress conditions (63).
Thioredoxin and redox regulation genes.
One of the objectives of our experiments was to map the functions of the different thioredoxins that have an impact on oxidative stress. Thioredoxins are small disulfide-containing redox proteins that serve as general protein disulfide oxidoreductases (54). We hypothesized that defects in a thioredoxin and the resulting inability to form disulfide bonds could have a negative impact on the cellular response to oxidative stress. The thioredoxin gene superfamily in Synechocystis sp. strain PCC 6803 is comprised of at least six genes, and attempts to construct mutants with mutations in some of these genes have been unsuccessful (42). The effects of the oxidizing agents H2O2 and methyl viologen on ΔtrxA1, ΔtrxM1, and other mutants used in this study are shown in Table 4. The ΔtrxA1 mutant was sensitive to both peroxide and methyl viologen, whereas the ΔtrxM1 mutant was sensitive only to methyl viologen. The response of the ΔperR mutant was similar to that of the wild type, whereas the ΔmrgA (slr1894) mutant was highly sensitive to peroxide but not to methyl viologen.
TABLE 4.
Effects of peroxide and methyl viologen on the exponential-phase growth of Synechocystis sp. wild-type strain PCC 6803 and mutants
| Strain | Concn ofa:
|
|
|---|---|---|
| H2O2 (mM) | Methyl viologen (μM) | |
| Wild type | 1.5 ± 0.6 | 2.0 ± 0.5 |
| ΔtrxA1 mutant | 1.0 ± 0.5 | 1.0 ± 0.25 |
| ΔtrxM1 mutant | 1.5 ± 0.6 | 1.0 ± 0.25 |
| ΔperR mutant | 1.5 ± 0.6 | 2.0 ± 0.5 |
| ΔmrgA mutant | 0.075 ± 0.025 | 2.0 ± 0.5 |
The values are the concentrations that permitted cellular growth that was ∼50% of the growth of the untreated controls. The values are means ± standard deviations (n ≥ 6).
Figure 1A shows the experimental loop design utilized to study the effect of peroxide stress in the ΔtrxA1 and ΔperR mutants. In general, the individual genes responded to peroxide qualitatively in the same way in the ΔtrxA1 mutant as in the wild type, but the changes in the ΔtrxA1 strain were generally smaller quantitatively. This was particularly true of genes encoding energy metabolism proteins, as shown in Table 3. In addition, the transcript levels of some other thioredoxin genes were decreased upon peroxide stress; the transcript levels of trxA1 (slr0623) and trxM2 (sll1057) both decreased approximately twofold. At the same time, three genes that have not been studied much but that appear to encode thioredoxin-like proteins were induced; these genes were slr1846 (ycf64), ssl2667 (cnfU), and sll0621 (dsbD-like). More importantly, the latter two genes appeared to be under control of PerR (see below).
Cellular processes, including chaperones: defining the heat shock regulon.
The chaperones represented one of the most prominent of the categories with many induced genes. We also analyzed the effect of heat shock on gene expression in Synechocystis sp. strain PCC 6803, and the responses of the chaperone genes to heat shock are shown in Table 5. The results of the heat shock experiment, in which cells were shifted from 35 to 45°C for 15 min, were analyzed on microarrays by using a separate loop. The experiment was performed with a mutant with a mutation in sigB, which may encode the heat shock sigma factor in Synechocystis sp. strain PCC 6803 (data not shown). From the results, it is evident that many stress-related genes were induced upon peroxide stress and that these genes in the peroxide stimulon were indirectly regulated by the specific stress.
TABLE 5.
Chaperone genes in Synechocystis sp. wild-type strain PCC 6803 and ΔperR and ΔtrxA1 mutants that are differentially regulated by peroxide stress and high temperature
| Gene | Function | P(treatment)a | Fold change
|
|||
|---|---|---|---|---|---|---|
| Wild type | ΔperR mutant | ΔtrxA1 mutant | Heat shockb | |||
| sll1514 | 16.6-kDa small Hsp | 8.0E-12 | 5.9 | 5.0 | 2.9 | 5.1 |
| sll1666 | DnaJ-like protein | 5.0E-10 | 5.1 | 3.8 | 3.5 | |
| slr2075 | 10-kDa chaperonin (groES) | 6.0E-10 | 3.6 | 3.5 | 3.8 | 8.7 |
| sll0416 | 60-kDa chaperonin 2 (groEL2) | 2.0E-08 | 4.0 | 2.5 | 6.1 | 11.6 |
| sll0430 | HtpG, Hsp90 | 4.0E-08 | 2.7 | 2.9 | 3.5 | 9.9 |
| slr2076 | 60-kDa chaperonin (groEL1) | 5.0E-06 | 3.2 | 2.4 | 2.1 | 8.2 |
| sll0897 | DnaJ protein, Hsp40 | 3.0E-04 | 1.8 | 1.8 | 2.9 | 2.2 |
| sll0170 | DnaK protein 2, Hsp70 | 7.0E-04 | 1.5 | 2.3 | 3.3 | 5.6 |
| slr0093 | DnaJ protein, Hsp40 | 5.0E-03 | 2.5 | 1.5 | 2.5 | 4.0 |
| sll0058 | DnaK protein 1, Hsp70 | 1.3E-02 | 1.4 | 1.2 | 1.4 | |
P(treatment) is the loop design for the ΔperR experiment.
In the heat shock experiment, the change after a shift from 35 to 45°C for 15 min was examined. All P values for this experiment were <0.05.
In addition to the chaperones, a number of genes coding for cellular processes were up- or downregulated by peroxide. Especially prominent were two of the ftsH genes (slr0228 and slr1604), both of which were upregulated approximately two- to fourfold. Conversely, the ftsZ (sll1633) and ftsW (slr1267) genes were downregulated approximately twofold. In addition, genes involved with pilins or motility were strongly downregulated (sll1533 and slr1928 to slr1931), as were many genes involved with the synthesis of surface polysaccharides (see Table S1 in the supplemental material). Also, transcription of the secDF genes and the leader peptidase gene increased after peroxide stress (see Table S1 in the supplemental material).
Regulatory genes.
The regulatory genes provided the richest panorama of changes in transcript levels of all the functional categories. The transcript levels of approximately 40 genes involved in regulatory and transcriptional activities were changed in the ΔtrxA1 mutant experiments, and the transcript levels of 24 of these genes were changed in both experiments (Table 3). The transcript levels of most of the regulatory genes decreased, whereas the transcript levels of one-half or more of the RNA synthesis genes increased (especially in the ΔtrxA1 mutant). Ten of the regulatory genes that displayed differential expression were genes encoding histidine kinases, and the transcript levels of all but one of these genes decreased; the transcript level of hik11 (slr1414) increased 1.4- to 2.2-fold in the two experiments. Marin et al. (37) demonstrated that Hik41 (sll1229) was involved in the response of Synechocystis sp. strain PCC 6803 to salt stress. The Hik41gene was upregulated (∼2.5-fold) in the ΔperR mutant experiment, as were slr0967 (∼5.5-fold), sll0939 (∼5.5-fold), and sll0938 (∼1.7-fold), the three genes shown to be under the control of Hik41 in response to salt stress. The genes encoding two group 2 sigma factors were strongly upregulated by peroxide in both experiments; these genes were sigD (sll2012; 8-fold upregulation; P = 9 × 10−11) and sigB (sll0306; ∼2.7-fold upregulation; P = 3 × 10−8).
Putative PerR regulon in Synechocystis sp. strain PCC 6803.
We used three criteria to tentatively determine the PerR regulon: the presence of a potential PerR binding motif, the effect of deleting PerR on gene transcription in the presence of peroxide, and an interaction term from the statistical analysis. The availability of the genome sequence of Synechocystis sp. strain PCC 6803 allowed preliminary prediction of the PerR regulon of this organism. We identified putative PerR binding sites in Synechocystis sp. strain PCC 6803 based on the 8-bp inverted repeat sequence ATAATTAT. A preliminary search revealed 66 sites that contained the 8-bp consensus sequence. Some of these candidate sites were found upstream of the coding sequences of genes whose protein products were likely to have a role in oxidative stress or metal ion storage, including genes that were strongly upregulated by peroxide stress (Table 6). The perR (slr1738) and ahpC (sll1621) genes share a divergent promoter with an excellent putative PerR box, and both of these genes are upregulated by peroxide. In addition, ahpC, which codes for a peroxiredoxin-like protein, is even more strongly induced by methyl viologen (a generator of singlet oxygen) than perR is (data not shown).
TABLE 6.
Putative genes that may be under PerR control
| Cyano designation | Gene name | Putative perR box | Position (bp)a | Function | Fold changeb
|
P(genoperoxide) | ||
|---|---|---|---|---|---|---|---|---|
| Wild type | ΔperR mutant | ΔtrxA1 mutant | ||||||
| slr1738 | perR | ATAATTATTCTATCTAATA | −186 | Peroxide regulon repressor | 7.0 | 1.3 | 4.0 | 4.0E-07 |
| sll1621 | ahpC | ATAATTATTCTATCTAATA | −150 | Alkyl hydroperoxide reductase | 13.1 | 1.0 | 2.3 | 5.0E-13 |
| sll1620 | Unknown | 5.8 | 1.0 | 4.7 | 7.0E-07 | |||
| slr1739 | psbW-like | Unknown | 1.7 | 1.0 | 1.7 | 6.0E-02 | ||
| ssl2667 | cnfU | NifU-like C terminal | 5.0 | 1.0 | 1.4 | 2.0E-07 | ||
| sll0621 | dsb-like | c-type cytochrome biogenesis | 3.1 | 1.0 | 1.2 | 5.0E-06 | ||
| slr0513 | idiA | AATAATTTATGGATTATT | −202 | IdiA homologue | 2.4 | 5.7 | 4.2 | 2.0E-05 |
| sll0247 | isiA | ATAAATTCTCATTTAT | −246 | Inducible Chl-binding protein | 11.3 | 10.0 | 5.8 | 1.6E-01 |
| sll1135 | TTAATTTTAATT | −28 | Unknown | 7.7 | 3.3 | 5.6 | 2.0E-02 | |
| sll1483 | ATAATTTGTCATAATA | −50 | Fasciclin-like domain | 15.3 | 14.4 | 6.5 | 4.3E-01 | |
| slr1894 | mrgA | ATATTATTTCTATCTAATAT | −188 | Metal-regulated gene | 2.2 | 2.0 | 2.3 | 9.7E-01 |
| slr1204 | htrA | HtrA protease | 2.6 | 1.2 | 3.0 | 1.6E-03 | ||
The position of the putative perR box is defined as the distance between the ATG and the first nucleotide of the putative box.
The changes for the wild type and the ΔperR and ΔtrxA1 mutants were calculated as described in Table 2 by dividing the normalized mean intensities of peroxide-treated strains by the normalized mean intensities of nontreated strains.
We utilized Northern blots to verify the microarray results and to determine transcriptional regulation of certain genes in the absence of PerR (Fig. 2). Strong induction by peroxide in the wild type was evident for isiA, perR, and ahpC, as was the approximately twofold increase in the level of slr0513. As reported previously, there was excellent quantitative and qualitative correspondence between the microarray and Northern blot data (57, 58). Differences in the induction pattern were evident between the ΔperR strain and the wild type. Transcription of isiA was similar in the wild type and the ΔperR mutant, and transcription of slr0513 was actually reduced in the ΔperR mutant in the absence of peroxide. This led to a greater increase compared to the wild type and was similar to the 2.4- and 5.7-fold changes revealed by the microarray results (Table 6). Transcription of perR and ahpC exhibited a different pattern. The results were consistent with the hypothesis that PerR is a transcription factor (a repressor) that regulates these two genes and that slr1738 and sll1621 are fully transcribed when PerR is not present. Analysis by using both microarrays and Northern blots in the ΔperR mutant experiment indicated that two genes that were contiguous to perR/ahpC (sll1620 and slr1739) also acted as if they were under PerR control, although neither had a PerR box motif in the promoter. Thus, this regulation might reflect a structural interaction involving PerR binding to the perR promoter on this neighboring set of genes.
FIG. 2.
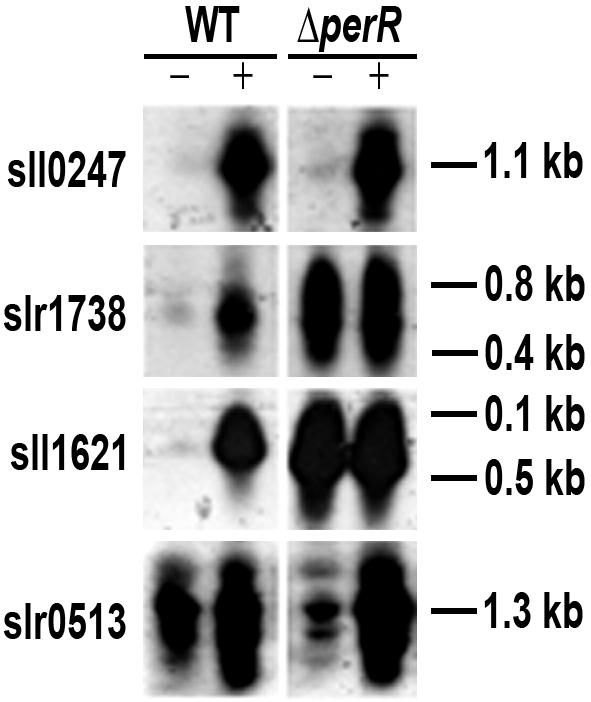
Northern blots showing the effect of peroxide on gene transcription in Synechocystis sp. wild-type strain PCC 6803 and the ΔperR mutant. RNA was isolated from cells that were treated with 1.5 mM peroxide for 30 min (+) or from untreated controls (−).
We then determined the set of genes for which there were large changes in the wild-type strain when there was peroxide stress but only approximately 1.0-fold changes in the ΔperR mutant. This was consistent with the action of a repressor; when PerR was absent, gene expression would be expected to be as high in the control as it is in the presence of peroxide (Table 6). This set of genes was then analyzed statistically for P(genoperoxide). This was the probability that there was an interaction between the stress and the mutant strain (i.e., that there was a significant change in the response of a particular gene to the stress in the mutant compared to the response in the wild type). Six genes fit these criteria and had P(genoperoxide) values of <2 × 10−5 (Table 6). These genes included slr1738 (perR) and sll1621 (ahpC), genes which have a good putative PerR box in a divergent promoter region. The neighboring genes sll1620 and slr1739 were also in this set of genes, although slr1739 did not have a good P(genoperoxide) value.
Based on these criteria, our current understanding of the putative PerR regulon is shown in Table 6. We tentatively propose that perR, ahpC, sll1620, slr1739, cnfU (ssl2667), sll0621, and sll1135 comprise the PerR regulon. In addition, there are four other genes that may be under PerR control. Of the genes in Table 5, perR, ahpC, mrgA, and the putative thioredoxin reductase gene ahp (sll1135) represent genes that have previously been identified as components of oxidative stress regulons in B. subtilis (19). The cnfU and dsbD-like (sll0621) genes code for proteins that may be involved in protein assembly and/or disulfide redox control. The CnfU protein has a thioredoxin motif (C-X-X-C), and sll0621 encodes a protein with four membrane-spanning segments that closely resembles DsbD from E. coli (54). However, isiA, sll1483, and slr0513 are genes that seem to be more specific to cyanobacteria. isiA and slr0513 were both first identified as inducible genes under iron-deficient conditions in a number of cyanobacteria and have some involvement in photosynthesis (58, 73). The sll1483 gene has not been studied previously, and it appears to have repeats of a fascicilin domain and may encode a protein involved with the cell envelope. Since induction of slr0513 actually increased in the ΔperR mutant and the transcript levels of isiA and sll1483 changed very little in the ΔperR mutant, we tentatively describe these genes as part of the regulon based on the presence of a PerR box and their strong induction by peroxide stress. The transcript levels of mrgA did not change much in either the ΔperR or ΔtrxA1 mutant, but there are additional reasons for placing this gene in the PerR regulon, as discussed below.
DISCUSSION
Reactive oxygen species, such as hydrogen peroxide, superoxide, and hydroxyl radicals, are inevitably produced during growth of organisms that carry out aerobic respiration and/or oxygenic photosynthesis. Survival of these organisms depends on the efficient scavenging of these oxygen species. In the present work, we utilized a full genome cDNA microarray to identify the genes that are regulated in response to hydrogen peroxide stress in the cyanobacterium Synechocystis sp. strain PCC 6803, which can carry out photosynthesis and respiration simultaneously. The effect of H2O2 on gene expression in the wild type was compared with the effect in two strains that lacked either a thioredoxin (TrxA1) or a transcriptional regulator (PerR). Whereas thioredoxin was chosen for its role in maintaining the cellular redox status of cells that might interfere with responses to H2O2, studies on PerR were based on our microarray results. The majority of genes regulated by peroxide stress in the three strains were similar, which strongly suggests that gene regulation occurred in response to peroxide stress rather than due to growth phase transitions.
The pathway leading to induction of the peroxide stimulon in cyanobacteria is not well understood. Our data demonstrated that the majority of genes that responded to oxidative stress are PerR and TrxA1 independent or are indirectly regulated by these two proteins. Interestingly, many genes that responded to peroxide stress also responded to high light (22), redox changes (23), high salinity (27), and high osmolality (27). For example, some of the pigment genes were downregulated under all of these stress conditions. The sll0306 (sigB), sll2012 (sigD), sll0790 (hik31), sll1621 (ahpC), slr1544 (unknown gene), heat shock, and chaperone genes are all induced under these conditions. The similar responses of these genes were expected, because these stresses often disrupt the cellular homeostasis of cells, leading to enhanced production of reactive oxygen species. It is also possible that the need to coordinate gene expression effectively in changing environmental conditions has led to the evolution of a set of signal transduction pathways which sense and respond to the various stress conditions. In this regard, it is possible that reactive oxygen species, such as hydrogen peroxide, can act as signaling molecules to mediate responses to these stress conditions. We do not yet know if H2O2 acts as a separate signal or as part of a systemic response in cyanobacteria. It is likely that the redox poise of cells may control cellular events through interactions with H2O2 and redox-sensing molecules, such as glutathione, thioredoxins, and peroxiredoxins. Thus, the largest single group of genes regulated by peroxide stress likely is part of a general stress regulon.
The heme and Chl biosynthesis genes represented a key difference between the oxidative stress responses in Synechocystis sp. strain PCC 6803 and B. subtilis. In Synechocystis sp. strain PCC 6803, genes in the first half of the heme pathway were transcribed similarly in the presence and absence of peroxide. However, many of the genes in the second half of the pathway, leading to Chl and bilin production, were significantly downregulated, as they were under low-iron growth conditions (Table 3; see Table S2 in the supplemental material). From the results in Table 3, it does not appear that PerR is directly involved in the regulation of these genes.
One objective of this study was to determine the importance of thioredoxins in oxidative stress. We found that the transcript levels of the genes encoding two putative thioredoxins, TrxA1 and TrxM2, decreased upon peroxide stress and that the ΔtrxA1 mutant was somewhat more sensitive to peroxide than the wild type. In fact, the increases in the transcript levels of many genes encoding proteins involved in the response to peroxide were smaller in the ΔtrxA1 mutant than in the wild type, and the genes acted as if TrxA1 provided a more oxidizing environment when it was present. This could be accomplished by other thioredoxin genes that play more active roles in the absence of TrxA1 and may explain the smaller increases in the transcript levels. For example, the E. coli Trx2 protein is required for the viability of cells that lack trxA and gshA (51). At the same time, two genes encoding proteins with thioredoxin or Dsb-like properties (ssl2667 and sll0621) were induced by peroxide and may be under PerR control (Table 6). We also pursued the possibility that the ΔtrxA1 mutant had higher transcript levels of many genes even prior to administration of the peroxide stress. However, a careful analysis of the intensity values for the genes listed in Table S1 in the supplemental material indicated that this was not generally true. The apparent lack of a difference in the responses to H2O2 in the wild type and the ΔtrxA1 mutant suggest that trxA1 might be functionally redundant. One objective for future studies is to determine the response of Synechocystis sp. strain PCC 6803 to disulfide stress and the relationship to peroxide stress, as was accomplished recently for B. subtilis (34).
To date, PerR binding sites have been identified in only B. subtilis (21), and the studies made good use of the PerR mutants. These binding studies confirmed the role of the proposed B. subtilis PerR binding site, 5′-TTATAATNATTATAA. A problem in predicting Fur and PerR boxes lies in the extreme similarity of the consensus sequences. Experimental evidence of Fur- or PerR-mediated regulation is necessary, although the analysis is complicated due to overlaps between the Fur and PerR regulons (24, 65). Our initial studies have shown that purified PerR binds to the promoters that carry a putative PerR binding site (data not shown). We utilized three criteria to identify the PerR regulon: induction in response to H2O2 treatment in the wild type and either the presence of a putative PerR binding motif or loss of inducibility in the PerR mutant. However, it is important to recognize that not all components of the PerR regulon in B. subtilis can be induced by peroxide and that the metalloregulation of different PerR regulon genes is distinct; some components can be repressed by either Mn or Fe, whereas others are Mn specific. In Staphylococcus aureus, PerR is selective for Mn(II) (25), whereas the Campylobacter jejuni orthologue responds to Fe (65). Thus, complete determination of the PerR regulon in Synechocystis sp. strain PCC 6803 will require additional experimentation.
One of the striking findings was that transcription of the operon that encoded the CP43-like iron stress-induced protein (IsiA) and a flavodoxin (IsiB) was highly induced by peroxide. Although these proteins were discovered almost two decades ago (5, 32, 33, 45), their precise functions are not completely understood. Recently, two groups of workers have shown that 18 copies of the IsiA protein form a ring around the PSI reaction center in Synechocystis sp. strain PCC 6803 (2, 3). These workers suggested that the additional chlorophylls in this antenna ring of a PSI supercomplex increased the light-harvesting capacity of the PSI reaction centers. This can be an important factor under long-term iron stress conditions (3 to 4 days of growth) when the numbers of PSI centers and phycobilisomes have been reduced. In addition to its response to iron limitation, IsiA of Synechocystis sp. strain PCC 6803 responded to salt stress (2, 3, 66, 67), and we show here that isiA and isiB are highly induced by short-term oxidative stress, even in the presence of low peroxide concentrations (data not shown). This implies that induction of isiA is initiated long before IsiA is used as an antenna for a smaller number of PSI units. We could not detect significant increases in the level of the IsiA protein under our oxidative stress conditions (data not shown), nor did we see the absorption changes that are typical of IsiA (5). Unexpectedly, ΔisiA or ΔisiB mutants were generally no more sensitive to peroxide than the wild type under normal growth conditions (50 μmol of photons m−2 s−1), although this was not the case at higher light intensities (100 μmol of photons m−2 s−1). These results suggested that IsiA has multiple functions; it may not only have a role as a structural protein but may also be involved in signaling.
Studies with the cyanobacterium Synechococcus elongatus strain PCC 7942 have important ramifications for understanding the cyanobacterial response to peroxide and the functions of proteins like IsiA. Yousef et al. (73) and Lundrigan et al. (36) presented evidence which suggested that there is a relationship between iron homeostasis and peroxide stress in this strain. In particular, they showed that many genes known to be iron inducible, including isiA and idiA, were also inducible by short-term peroxide or methyl viologen stress. They proposed that IdiA was involved in the protection of PSII and that an enhanced cyclic electron transport activity around PSI was part of the cyanobacterial adaptive response to oxidative stress. Interestingly, there are two potential IdiA orthologues in Synechocystis sp. strain PCC 6803, slr1295 and slr0513. Although slr1295 has the greatest similarity to the IdiA gene, this gene shows little responsiveness to changes in the iron concentration (58) or to peroxide (this study). On the other hand, slr0513 was strongly induced under both conditions, and thus the gene product might have the functional properties of IdiA. However, the differences indicate that caution must be used before such models are extrapolated to all cyanobacteria.
We constructed mutants with mutations in a number of the genes that were induced by peroxide and studied their sensitivity to this stress. Interestingly, many of these mutants showed little change in peroxide sensitivity, and some of the mutants were actually more resistant to peroxide; this was most noticeable for the ΔisiA mutant. However, the ΔmrgA mutant was hypersensitive to peroxide (Table 4) (∼20-fold more sensitive), and MrgA currently represents the most critical protein studied to date that is needed for peroxide protection. In B. subtilis, MrgA forms highly stable, multimeric protein-DNA complexes that accumulate in stationary-phase cells and protect against oxidative killing (15, 18). A Per box was identified in the promoter region, suggesting that mrgA is a member of the B. subtilis peroxide regulon (6, 7). In Synechococcus sp. strain PCC 7942, dpsA (the mrgA homologue) transcripts accumulated in the stationary phase and under most nutrient stress conditions (13, 46, 56), and a strain lacking dpsA was more sensitive to peroxide (14). The mrgA gene in Synechocystis sp. strain PCC 6803 has a good putative PerR box (Table 6), although the transcript levels were about the same after peroxide treatment in the wild type and the ΔperR mutant. Thus, although we cannot definitively place mrgA in the PerR regulon, the sensitivity of the ΔmrgA mutant to peroxide and its relationship to orthologous genes in other bacteria emphasize its importance to the response of Synechocystis sp. strain PCC 6803 to peroxide stress.
These microarray experiments provided a vast amount of new data on the response of Synechocystis sp. strain PCC 6803 to short-term, exogenous peroxide stress. A number of key regulatory genes were induced, and understanding the induction of these genes and identifying the genes that they control are important tasks. Toward this goal, we studied the effect of loss of PerR on the H2O2 response and showed that the PerR regulon controls only a small subset of genes. The strong induction of sigD by peroxide suggested that this gene could play an important role in peroxide protection, and evaluating the importance of SigD in the response to peroxide stress is one of our short-term goals. Since approximately 40% of the peroxide-regulated genes have unknown functions, this system should help us answer some of the most pertinent questions related to oxidative stress in cyanobacteria, including what regulatory networks control the production and scavenging of reactive oxygen species. For example, are there novel scavenging mechanisms that operate in cyanobacteria? It is interesting that Perelman et al. (48) identified a novel H2O2 detoxification activity in Synecochoccus sp. strain PCC 7942. Tichy and Vermaas (63) also showed that the absence of catalase led to a 30-fold decrease in H2O2 decomposition, although the mutant strain had a normal phenotype and the same resistance to H2O2 and methyl viologen as the wild type. Identification of such novel detoxification pathways and additional regulatory networks and characterization of the redox-sensing mechanisms are some of the important directions for future studies.
Supplementary Material
Acknowledgments
We thank Elsie Grace and Lisa Bono for help with the statistical analysis of the data.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 2.Bibby, T. S., J. Nield, and J. Barber. 2001. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412:743-745. [DOI] [PubMed] [Google Scholar]
- 3.Boekema, E. J., A. Hifney, A. E. Yakushevska, M. Piotrowski, W. Keegstra, S. Berry, K. P. Michel, E. K. Pistorius, and J. Kruip. 2001. A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412:745-748. [DOI] [PubMed] [Google Scholar]
- 4.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Burnap, R. L., T. Troyan, and L. A. Sherman. 1993. The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43′) is encoded by the isiA gene. Plant Physiol. 103:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., and J. D. Helmann. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol. Microbiol. 18:295-300. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 9.Churchill, G. A. 2002. Fundamentals of experimental design for cDNA microarrays. Nat. Genet. 32(Suppl.):490-495. [DOI] [PubMed] [Google Scholar]
- 10.Colon-Lopez, M. S., D. M. Sherman, and L. A. Sherman. 1997. Transcriptional and translational regulation of nitrogenase in light-dark- and continuous-light-grown cultures of the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 179:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doerge, R. W., and G. A. Churchill. 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drenth, J. P., R. H. te Morsche, R. Smink, J. S. Bonifacino, and J. B. Jansen. 2003. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat. Genet. 33:345-347. [DOI] [PubMed] [Google Scholar]
- 13.Durham, K. A., and G. S. Bullerjahn. 2002. Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Basic Microbiol. 42:367-372. [DOI] [PubMed] [Google Scholar]
- 14.Dwivedi, K., A. Sen, and G. S. Bullerjahn. 1997. Expression and mutagenesis of the dpsA gene of Synechococcus sp. PCC 7942, encoding a DNA-binding protein involved in oxidative stress protection. FEMS Microbiol. Lett. 155:85-91. [Google Scholar]
- 15.Frenkiel-Krispin, D., S. Levin-Zaidman, E. Shimoni, S. G. Wolf, E. J. Wachtel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 2001. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 20:1184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. USA 99:6690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant, R. A., D. J. Filman, S. E. Finkel, R. Kolter, and J. M. Hogle. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5:294-303. [DOI] [PubMed] [Google Scholar]
- 19.Halliwell, B., and J. M. Gutteridge. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849-859. [DOI] [PubMed] [Google Scholar]
- 22.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hihara, Y., K. Sonoike, M. Kanehisa, and M. Ikeuchi. 2003. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel, and G. Gibson. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29:389-395. [DOI] [PubMed] [Google Scholar]
- 27.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 28.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerr, M. K., and G. A. Churchill. 2001. Experimental design for gene expression microarrays. Biostatistics 2:183-201. [DOI] [PubMed] [Google Scholar]
- 30.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77:123-128. [DOI] [PubMed] [Google Scholar]
- 31.Klughammer, B., M. Baier, and K. J. Dietz. 1998. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol. Plant. 104:699-706. [Google Scholar]
- 32.Laudenbach, D. E., M. E. Reith, and N. A. Straus. 1988. Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J. Bacteriol. 170:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laudenbach, D. E., and N. A. Straus. 1988. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J. Bacteriol. 170:5018-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 36.Lundrigan, M. D., J. E. Arceneaux, W. Zhu, and B. R. Byers. 1997. Enhanced hydrogen peroxide sensitivity and altered stress protein expression in iron-starved Mycobacterium smegmatis. Biometals 10:215-225. [DOI] [PubMed] [Google Scholar]
- 37.Marin, K., I. Suzuki, K. Yamaguchi, K. Ribbeck, H. Yamamoto, Y. Kanesaki, M. Hagemann, and N. Murata. 2003. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 100:9061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIntyre, L. M., E. R. Martin, K. L. Simonsen, and N. L. Kaplan. 2000. Circumventing multiple testing: a multilocus Monte Carlo approach to testing for association. Genet. Epidemiol. 19:18-29. [DOI] [PubMed] [Google Scholar]
- 39.Meunier, P. C., M. S. Colon-Lopez, and L. A. Sherman. 1997. Temporal changes in state transitions and photosystem organization in the unicellular, diazotrophic cyanobacterium Cyanothece sp. ATCC 51142. Plant Physiol. 115:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 41.Moody, D., B. Fadlia, A. Singh, S. Shah, and L. M. McIntyre. 2002. Quantitative comparison of image analysis software, p. 155-166. In S. Shah and G. Kamberova (ed.), Microarray image analysis—nuts & bolts. DNA Press, Eagleville, Pa.
- 42.Navarro, F., and F. J. Florencio. 1996. The cyanobacterial thioredoxin gene is required for both photoautotrophic and heterotrophic growth. Plant Physiol. 111:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiyama, Y., H. Yamamoto, S. I. Allakhverdiev, M. Inaba, A. Yokota, and N. Murata. 2001. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 20:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oleksiak, M. F., G. A. Churchill, and D. L. Crawford. 2002. Variation in gene expression within and among natural populations. Nat. Genet. 32:261-266. [DOI] [PubMed] [Google Scholar]
- 45.Pakrasi, H. B., H. C. Riethman, and L. A. Sherman. 1985. Organization of pigment proteins in the photosystem II complex of the cyanobacterium Anacystis nidulans R2. Proc. Natl. Acad. Sci. USA 82:6903-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pena, M. M., and G. S. Bullerjahn. 1995. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J. Biol. Chem. 270:22478-22482. [DOI] [PubMed] [Google Scholar]
- 47.Peng, X., C. L. Wood, E. M. Blalock, K. C. Chen, P. W. Landfield, and A. J. Stromberg. 2003. Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinformatics 4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perelman, A., A. Uzan, D. Hacohen, and R. Schwarz. 2003. Oxidative stress in Synechococcus sp. strain PCC 7942: various mechanisms for H2O2 detoxification with different physiological roles. J. Bacteriol. 185:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 50.Postier, B. L., H. L. Wang, A. Singh, L. Impson, H. L. Andrews, J. Klahn, H. Li, G. Risinger, D. Pesta, M. Deyholos, D. W. Galbraith, L. A. Sherman, and R. L. Burnap. 2003. The construction and use of bacterial DNA microarrays based on an optimized two-stage PCR strategy. BMC Genomics 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 52.Reddy, K. J., R. Webb, and L. A. Sherman. 1990. Bacterial RNA isolation with one hour centrifugation in a table-top ultracentrifuge. BioTechniques 8:250-251. [PubMed] [Google Scholar]
- 53.Regelsberger, G., C. Jakopitsch, L. Plasser, H. Schwaiger, P. G. Furtmuller, G. A. Peschek, M. Zamocky, and C. Obinger. 2002. Occurence and biochemistry of hydroperoxidases in oxygenic phototrophic prokaryotes (cyanobacteria). Plant Physiol. Biochem. 40:479-490. [Google Scholar]
- 54.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 55.Schroder, E., and C. P. Ponting. 1998. Evidence that peroxiredoxins are novel members of the thioredoxin fold superfamily. Protein Sci. 7:2465-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen, A., K. Dwivedi, K. A. Rice, and G. S. Bullerjahn. 2000. Growth phase and metal-dependent regulation of the dpsA gene in Synechococcus sp. strain PCC 7942, USA. Arch. Microbiol. 173:352-357. [DOI] [PubMed] [Google Scholar]
- 57.Singh, A. K., H. Li, and L. A. Sherman. 2004. Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol. Plant. 120:27-35. [DOI] [PubMed] [Google Scholar]
- 58.Singh, A. K., L. M. McIntyre, and L. A. Sherman. 2003. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 132:1825-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh, A. K., and L. A. Sherman. 2002. Characterization of a stress-responsive operon in the cyanobacterium Synechocystis sp. strain PCC 6803. Gene 297:11-19. [DOI] [PubMed] [Google Scholar]
- 60.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 61.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response. ASM Press, Washington, D.C.
- 62.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tichy, M., and W. Vermaas. 1999. In vivo role of catalase-peroxidase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 181:1875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tottey, S., M. A. Block, M. Allen, T. Westergren, C. Albrieux, H. V. Scheller, S. Merchant, and P. E. Jensen. 2003. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Vliet, A. H., M. L. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vinnemeier, J., and M. Hagemann. 1999. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch. Microbiol. 172:377-386. [DOI] [PubMed] [Google Scholar]
- 67.Vinnemeier, J., A. Kunert, and M. Hagemann. 1998. Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 169:323-330. [DOI] [PubMed] [Google Scholar]
- 68.Wayne, M. L., and L. M. McIntyre. 2002. Combining mapping and arraying: an approach to candidate gene identification. Proc. Natl. Acad. Sci. USA 99:14903-14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westfall, P. H., and S. S. Young. 1993. Resampling-based multiple testing: examples and methods for p-value adjustment. John Wiley & Sons, Inc., Somerset, N.J.
- 70.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto, H., C. Miyake, K. J. Dietz, K. Tomizawa, N. Murata, and A. Yokota. 1999. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 447:269-273. [DOI] [PubMed] [Google Scholar]
- 72.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
- 73.Yousef, N., E. K. Pistorius, and K. P. Michel. 2003. Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch. Microbiol. 180:471-483. [DOI] [PubMed] [Google Scholar]
- 74.Yue, H., P. S. Eastman, B. B. Wang, J. Minor, M. H. Doctolero, R. L. Nuttall, R. Stack, J. W. Becker, J. R. Montgomery, M. Vainer, and R. Johnston. 2001. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29:e41. [DOI] [PMC free article] [PubMed]
- 75.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.