Abstract
We evaluated the protective effects of fermented dairy products (FDPs) in an infection model, using the mouse pathogen Citrobacter rodentium (CR). Treatment of mice with FDP formulas A, B, and C or a control product did not affect CR colonization, organ specificity, or attaching and effacing lesion formation. Fermented dairy product A (FDP-A), but neither the supernatant from FDP-A nor β-irradiated (IR) FDP-A, caused a significant reduction in colonic crypt hyperplasia and CR-associated pathology. Profiling the gut microbiota revealed that IR-FDP-A promoted higher levels of phylotypes belonging to Alcaligenaceae and a decrease in Lachnospiraceae (Ruminococcus) during CR infection. Conversely, FDP-A prevented a decrease in Ruminococcus and increased Turicibacteraceae (Turicibacter). Importantly, loss of Ruminococcus and Turicibacter has been associated with susceptibility to dextran sodium sulfate–induced colitis. Our results demonstrate that viable bacteria in FDP-A reduced CR-induced colonic crypt hyperplasia and prevented the loss of key bacterial genera that may contribute to disease pathology.
Keywords: probiotic, C. rodentium, fermented dairy products, bioluminescence imaging, microbiota, DLIT-µCT
Probiotics are increasingly being used as alternative therapies for inflammatory bowel diseases, such as ulcerative colitis and Crohn's disease, or as a treatment for antibiotic-resistant gastrointestinal infections [1–3]. Probiotic bacteria are typically consumed daily in the form of fermented dairy products (FDPs), supplements, or medical foods [1, 3], and are defined as live microorganisms that confer a health benefit to the host, aside from general nutrition [4]. Typically, probiotic bacteria are from the Lactobacillus and Bifidobacterium genera, and the majority of commercially available probiotic products contain multiple bacterial species in the form of supplements and FDPs. Members of the Lactobacillus casei group (Lactobacillus paracasei subspecies paracasei [L. paracasei] and Lactobacillus rhamnosus [L. rhamnosus]) are among the most widely marketed and characterized probiotic species. Few Lactobacillus species are likely to be permanent residents of the human gastrointestinal microbiota and are often transient colonizers found in fermented foods [5–8]. Therefore, dosing regimens and the activity of probiotic Lactobacillus strains during transit through the gastrointestinal tract is of paramount importance [6–8].
The innate ability of the intestinal microbiota to outcompete invading bacterial species, known as competitive exclusion or colonization resistance, is executed by a variety of mechanisms including the production of antimicrobial peptides [9] and compounds such as organic acids [10], enhancement of epithelial barrier function [11], antagonism of receptor sites present on the mucosal epithelium [12], competition for nutrients within the lumen of the gastrointestinal tract [13], inhibition of quorum sensing systems in pathogenic bacteria [14], and direct modulation of the host's immune system (including innate defenses such as mucin or defensin production) [15, 16]. However, the specific molecular mechanisms underlying these well-documented protective effects and the probiotic “effectors” that confer these effects are largely unknown.
Citrobacter rodentium (CR) is a mouse-specific pathogen that shares virulence factors and mechanism of colonization via attaching and effacing (A/E) lesions with the human pathogens enteropathogenic and enterohemorrhagic Escherichia coli [17, 18]. CR causes a self-limiting disease in C57Bl/6 mice, colonic epithelial cell hyperplasia, and transmissible colitis delineated by Th1 and Th17 immune responses [19]. The CR model has been widely adopted to study if probiotics can confer colonization resistance, or modulate colitis [15, 20–26]. These studies demonstrated that pretreatment of neonatal and adult mice with individual probiotic strains such as Lactobacillus acidophilus, L. rhamnosus, and Lactobacillus helveticus resuspended in phosphate-buffered saline (PBS) ameliorated CR-induced inflammation and colonic crypt hyperplasia [15, 16, 23], whereas Wu et al [26] showed that CR virulence was modulated if mice were pretreated with the probiotic yeast Saccharomyces boulardii. In this study, we evaluated the impact of formulations of FDPs on host–pathogen interactions and the intestinal microbiota in the context of CR infection.
METHODS
Bacterial Strains and Products
The bacterial strains (Table 1) used to make the FDPs at Danone Research were isolated from traditional dairy products and selected through in vitro screening of anti-infectious and immunomodulatory properties. Short and long fermentations were separated by at least 12 hours. The pH of each product was not buffered and was measured at the end of fermentation. The appearance, taste, and nutritional composition of the products and control were identical. FDPs were stored at 4°C. CR was grown as described [27]. Supernatant from FDP-A (S-FDP-A) was obtained by centrifugation (4000g, 4°C, 10 minutes) of 100 mL of the product and stored at −20°C. Fresh FDP-A was β-irradiated (IR-FDP-A) with a dosage of 20 kGy and stored at 4°C. Loss of viability was confirmed by plating.
Table 1.
Composition of Fermented Dairy Products
| Product | Product pH | Fermentation Time | Bacterial Strains |
|---|---|---|---|
| FDP-A | pH 3.8 | Long | L. paracasei CNCM I-1518 |
| L. paracasei CNCM I-3689 | |||
| L. rhamnosus CNCM I-3690 | |||
| L. delbrueckii subsp bulgaricus | |||
| S. thermophilus | |||
| FDP-B | pH 4.1 | Short | L. paracasei CNCM I-1518 |
| L. paracasei CNCM I-3689 | |||
| L. rhamnosus CNCM I-3690 | |||
| L. delbrueckii subsp bulgaricus | |||
| S. thermophilus | |||
| FDP-C | pH 3.8 | Long | L. paracasei CNCMI-1518 |
| L. delbrueckii subsp bulgaricus | |||
| S. thermophilus | |||
| Control producta | pH 3.8 | NA | None |
Abbreviations: FDP, fermented dairy product; NA, not applicable.
a The control product was a sweetened nonfermented, acidified milk. The appearance, taste, and nutritional composition (proteins, carbohydrates, lipids, and energy) of the test products and control were identical.
Treatment of Mice With FDPs and CR Infection
Pathogen-free female C57Bl/6 mice weighing 18–20 g were housed in HEPA-filtered cages with sterile bedding, food, and water. Experiments were repeated on 2 or 3 separate occasions with 6–8 mice per group. In separate experiments, mice were gavaged daily between 9 and 10 am without anesthesia with 200 µL of one of the FDPs, IR-FDP-A, S-FDP-A, control product [CP] or PBS for 10 days. At 2 pm on day 10 after initiation of FDP treatment, mice were infected by oral gavage with 200 µL of CR cultured overnight in Luria Bertani broth [20], and were given the FDPs for an additional 8 days. All animal experiments were performed in accordance with the Animals (Scientific Procedures) Act 1986 and were approved by the local ethical review committee.
In Vivo Optical Imaging of C. rodentium–Infected Mice
Whole-animal bioluminescence imaging (BLI) was performed on days 4 and 8 postinfection (p.i.) using an IVIS 50 or IVIS Spectrum CT (PerkinElmer) [20, 28]. Regions of interest were identified and quantified, photons s−1 (total photon flux), using the appropriate version of Living Image software (PerkinElmer). Two representative mice from each group were imaged daily using diffuse light imaging tomography with integrated µCT imaging [20, 28].
Sample Processing and Histological Analysis
Colonization was monitored by daily enumeration of viable bacteria per gram of feces [20]. At day 8 p.i., segments of terminal colon were removed and fixed in 10% buffered formalin [20]. Crypt hyperplasia was calculated by measuring the lengths of at least 20 well-oriented crypts from each section, from all of the mice. Additional colonic segments were fixed in 2.5% glutaraldehyde for transmission electron microscopy (TEM) and scanning electron microscopy (SEM). Histological damage scoring was determined as described elsewhere [26]. Five independent fields of view from one representative tissue section was graded from all of the mice and averaged to obtain a mean histological score. All histological sections were evaluated blindly.
Indirect Immunofluorescence Staining and Electron Microcopy
Rabbit polyclonal antiserum was raised against β-irradiated L. rhamnosus CNCM I-3690 by Covalab. Indirect immunofluorescence was performed following heat-induced epitope retrieval, on formalin-fixed, paraffin-embedded sections. Chicken polyclonal anti-β-intimin and anti–L. rhamnosus antibodies were used to visualize CR and L. rhamnosus, respectively; DNA was counterstained with Hoescht 33342. Colonic tissues were processed for TEM and SEM as described previously [29].
Fecal DNA Extraction
Forty-two fecal samples were collected from FDP-A– and IR-FDP-A–treated groups at the following time points: prior to FDP treatment (day –10), the day of CR infection (day 0), and 8 days after CR infection (day 8). Fecal pellets were transferred to an RNAlater solution (Ambion), homogenized, and volume-adjusted to a final fecal dilution of 1:10 (wt/vol). Two hundred microliters was added to 1 mL PBS and centrifuged for 5 minutes at 5000g. Supernatant was discarded and pellets stored at −80°C until extraction as described previously [30]. DNA was analyzed by quantitative polymerase chain reaction (qPCR) and 16S amplicon pyrosequencing.
Quantification of Bacteria by qPCR
Primers and target loci are described in Supplementary Table 1. The Yakult Intestinal Flora-SCAN technology was used to perform qPCR as described previously [30, 31]. Reference strains (outlined in Supplementary Table 1) were used to establish standard curves and test primer specificity.
Gut Microbiota 454 Analysis
V5 and V6 hypervariable 16S ribosomal RNA (rRNA) regions were amplified using primers 784F and 1061R (Supplementary Table 1) [32]. Sequencing was performed by DNAVision SA (Charleroi) on a 454 Life Sciences Genome Sequencer FLX instrument (Roche) using titanium chemistry and primer A.
16s rRNA Pyrosequencing Analysis
Analyses were performed using QIIME version 1.6 [33]. A total of 306 393 reads were obtained from 5 Multiplex 454 FLX regions and assigned to 42 samples after filtering according to the following quality criteria: size between 150 and 500 nt, quality >25 over a 50 base-pair window, no mismatch authorized in primers and barcode sequences, and absence of polymers >6 nt. A total of 225 096 reads were clustered into operational taxonomic units (OTUs) defined at 97% identity using cd-hit [34], and representative sequences for each OTU were aligned and taxonomically assigned using Greengenes version 11_04 database. ChimeraSlayer [35] was used to discard potential chimeric sequences, leading to a mean of 5039 ± 1213 (SD) reads per sample.
For α and β diversity, samples were rarefied to 3000 sequences per sample. α-Diversity (that measures diversity within samples) was assessed using rarefaction curves for richness (Chao1), and evenness (Shannon index) and numbers of observed OTUs (Supplementary Figure 2A–C); β-diversity (that measures diversity between samples) was performed on both weighted and unweighted Unifrac distances. Jackknife randomization for robustness evaluation was performed 10 times using 2700 randomly chosen sequences for each sample.
Statistical Analyses
For the microbiota analysis, a selection of discriminant bacterial genera between different treatment groups was identified using an extension of a multivariate statistical analysis, sparse partial least squares discrimination analysis (sPLS-DA) [36]. To determine the difference between bacterial genera between 2 time points, the value of the last time point minus the value of the first time point for each population was used. Nonparametric Kruskal–Wallis test was then performed on the subspace of selected genera to confirm their differences between populations with a Benjamini–Hochberg multiple testing correction [37]. Wilcoxon Mann–Whitney tests without multiple testing correction were used to identify discriminant phyla and families and for qPCR analysis.
A one-way analysis of variance with group-specific variance and Tukey multiple-comparison posttest was used to analyze all other data, using commercially available software (GraphPad 5 and SAS 9.2); a P value of <.05 was taken to be significant.
RESULTS
The Effect of FDPs on Host–Pathogen Interactions
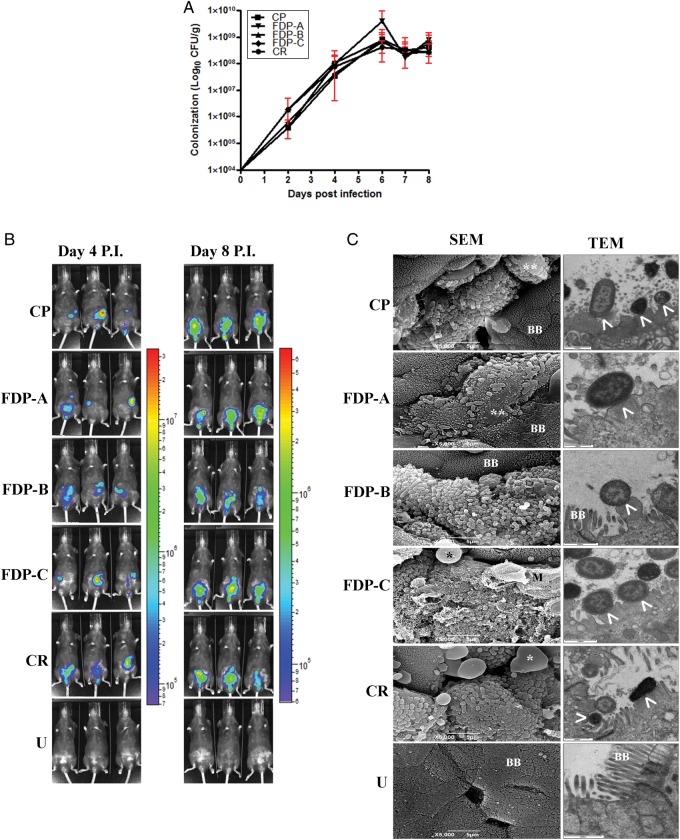
We determined whether treatment with FDPs A–C could competitively exclude CR. None of the FDPs caused any significant reduction in CR colonization when evaluated by bacterial enumeration (Figure 1A) or BLI (Figure 1B), compared with the CP-treated group or the untreated control (CR). The spatial distribution of bioluminescent (BL) CR within infected mice was heterogeneous within a treatment group, due to the dynamic nature of the intestines within a live mouse (Figure 1B). The organ specificity of infection monitored by BLI was in line with previous reports [27], with the cecum heavily colonized at day 4 p.i. and cecum, colon, and rectum at day 8 p.i. (Figure 1B). None of the FDP treatments affected the organ specificity of infection (Figure 1B) or A/E lesion formation (Figure 1C).
Figure 1.
A, Quantification of Citrobacter rodentium (CR) colony-forming units (CFU) from stools over 8 days postinfection (p.i.) in the different treatment groups. B, In vivo optical imaging of a bioluminescent (BL) CR infection from 3 representative mice per treatment at days 4 and 8 p.i. C, Electron microscopy of terminal colon at day 8 p.i. reveals epithelial cell death (*) and attaching and effacing (A/E) lesions (arrowheads), irrespective of the fermented dairy product (FDP) treatment used. Abbreviations: BB, brush border; CFU, colony-forming units; CP, control product; SEM, scanning electron microscopy; TEM, transmission electron microscopy; U, untreated and uninfected.
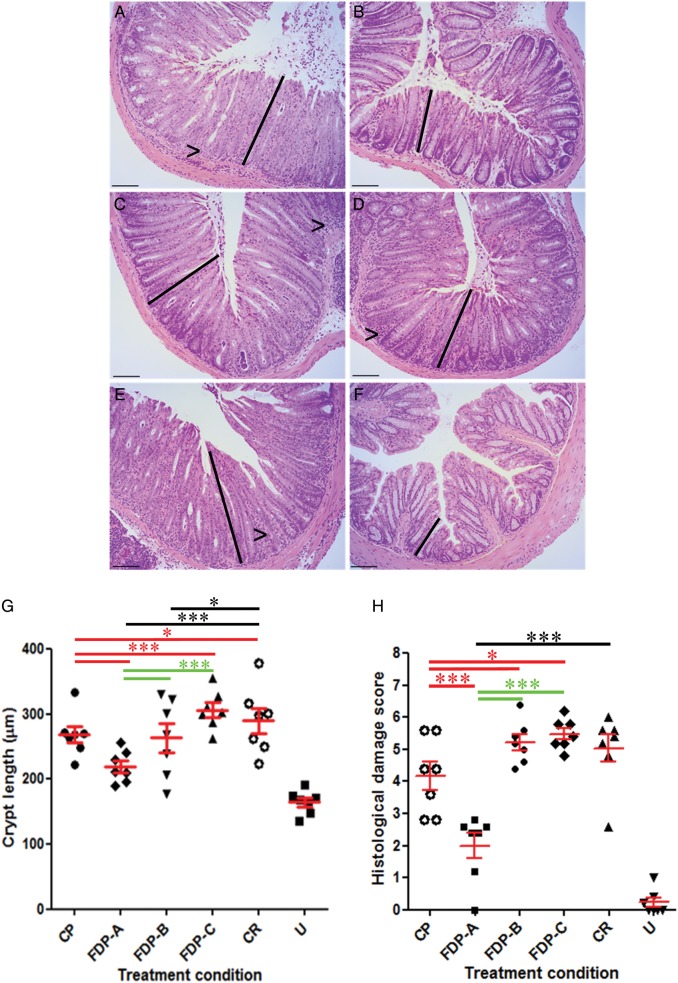
Quantification of colonic crypt hyperplasia at day 8 p.i. demonstrated that CP (P < .0001) and FDP-B (P < .01) significantly reduced colonic crypt length (Figure 2A–H). However, treatment with FDP-A was significantly more effective than CP and FDP-B (Figure 2G and 2H) and was therefore selected for further study.
Figure 2.
Histological analysis of Citrobacter rodentium (CR)–infected mice following treatment with (A) control product (CP), (B) fermented dairy product (FDP) A, (C) FDP-B, (D) FDP-C, (E) no treatment (Citrobacter rodentium [CR]), and (F) untreated and uninfected (U). FDP-A and FDP-B reduced lymphocyte accumulation in the lamina propria (arrowheads). Scale bar = 100 µm. G, Treatment of mice with FDP-A significantly reduced crypt hyperplasia compared with CP-treated mice (***P < .0001). H, Histological damage score demonstrating that FDP-A significantly reduced disease pathology compared with CR- and CP-treated groups (***P < .0001). FDP-A significantly reduced CR-associated pathology compared with FDP-B (***P < .0001). P values in red have been calculated compared with CP; P values in black have been compared to the CR group (*P < .05); P values in green are comparisons between FDPs.
Viable Bacteria in FDP-A Reduce CR-Associated Pathology
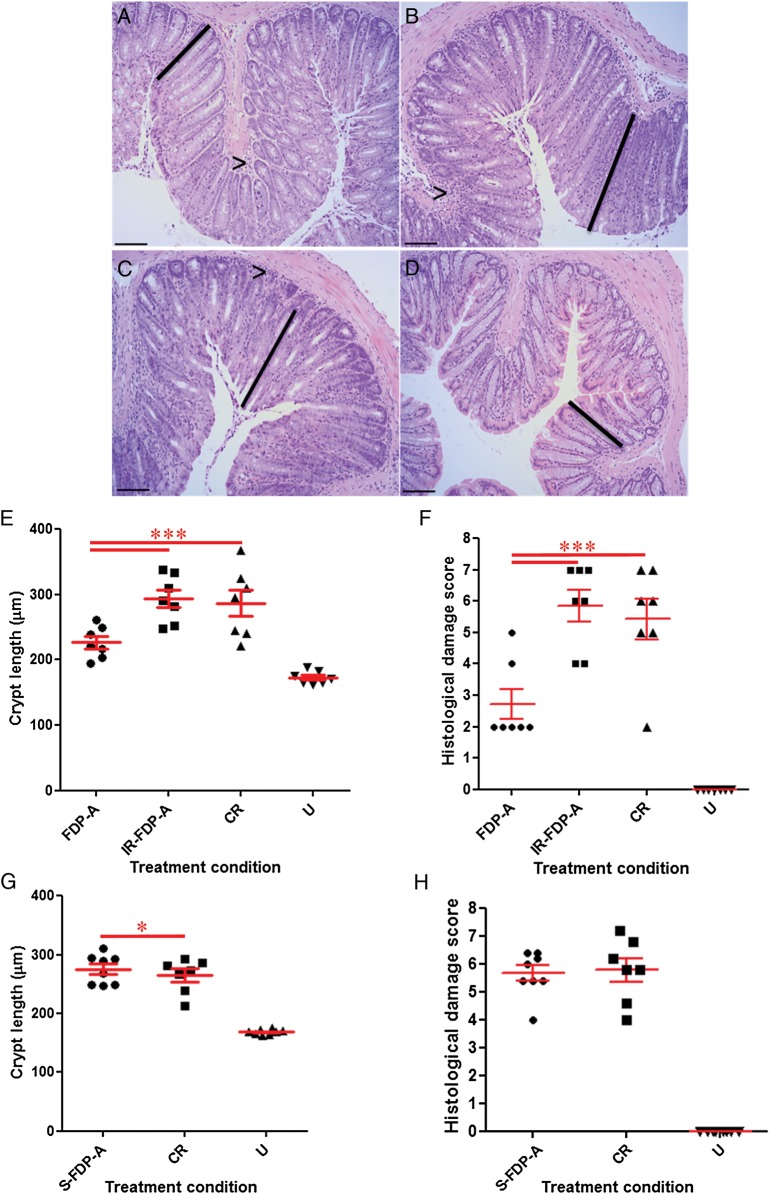
To determine the functional component of FDP-A responsible for causing a reduction in colonic crypt hyperplasia, mice were pretreated for 10 days with FDP-A, IR-FDP-A, or S-FDP-A; infected with CR; and administered the products daily for the duration of the study. Neither treatment caused a reduction in colonization or organ specificity of infection (Supplementary Figure 1). Quantification of colonic crypt hyperplasia demonstrated that FDP-A but not IR-FDP-A or S-FDP-A significantly reduced crypt length (P < .0001; Figure 3A–H). Moreover, the pathology score of mice treated with IR-FDP-A or S-FDP-A was not significantly reduced (P > .05; Figure 3A and 3C), demonstrating that health benefits require viable bacteria in the FDP.
Figure 3.
Quantification of crypt hyperplasia following treatment of mice with (A) fermented dairy product (FDP) A or (B) β-irradiated (IR) FDP-A, (C) no treatment (Citrobacter rodentium [CR]), and (D) untreated uninfected (U). E, FDP-A significantly reduced crypt hyperplasia compared with IR-FDP-A (***P < .0001) and qualitatively reduced lymphocyte accumulation in the lamina propria (arrowheads). IR-FDP-A treatment did not significantly reduce crypt hyperplasia compared with CR-infected mice (P > .05). F, Histological damage score demonstrating that FDP-A, but not IR-FDP-A, significantly reduced CR-associated pathology (***P < .0001). G, Treatment with supernatant from S-FDP-A significantly increased crypt hyperplasia compared with CR-treated or untreated and uninfected (U) mice (*P < .05). H, Histological damage score demonstrated that S-FDP-A treatment did not significantly alter CR associated pathology compared with CR-treated mice (P > .05). Scale bar = 100 µm.
Lactobacillus rhamnosus Present in FDP-A Is Associated With Colonic Epithelial Cells
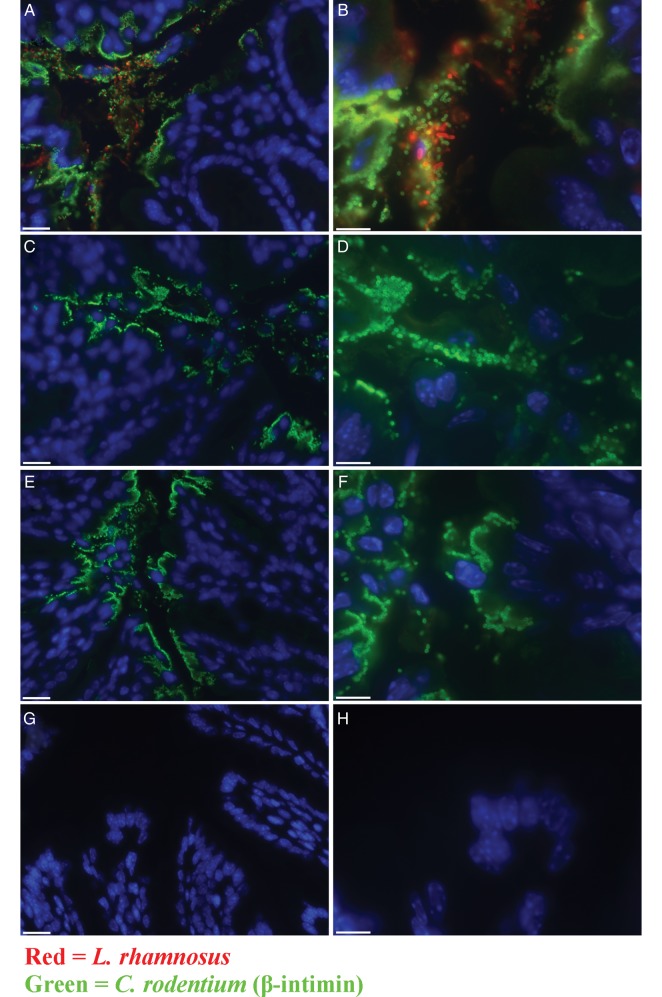
We visualized the distribution of L. rhamnosus on the colonic mucosa following treatment with FDP-A or IR-FDP-A and CR infection (day 8 p.i.). L. rhamnosus could only be found in FDP-A–treated mice, but not in IR-FDP-A–treated mice (Figure 4A and 4B). Qualitative assessment of L. rhamnosus distribution showed multiple bacteria associated with colonic epithelial cells lining the lumen and colocalization with CR on infected epithelial cells (Figure 4A and 4B). Small numbers of L. rhamnosus could be visualized at the bottom of the crypts and within goblet cells (data not shown). L. rhamnosus was also present in high numbers within the intestinal lumen (Figure 4A and 4B). Evaluating whether the treatment altered CR distribution within the colonic mucosa revealed that neither FDP-A nor IR-FDP-A affected the pattern of CR colonization of epithelial cells lining the colonic lumen and crypts (Figure 4A–G).
Figure 4.
Indirect immunofluorescence using anti β-intimin (green) and anti–Lactobacillus rhamnosus (red). A and B, Fermented dairy product (FDP)–A treated and Citrobacter rodentium (CR) infected. (C and D), Irradiated FDP-A treated and CR infected. (E and F), No treatment and CR-infected and (G and H) untreated and uninfected (U). CR and Lactobacillus rhamnosus were observed associated to the epithelial layer lining the lumen of the colon. Lactobacillus rhamnosus was only found in the intestinal lumen, or associated with colonic epithelial cells following FDP-A treatment. The distribution of CR associated with the colonic mucosa was not affected by FDP treatment. Scale bars = 20 µm (left column) and 10 µm (right column).
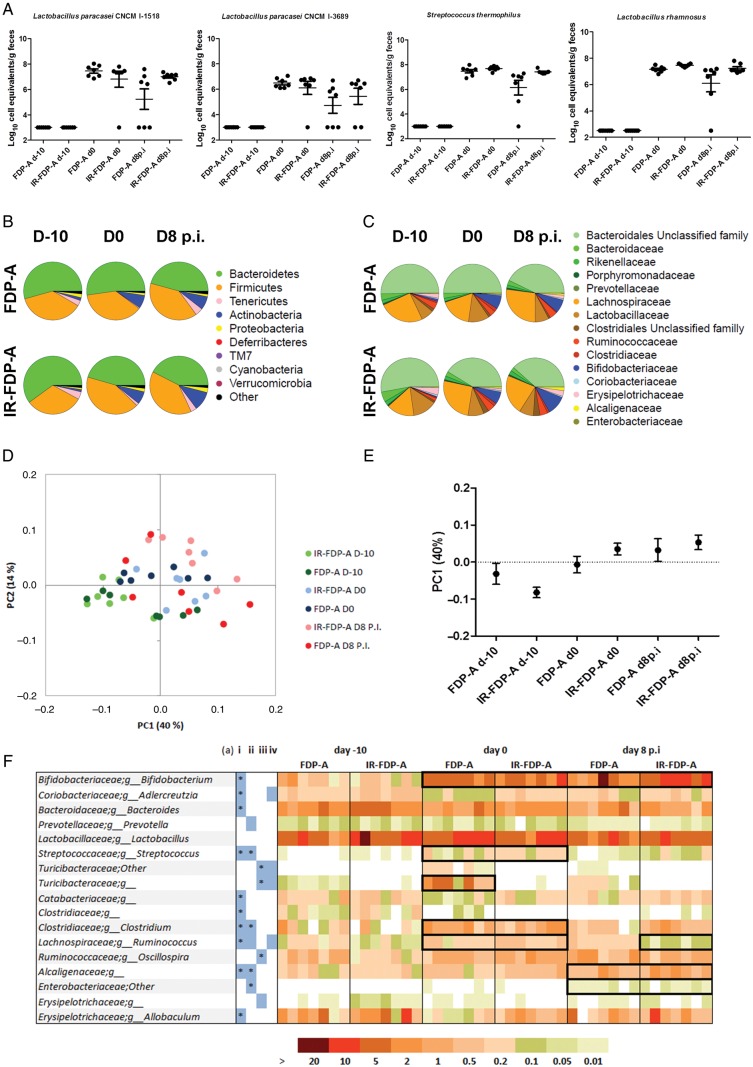
Quantification of FDP-A Bacteria in Feces and Modulation of the Intestinal Microbiota
We quantified the levels of 4 bacterial strains present in FDP-A by qPCR prior to treatment with the FDPs (day –10), 10 days after treatment with FDP-A or IR-FDP-A (day 0), and 8 days following CR infection (day 8 p.i.). This was done at either the strain level (L. paracasei strains CNCM I-1518 and CNCM I-3689) or species level (L. rhamnosus and Streptococcus thermophilus [S. thermophilus]) (Figure 5A). None of the bacterial species found in FDP-A were detected in feces before administration of the product. We detected similar levels of the bacterial species in the FDP-A– and IR-FDP-A–treated groups on days 0 and 8 after CR infection, suggesting that DNA from dead bacteria in the IR-FDP-A group was detected, with levels of different strains ranging from 5 × 106 to 1 × 108 cell equivalents per gram of feces.
Figure 5.
Microbiota profiling of stool samples by 16S rRNA gene pyrosequencing and quantitative polymerase chain reaction (qPCR). Samples were taken before treatment (day –10 [d-10]), before infection (day 0 [d0]), and 8 days postinfection (d8 p.i.). A, Evaluation of colonization of 4 strains present within fermented dairy product (FDP) A and β-irradiated (IR) FDP-A by qPCR. Taxa abundance profiles at (B) the phylum level and (C) the family level. D and E, Representation of 2D and 1D principal coordinates analysis of weighted Unifrac distances. F, Discriminant genera identified by sparse partial least squares multivariate analyses. Blue-colored boxes indicate discriminant genera in the corresponding comparisons (i) between day –10 and day 0 for both products, (ii) between day 0 and day 8 p.i. for both products, (iii) between FDPs for change after consumption (day 0 – day –10), or (iv) after infection (day 8 p.i. – day 0). Bold black frames focus on results described in the text, and stars in the blue boxes indicates significant population differences identified using a nonparametric Kruskal–Wallis test. “Other” indicates sequences that could not be attributed to a lower taxonomic level; g_ indicates sequences assigned to bacteria unclassified at the genus level.
We hypothesized that viable bacteria present in FDP-A may alter the intestinal microbiota and that these changes could be responsible for the observed reduction in CR-induced colonic hyperplasia and pathology. To test this, fresh feces were collected at the different time points, and the microbial community present was profiled by 16S rRNA gene pyrosequencing (Figure 5B–F) and qPCR (Figures 5A and 6). The 16S rRNA gene sequences were clustered into OTUs (97% identity), and representative sequences were aligned and used to calculate Unifrac distances between each sample pairs. Principal coordinates analysis of the weighted Unifrac distances (Figure 6D) indicated that samples from day –10 (baseline) clustered together and were more similar than samples from later time points (day 0 and day 8 p.i.). However, neither IR-FDP-A nor FDP-A were identified as discriminating factors following principal coordinates analysis, despite their good representation of the total variability (54%; Figure 5E). Therefore, the major modification of the intestinal microbiota in this study occurred between day –10 and day 0 (the administration phase).
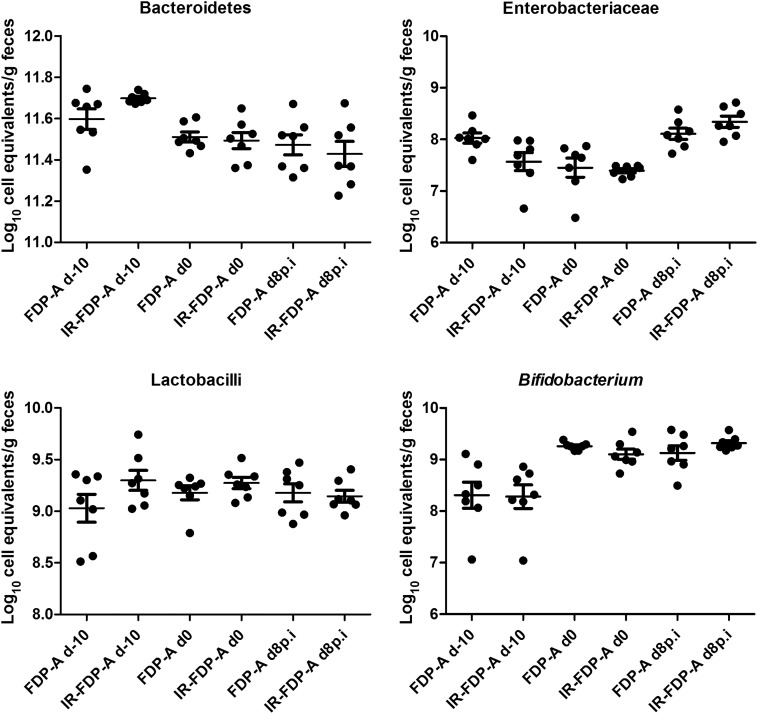
Figure 6.
Taxa abundance profiles according to treatment groups and time points in cell equivalents per gram of feces were calculated using quantitative polymerase chain reaction. Abbreviations: FDP, fermented dairy product; IR, β-irradiated; p.i. postinfection.
Prior to FDP treatment (day –10), the microbiota profiles from the phylum level of the 2 treatment groups were similar, apart from Proteobacteria representing 2% (±1%) in the FDP-A group and 1% (±0.28%) in the IR-FDP-A group (Figure 5B). Treatment of mice for 10 days with FDP-A or IR-FDP-A caused a significant increase in the proportion of Actinobacteria in both treatment groups, from a mean of 1.65% (±1.35%) at day –10 to 7.28% (±1.63%) at day 0 in the FDP-A group and from 1.29% (±0.37%) at day –10 to 7.1% (±3.69%) at day 0 in the IR-FDP-A group. At a lower taxonomic level, there was an increase at day 0 in the Bifidobacteriaceae family detected by 16S rRNA sequencing for both groups (P = .002) and in the Bifidobacterium genus detected by qPCR (P = .002 and P = .004 for FDP-A and IR-FDP-A, respectively) (Figures 5B, 5C, and 5F and 6). The increase in Bifidobacterium species persisted at day 8 p.i. in both treatment groups. In addition, both sequencing analysis and qPCR indicated a significant (P = .0049 and P = .0022) decrease in the relative abundance of Bacteroidetes following intake of IR-FDP-A (Figures 5B and 6), whereas Proteobacteria and Actinobacteria were higher in IR-FDP-A at day 8 p.i.
At the family level, Bifidobacteriaceae and Clostridiaceae increased following the consumption period, with an increase in Alcaligenaceae and Enterobacteriaceae observed in both groups during infection. However, Alcaligenaceae reached significantly (P = .04) higher levels at day 8 p.i. in IR-FDP-A (1.9% [±0.93%] vs 1% [±0.73%] in FDP-A; Figure 5C and 5F).
FDPs Induce Discriminant Changes at Genus Level During CR Infection
Multivariate analysis (sPLS-DA) was performed to investigate global microbiota changes between day –10 and day 0 (Supplementary Figure 2D and 2F and Figure 5F) and then between day 0 and day 8 p.i. (Supplementary Figure 2E and 2G and Figure 5F) to identify bacterial phylotypes moving either concomitantly or specifically in both FDP-treated groups. All analysis achieved at least 85% accuracy in discriminating 2 different groups.
Consumption of FDPs
During the administration period (day –10 to day 0), for both FDPs, Bifidobacterium, Clostridium, unclassified genera from Alcaligenaceae, Streptococcus, and Ruminococcus were increased, whereas Bacteroides, Allobaculum, unclassified Clostridiaceae, and Adlercreutzia decreased (Figure 5F, i). An unclassified Turicibacteraceae was the only phylotype that discriminated between FDP-A and IR-FDP-A during this period. This family was almost absent from the IR-FDP group, but was detected in FDP-A–treated mice at very low levels at day –10 (0.07% corresponding to 1 or 2 reads).
CR Infection
In both treatment groups, we observed an increase in unclassified Alcaligenaceae (Figure 5F, ii) and a decrease in Streptococcus and Clostridium. Importantly, Ruminococcus (Lachnospiraceae family) and, to a lesser extent, Adlercreutzia, were discriminant between the 2 treatment groups between day 0 and day 8 p.i. (Figure 5F and Supplementary Figure 2G), and these 2 taxa decreased in IR-FDP-A at day 8 p.i. and were either stable or slightly increased in FDP-A at the same time point.
DISCUSSION
This study is the first to evaluate whether a range of FDPs could affect the outcome of infectious colitis using a dosing regimen that mimics typical human consumption, in a murine infection model. In an initial screen of FDPs, our results demonstrated that daily prophylactic administration of FDP-A, FDP-B, and FDP-C did not prevent bacterial colonization or A/E lesion formation, or alter tissue specificity within the gastrointestinal tract. Studies using prophylactic administration of single strains of probiotic bacteria, or yeast resuspended in PBS, prior to CR infection significantly reduced the total bacterial load during infection [16, 22, 23, 26, 38]. These differences could be due to the different formulations, strain compositions, dosing regimens, and mouse strains employed.
In addition, probiotic bacteria have been demonstrated to alter the intestinal inflammatory response following chemical- or CR-induced colitis [15, 16, 23–26]. Pretreatment of mice with FDP-A and -B significantly reduced colonic hyperplasia, which is routinely used as a marker of intestinal inflammation in the CR model. Importantly, L. rhamnosus GG has been demonstrated to reduce cell cycle progression in cancer cells in vitro and, following a single treatment, induces epithelial cell proliferation in Drosophila melanogaster, mice, and humans [39–41]. Collectively, these data suggest that treatment of mice with Lactobacillus species (including L. rhamnosus) in the form of FDP-A can antagonize colonic epithelial cell hyperproliferation.
Secreted proteins p40 and p75 from L. rhamnosus GG have been shown to modify dextran sodium sulphate– or oxazolone-induced colitis and reduce colonic hyperplasia [42]. In this study, prophylactic treatment with FDP-A and -B demonstrated varying efficacy in the reduction of colonic hyperplasia, with FDP-A being the most effective. FDP-A and FDP-B contain exactly the same bacterial cultures and differ only in their fermentation time and culture pH, suggesting that the phenotype might involve either secreted metabolites or the physiology of the strains. However, as the supernatant from FDP-A did not reduce colonic hyperplasia or CR-associated pathology, the beneficial component of FDP-A is unlikely a secreted compound, unless it has a short half-life. Therefore, the fact that longer fermentation is needed to obtain greater protection suggests that protection is conferred by metabolic or proteomic/glycomic changes to the surface of bacteria in the product.
Quantification of bacteria present in the products in fecal samples demonstrated that Lactobacillus species and S. thermophilus were found in mice treated with either active or irradiated products following CR infection, indicating that DNA was still detected in the feces when mice were fed with dead bacteria. In contrast with our findings, it was reported that Lactobacillus species were decreased during CR infection [43]. However, it is likely that the daily administration of lactobacilli in FDPs masked the loss of endogenous Lactobacillus species. Importantly, we found that bacteria present in FDP-A, but not IR-FDP-A, were associated with CR-infected epithelial cells, which might be important for the protective effect of FDP-A.
Currently, there is much conjecture regarding what constitutes a “healthy” microbiota; however, it is well established that the loss of key bacterial genera can modify intestinal homeostasis and alter the immune response to gastrointestinal infections [44]. Profiling changes to the composition of the intestinal microbiota following FDP treatment demonstrated a large increase of Bifidobacterium species for both FDP-A– and IR-FDP-A–treated mice, suggesting that the composition of FDP-A, and not viable bacteria, is responsible; a similar phenotype was described in a humanized rat model fed L. paracasei–fermented milk [45]. We hypothesized that the reduced colonic hyperplasia in FDP-A–treated mice was associated with changes to key bacterial taxa that modulate mucosal homeostasis. Consumption of IR-FDP-A promoted higher levels of phylotypes belonging to Alcaligenaceae and a decrease in Lachnospiraceae (Ruminococcus) during CR infection, which was prevented in the FDP-A–treated group. Consumption of FDP-A induced a strong increase in Turicibacteraceae (Turicibacter), which decreased during CR infection but was below the detection limit in IR-FDP-A–treated mice. Importantly, loss of Ruminococcus and Turicibacter has been associated with susceptibility to dextran sodium sulphate–induced colitis [46]. In addition, we observed an increase in the Alcaligenaceae family in IR-FDP-A–treated mice, which has been associated with immunomodulation in mice [47]. In future studies, we intend to use germ-free mice, or mice with a reduced complexity microbiota, to investigate how genera identified through microbial profiling contribute to colonic hyperplasia during CR infection and FDP treatment.
Collectively, these data demonstrate that FDP-A modifies murine intestinal homeostasis following CR infection through an active process, which requires live bacteria and is independent of stable secreted bacterial molecule(s), highlighting the importance of consuming viable fermented products. Moreover, the reduction in colonic hyperplasia is not caused by FDP-C, and therefore the protective effects are unique to the combination of strains within FDP-A, demonstrating the importance of using fermented products containing multiple probiotic species.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Chloé Beal for performing qPCR analysis and providing bacterial cultures for antibody production. We also thank Christophe Daval for the production of FDPs.
Financial support. This work was supported by a grant from Danone Nutricia Research (to J. W. C.) and by the Wellcome Trust (A. K., B. R., V. F. C., G. F.).
Potential conflicts of interest. C. C., M. D., R. B., and I. C. are employees of Danone Nutricia Research. G. F. has received research grants from Danone Nutricia Research. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Sanders ME, Guarner F, Guerrant R, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–96. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huebner ES, Surawicz CM. Probiotics in the prevention and treatment of gastrointestinal infections. Gastroenterol Clin North Am. 2006;35:355–65. doi: 10.1016/j.gtc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterol. 2011;140:8–14. doi: 10.1053/j.gastro.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.United Nations Food and Agriculture Organization. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. http://www.who.int/foodsafety/publications/fsmanagement/en/probiotics.pdf . Accessed 16 June 2009.
- 5.Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–96. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oozeer R, Leplingard A, Mater DD, et al. Survival of Lactobacillus casei in the human digestive tract after consumption of fermented milk. Appl Environ Microbiol. 2006;72:5615–7. doi: 10.1128/AEM.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oozeer R, Goupil-Feuillerat N, Alpert CA, et al. Lactobacillus casei is able to survive and initiate protein synthesis during its transit in the digestive tract of human flora-associated mice. Appl Environ Microbiol. 2002;68:3570–4. doi: 10.1128/AEM.68.7.3570-3574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochet V, Rigottier-Gois L, Sutren M, et al. Effects of orally administered Lactobacillus casei DN-114 001 on the composition or activities of the dominant faecal microbiota in healthy humans. Br J Nutr. 2006;95:421–9. doi: 10.1079/bjn20051625. [DOI] [PubMed] [Google Scholar]
- 9.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104:7617–21. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernet-Camard MF, Lievin V, Brassart D, Neeser JR, Servin AL, Hudault S. The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl Environ Microbiol. 1997;63:2747–53. doi: 10.1128/aem.63.7.2747-2753.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterol. 2001;121:580–91. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Xu J, Shuai J, Chen J, Zhang Z, Fang W. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int J Food Microbiol. 2007;115:307–12. doi: 10.1016/j.ijfoodmicro.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Fuller R. Probiotics: the scientific basis. London: Chapman & Hall; 1992. [Google Scholar]
- 14.Medellin-Pena MJ, Wang H, Johnson R, Anand S, Griffiths MW. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–67. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gareau MG, Wine E, Reardon C, Sherman PM. Probiotics prevent death caused by Citrobacter rodentium infection in neonatal mice. J Infect Dis. 2010;201:81–91. doi: 10.1086/648614. [DOI] [PubMed] [Google Scholar]
- 16.Chen CC, Chiu CH, Lin TY, Shi HN, Walker WA. Effect of probiotics Lactobacillus acidophilus on Citrobacter rodentium colitis: the role of dendritic cells. Pediatr Res. 2009;65:169–75. doi: 10.1203/PDR.0b013e31818d5a06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 18.Wong AR, Pearson JS, Bright MD, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–38. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 19.Symonds EL, Riedel CU, O'Mahony D, Lapthorne S, O'Mahony L, Shanahan F. Involvement of T helper type 17 and regulatory T cell activity in Citrobacter rodentium invasion and inflammatory damage. Clin Exp Immunol. 2009;157:148–54. doi: 10.1111/j.1365-2249.2009.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins JW, Akin AR, Kosta A, et al. Pre-treatment with Bifidobacterium breve UCC2003 modulates Citrobacter rodentium-induced colonic inflammation and organ specificity of infection. Microbiology. 2012;158:2826–34. doi: 10.1099/mic.0.060830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Arienzo R, Maurano F, Mazzarella G, et al. Bacillus subtilis spores reduce susceptibility to Citrobacter rodentium-mediated enteropathy in a mouse model. Res Microbiol. 2006;157:891–7. doi: 10.1016/j.resmic.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Fanning S, Hall LJ, Cronin M, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A. 2012;109:2108–13. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson-Henry KC, Nadjafi M, Avitzur Y, et al. Amelioration of the effects of Citrobacter rodentium infection in mice by pretreatment with probiotics. J Infect Dis. 2005;191:2106–17. doi: 10.1086/430318. [DOI] [PubMed] [Google Scholar]
- 24.Jones SE, Knight KL. Bacillus subtilis-mediated protection from Citrobacter rodentium-associated enteric disease requires espH and functional flagella. Infect Immun. 2012;80:710–9. doi: 10.1128/IAI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues DM, Sousa AJ, Johnson-Henry KC, Sherman PM, Gareau MG. Probiotics are effective for the prevention and treatment of Citrobacter rodentium-induced colitis in mice. J Infect Dis. 2012;206:99–109. doi: 10.1093/infdis/jis177. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Vallance BA, Boyer L, et al. Saccharomyces boulardii ameliorates Citrobacter rodentium-induced colitis through actions on bacterial virulence factors. Am J Physiol Gastrointest Liver Physiol. 2008;294:G295–306. doi: 10.1152/ajpgi.00173.2007. [DOI] [PubMed] [Google Scholar]
- 27.Wiles S, Clare S, Harker J, et al. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 2004;6:963–72. doi: 10.1111/j.1462-5822.2004.00414.x. [DOI] [PubMed] [Google Scholar]
- 28.Collins JW, Meganck J, Kuo C, Francis KP, Frankel G. 4D Multimodality imaging of Citrobacter rodentium infections in mice. J Vis Exp. 2013 doi: 10.3791/50450. 78. doi:10.3791/50450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard F, Dziva F, van Diemen P, Phillips AD, Stevens MP, Frankel G. Adherence of enterohemorrhagic Escherichia coli O157, O26, and O111 strains to bovine intestinal explants ex vivo. Appl Environ Microbiol. 2007;73:3084–90. doi: 10.1128/AEM.02893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuki T, Watanabe K, Fujimoto J, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–73. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol. 2009;75:1961–9. doi: 10.1128/AEM.01843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinfor. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 35.Haas BJ, Gevers D, Earl AM, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Gen Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lê Cao K-A, Rossouw D, Robert-Granié C, Besse P. A sparse PLS for variable selection when integrating Omics data. Stat App Gen Mol Biol. 2008;7 doi: 10.2202/1544-6115.1390. Article 35. doi:10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 38.Chen CC, Louie S, Shi HN, Walker WA. Preinoculation with the probiotic Lactobacillus acidophilus early in life effectively inhibits murine Citrobacter rodentium colitis. Pediatr Res. 2005;58:1185–91. doi: 10.1203/01.pdr.0000183660.39116.83. [DOI] [PubMed] [Google Scholar]
- 39.Vielfort K, Weyler L, Soderholm N, Engelbrecht M, Lofmark S, Aro H. Lactobacillus decelerates cervical epithelial cell cycle progression. PLoS One. 2013;8:e63592. doi: 10.1371/journal.pone.0063592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RM, Luo L, Ardita CS, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–28. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Baarlen P, Troost F, van der Meer C, et al. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4562–9. doi: 10.1073/pnas.1000079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F, Cao H, Cover TL, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121:2242–53. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann C, Hill DA, Minkah N, et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–78. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–90. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djouzi Z, Andrieux C. Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br J Nutr. 1997;78:313–24. doi: 10.1079/bjn19970149. [DOI] [PubMed] [Google Scholar]
- 46.Zenewicz LA, Yin X, Wang G, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–12. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunisawa J, Kiyono H. Alcaligenes is commensal bacteria habituating in the gut-associated lymphoid tissue for the regulation of intestinal IgA responses. Front Immunol. 2012;3:65. doi: 10.3389/fimmu.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.