Abstract
Two trials of clinically approved human papillomavirus (HPV) vaccines, Females United to Unilaterally Reduce Endo/Ectocervical Disease (FUTURE I/II) and the Papilloma Trial Against Cancer in Young Adults (PATRICIA), reported a 22% difference in vaccine efficacy (VE) against cervical intraepithelial neoplasia grade 2 or worse in HPV-naïve subcohorts; however, serological testing methods and the HPV DNA criteria used to define HPV-unexposed women differed between the studies. We applied previously described methods to simulate these HPV-naïve subcohorts within the Costa Rica HPV16/18 Vaccine Trial and assessed how these criteria affect the estimation of VE. We applied 2 enzyme-linked immunosorbent assay (ELISA) thresholds for HPV16 and HPV18 seropositivity (8 and 7 ELISA units/mL, respectively, for PATRICIA; 54 and 65 ELISA units/mL, respectively, for FUTURE I/II (to approximate the competitive Luminex immunoassay)) and 2 criteria for HPV DNA positivity (12 oncogenic HPV types, plus HPV66 and 68/73 for PATRICIA; or plus HPV6 and 11 for FUTURE I/II). VE was computed in the 2 naïve subcohorts. Using the FUTURE I/II and PATRICIA criteria, VE estimates against cervical intraepithelial neoplasia grade 2 or worse, regardless of HPV type, were 69.0% (95% confidence interval: 40.3%, 84.9%) and 80.8% (95% confidence interval: 52.6%, 93.5%), respectively (P = 0.1). Although the application of FUTURE I/II criteria to our cohort resulted in the inclusion of more sexually experienced women, methodological differences did not fully explain the VE differences.
Keywords: human papillomavirus, methodological differences, naïve population, vaccine efficacy
Gardasil (Merck & Co., Inc., West Point, Pennsylvania) and Cervarix (GlaxoSmithKline Vaccines, Rixensart, Belgium) are 2 highly efficacious prophylactic human papillomavirus (HPV) vaccines. Gardasil is a quadrivalent vaccine containing the recombinant L1 major capsid proteins of oncogenic HPV16 and HPV18 and low-risk HPV6 and HPV11, and Cervarix is a bivalent vaccine containing the recombinant L1 major capsid proteins of HPV16 and HPV18 (1).
HPV vaccination is recommended for young adolescents before the initiation of sexual activity and exposure to the virus (2). However, the phase III clinical trials that led to vaccine licensure were conducted among young adult women, many of whom were sexually active and potentially HPV exposed. A combination of HPV DNA and serological testing was used to define a subcohort of women likely to be HPV naïve before vaccination to estimate vaccine efficacy (VE) in an HPV-unexposed target population. Published estimates of VE against cervical intraepithelial neoplasia grade 2 or worse (CIN2+), irrespective of HPV DNA typing, were 42.7% (95% confidence interval (CI): 23.7%, 57.3%) for Gardasil and 64.9% (95% CI: 52.7%, 74.2%) for Cervarix in the naïve-population subanalyses (3, 4). If the observed difference in estimated VE among HPV-naïve recipients of Gardasil and Cervarix reflects vaccine performance, it may have important public health implications.
Alternatively, these differences in VE estimates might reflect differences in the approaches used to simulate an HPV-unexposed group in the 2 trials—Females United to Unilaterally Reduce Endo/Ectovervical Disease (FUTURE I/II) and the Papilloma Trial Against Cancer in Young Adults (PATRICIA). In both trials, the criteria used to define HPV-unexposed groups included negative HPV DNA tests of cervical cells and negative serological results for HPV-related antibodies. Specifically, in both trials, subjects who tested HPV DNA positive for any of 12 oncogenic HPV types (type 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, or 59) were excluded from the HPV-naïve group. PATRICIA additionally excluded subjects on the basis of HPV DNA positivity for HPV types 66 and 68/73, whereas FUTURE I/II excluded subjects on the basis of HPV DNA positivity for HPV types 6 and 11 (3, 4). For the serological analyses, PATRICIA used a viruslike particle (VLP)–based direct enzyme-linked immunosorbent assay (ELISA), which detects the presence of both neutralizing and nonneutralizing HPV antibodies (4). In contrast, a VLP-based competitive Luminex immunoassay (cLIA) was used in FUTURE I/II, which is designed to detect neutralizing HPV antibodies specifically directed against the V5 epitope of HPV16 and the J4 epitope of HPV18 (3). Previous analyses have demonstrated that ELISA is more sensitive than cLIA (5). Thus, the use of cLIA may have resulted in the inclusion of more HPV-exposed women within the naïve subanalysis in FUTURE I/II compared with the use of the ELISA in PATRICIA.
The objective of the present analysis was to investigate the extent to which methodological differences in estimating VE within HPV-naïve subcohorts contributes to differences in VE estimates. To address our objective, we applied the FUTURE I/II and PATRICIA criteria within the Costa Rica HPV16/18 Vaccine Trial (CVT) to approximate the 2 naïve subcohorts and to estimate VE for the following 3 endpoints: 1) CIN2+ regardless of type of HPV infection, 2) HPV type–specific CIN2+, and 3) HPV type–specific 12-month persistence.
METHODS
Participants and study design
Women included in this study are participants in both arms of the CVT, a double-blind, controlled, randomized, phase III clinical trial of the efficacy of the bivalent HPV16/18 VLP vaccine conducted in Costa Rica with enrollment in 2004–2005. The CVT was designed to evaluate efficacy against persistent cervical HPV16/18 infection and precancerous lesions, as previously described (6, 7). The main eligibility requirements were as follows: aged 18–25 years (inclusive), in good general health as determined by medical history and a physical examination, not pregnant or breastfeeding, using contraception during the vaccination period, and willing to provide written informed consent. The trial was reviewed and approved by human subjects review committees of the Instituto Costarricense de Investigacion y Enseñanza en Nutrición y Salud (Tres Ríos, Cartago, Costa Rica) and the National Cancer Institute (Bethesda, Maryland). The CVT is registered with clinicialtrials.gov (number NCT00128661).
Procedure
At enrollment, women reporting prior sexual experience underwent a pelvic examination, with collection of exfoliated cervical cells for liquid-based cytological testing and HPV DNA testing and collection of blood for HPV16/18 serological testing. Women were randomized to receive 3 doses of the HPV bivalent vaccine Cervarix (GlaxoSmithKline Vaccines) or a control hepatitis A vaccine (modified Havrix, GlaxoSmithKline Vaccines) over a 6-month period.
At annual follow-up visits, clinicians collected cervical cells for cytology and HPV testing from sexually active women. Women with low-grade cytological abnormalities were evaluated semiannually, and those with high-grade or persistent low-grade abnormalities were referred for colposcopy and treatment as needed (for more details on study design, see the article by Herrero, et al. (6)).
Specimen collection
Exfoliated cells for cytology, HPV DNA, Chlamydia trachomatis DNA, Neisseria gonorrhea DNA, and other testing were collected with a Cervex-Brush (Rovers Medical Devices, B.V., Lekstraat, the Netherlands) during pelvic examination as previously described (6, 7). Cervical cells were placed in a liquid preservation medium; aliquots were frozen in liquid nitrogen until HPV polymerase chain reaction (PCR) testing as described below.
HPV DNA SPF10-DEIA and LiPA25
HPV DNA detection and genotyping were conducted at DDL Diagnostic Laboratory as previously described (8, 9). Briefly, DNA was extracted from cervical cells and SPF10 primer sets were used to PCR-amplify HPV-specific DNA. HPV genotype of SPF10-DNA enzyme immunoassay (DEIA)–positive samples was identified by reverse hybridization on a line probe assay (LiPA) (SPF10-DEIA/HPVLiPA25, version 1, Labo Bio-Medical Products, B.V., Rijswijk, the Netherlands), which detects 25 HPV genotypes (HPV6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70, and 74). The sensitivity of HPV16 and HPV18 detection was improved via PCR with type-specific primer sets for specimens testing SPF10-DEIA positive but LiPA25 negative for HPV16 and/or HPV18.
VLPs and ELISA
Serum collected at enrollment was used to determine HPV16- and HPV18-specific immunoglobulin G serostatus at GlaxoSmithKline Vaccines using a VLP-based direct ELISA as previously described (10). Briefly, serial dilutions of serum samples and standards were added to ELISA microtiter plates coated with HPV VLPs. A peroxidase-conjugated antihuman polyclonal antibody was added, followed by enzyme substrate and chromogen. Reactions were stopped, and optical density at 620 nm (background) was subtracted from optical density at 450 nm. Antibody levels, expressed as ELISA units/mL, were calculated by the interpolation of optical density values from the standard curve by averaging the calculated concentrations from all dilutions that fell within the working range of the reference curve. The seropositivity cutoff points were determined by GlaxoSmithKline Vaccines and calculated on the basis of the limit of detection (95th percentile from a virgin population) and the lower limit of quantitation of the assay (10, 11). The variability of this assay within the testing laboratory is low (mean coefficient of variation = 12.31% (10)).
Statistical analysis
Participants who received at least 1 vaccine dose, who had normal cytology, or who were virgins and had a valid ELISA result at enrollment were included in this analysis. Among eligible women, we defined the following 2 analytical cohorts on the basis of HPV DNA and HPV16/18 antibody positivity: naïve cohort 1 (using the PATRICIA-like criteria) and naïve cohort 2 (using the FUTURE I/II-like criteria). Follow-up began the day after administration of the first vaccine dose and ended at the time each outcome occurred or at the last study visit.
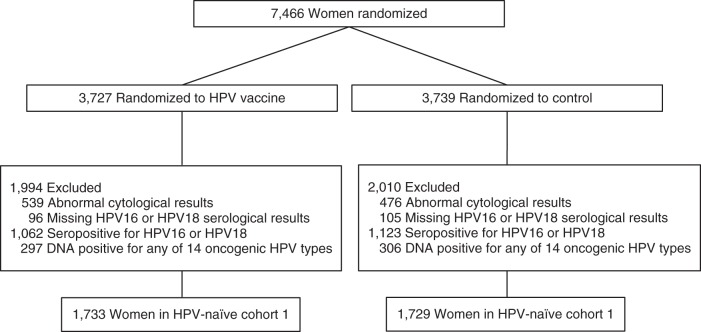
The criteria used to define naïve cohort 1 were identical to those used in the naïve subcohort analysis of PATRICIA (4). Naïve cohort 1 excluded women who were DNA positive at the cervix for the following HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68/73. It further excluded women who were seropositive for HPV16 or HPV18 using the standard ELISA cutoffs of 8 ELISA units/mL and 7 ELISA units/mL, respectively. The final analytical naïve cohort 1 included 3,462 women (1,733 in the vaccine arm and 1,729 in the control arm). Exclusions are shown in Figure 1.
Figure 1.
CONSORT diagram for naïve cohort 1 in the Costa Rica HPV16/18 Vaccine Trial, 2004–2005. Women were excluded on the basis of 1) human papillomavirus (HPV) type 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68/73 DNA positivity and 2) HPV16 or HPV18 seropositivity defined by standard enzyme-linked immunosorbent assay (ELISA) cutoffs of 8 ELISA units/mL or 7 ELISA units/mL, respectively.
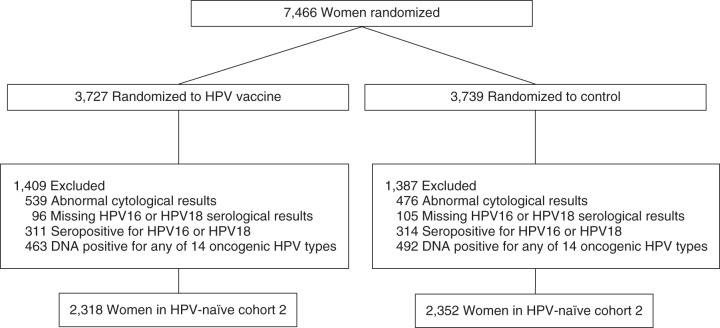
Naïve cohort 2 was defined to approximate the HPV-naïve cohort in FUTURE I/II. Naïve cohort 2 excluded women who were DNA positive at the cervix for the following HPV types: 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, or 59. It further excluded women who were seropositive for HPV16 or HPV18, as defined by modified ELISA cutoffs of 54 ELISA units/mL and 65 ELISA units/mL, respectively. The modified ELISA cutoffs were based on previous findings from our group that showed that increasing HPV16 and HPV18 ELISA cutoffs to 54 ELISA units/mL and 65 ELISA units/mL, respectively, maximized ELISA's agreement with cLIA (overall agreement = 97%, positive agreement = 78%) (5). The final analytical naïve cohort 2 included 4,670 women (2,318 in the vaccine arm and 2,352 in the control arm). Exclusions are shown in Figure 2.
Figure 2.
CONSORT diagram for naïve cohort 2 in the Costa Rica HPV16/18 Vaccine Trial, 2004–2005. Women were excluded on the basis of 1) human papillomavirus (HPV) type 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, or 59 DNA positivity and 2) HPV16 or HPV18 seropositivity, defined by modified enzyme-linked immunosorbent assay (ELISA) cutoffs of 54 ELISA units/mL or 65 ELISA units/mL, respectively.
The following 3 outcomes were evaluated: 1) persistence, defined as detection of same-type HPV (HPV16 or HPV18) in samples collected at 2 consecutive visits at least 10 months apart without intervening negative results; 2) CIN2+ regardless of HPV type, defined on the basis of masked review by a Costa Rican and a US pathologist, with masked review by a second US pathologist in instances when the first 2 reviewers disagreed; and 3) HPV16/18 type–specific CIN2+, in which a CIN2+ lesion was attributed to HPV16 or HPV18 if either of these HPV types was detected by PCR in the directly preceding cervical cytology specimen that led to colposcopy referral. For CIN2+ type–attribution in instances of multiple infections, we considered evidence of HPV persistence.
Event proportions were calculated by dividing the number of cases by the number of women for each vaccine arm and were expressed as the number of events per 1,000 women. VE was defined as the percentage reduction in the frequency of the endpoint related to vaccine administration, estimated as the complement of the ratio of the cumulative attack proportions in the HPV and control arms. The attack proportion is the percentage of women in the cohort who experience the endpoint. The confidence interval for VE is derived from the corresponding confidence interval for the risk ratio. The exact conditional test was used for analyses of VE.
A total of 4,689 participants were included in either 1 or both of the analytical cohorts. These women were further subdivided into 5 mutually exclusive subgroups on the basis of HPV antibody and DNA positivity (Web Figure 1 and Web Table 1, available at http://aje.oxfordjournals.org/). Groups A and B contributed to naïve cohort 1, and groups B, C, D, and E contributed to naïve cohort 2. We examined whether VE estimates differed by cohort definition. Because of the overlap of participants eligible to contribute to both cohorts (n = 3,443), we calculated P values in 2 different ways. First, we estimated a mid-P corrected exact P value by excluding group A (n = 19) from the comparison and compared group B (n = 3,443) with groups C, D, and E combined (n = 1,227). Second, we estimated a mid-P corrected exact P value for groups A and B combined (n = 3,462) versus groups C, D, and E combined (n = 1,227). We observed similar results and present the second set of P values. Statistical analyses were performed using SAS, version 9.2, software (SAS Institute, Inc., Cary, North Carolina).
RESULTS
Characteristics of the HPV-naïve subcohorts
A total of 3,443 women were included in both naïve cohort 1 (PATRICIA-like criteria) and naïve cohort 2 (FUTURE I/II-like criteria). A subset of 1,227 women was excluded from naïve cohort 1 but included in naïve cohort 2 (Web Figure 1). The subset of 1,227 women unique to cohort 2 was older and more sexually experienced, as indicated by younger age of sexual debut, greater number of years of sexual activity, higher total number of lifetime sexual partners, and enrollment positivity for low-risk HPV types. Thus, this subset of women appeared more likely than naïve cohort 1 to have been exposed to HPV on the basis of their risk profile; and they more closely resembled the subset of 2,777 women excluded from both analyses (Table 1).
Table 1.
Characteristics of Participants in Naïve Cohort 1, the Subset of Individuals Who Were Excluded From Naïve Cohort 1 but Included in Naïve Cohort 2, and the Subset of Individuals Who Were Excluded from Both Cohorts, Costa Rica HPV16/18 Vaccine Trial, 2004–2005
| Characteristic | Naïve Cohort 1 (n = 3,462) |
Individuals Excluded From Naïve Cohort 1 But Included in Naïve Cohort 2 (n = 1,227) |
Individuals Excluded From Both Cohorts (n = 2,777) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Median (IQR) | No. | % | Median (IQR) | No. | % | Median (IQR) | No. | % | |
| Age, years | 20 (19–23) | 21 (19–23) | 20 (19–23) | ||||||
| 17–19 | 1,357 | 39.2 | 329 | 26.8 | 751 | 27.0 | |||
| 20–21 | 826 | 23.9 | 321 | 26.2 | 704 | 25.4 | |||
| 22–23 | 695 | 20.0 | 290 | 23.6 | 681 | 24.5 | |||
| 24–27 | 584 | 16.9 | 287 | 23.4 | 641 | 23.1 | |||
| P value | <0.0001 | ||||||||
| Educational level | |||||||||
| ≤6 Years | 900 | 26.0 | 397 | 32.4 | 819 | 29.5 | |||
| 7–9 Years | 730 | 21.1 | 319 | 26.0 | 649 | 23.4 | |||
| ≥10 Years technicala | 1,213 | 35.1 | 340 | 27.7 | 865 | 31.2 | |||
| Any university | 614 | 17.8 | 170 | 13.9 | 440 | 15.9 | |||
| P value | <0.0001 | ||||||||
| Marital status | |||||||||
| Single | 2,122 | 61.4 | 616 | 50.2 | 1,443 | 52.0 | |||
| Married or living as married | 1,288 | 37.2 | 572 | 46.7 | 1,217 | 43.9 | |||
| Divorced/separated/ widowed | 49 | 1.4 | 38 | 3.1 | 113 | 4.1 | |||
| P value | <0.0001 | ||||||||
| Smoking status | |||||||||
| Never | 3,150 | 91.1 | 1,056 | 86.1 | 2,235 | 80.6 | |||
| Former | 149 | 4.3 | 71 | 5.8 | 207 | 7.5 | |||
| Current | 160 | 4.6 | 99 | 8.1 | 331 | 11.9 | |||
| P value | <0.0001 | ||||||||
| Age at first vaginal intercourse, yearsb | 17 (15–18) | 16 (15–18) | 16 (15–18) | ||||||
| Virgin | 1,281 | 37.0 | 228 | 18.6 | 83 | 3.0 | |||
| ≥19 | 397 | 11.5 | 145 | 11.8 | 419 | 15.1 | |||
| 17–18 | 816 | 23.6 | 342 | 27.9 | 903 | 32.6 | |||
| ≤16 | 965 | 27.9 | 511 | 41.7 | 1,367 | 49.3 | |||
| P value | <0.0001 | ||||||||
| Total no. of years sexually activeb | 4 (2–6) | 5 (3–7) | 5 (3–7) | ||||||
| Virgin | 1,281 | 37.0 | 228 | 18.6 | 83 | 3.0 | |||
| 0–7 | 1,815 | 52.5 | 770 | 62.7 | 2,153 | 77.7 | |||
| 8–10 | 307 | 8.9 | 198 | 16.2 | 461 | 16.6 | |||
| ≥11 | 56 | 1.6 | 30 | 2.5 | 75 | 2.7 | |||
| P value | <0.0001 | ||||||||
| Lifetime no. of sexual partnersb | 1 (1–2) | 2 (1–3) | 2 (1–3) | ||||||
| 0–1 | 2,514 | 72.7 | 648 | 52.9 | 904 | 32.7 | |||
| 2–3 | 765 | 22.1 | 421 | 34.4 | 1,226 | 44.4 | |||
| ≥4 | 179 | 5.2 | 156 | 12.7 | 634 | 22.9 | |||
| P value | <0.0001 | ||||||||
| Use of oral contraceptives | |||||||||
| Never | 520 | 15.0 | 208 | 17.0 | 632 | 22.8 | |||
| Virgin | 1,281 | 37.0 | 228 | 18.6 | 83 | 3.0 | |||
| In the past | 451 | 13.0 | 254 | 20.7 | 669 | 24.2 | |||
| Current (past month) | 1,206 | 35.0 | 536 | 43.7 | 1,383 | 50.0 | |||
| P value | <0.0001 | ||||||||
| Enrollment positivity for low-risk HPV typesc | |||||||||
| Virgin | 1,282 | 37.0 | 228 | 18.6 | 85 | 3.1 | |||
| Negative | 2,042 | 59.0 | 899 | 73.3 | 2,151 | 77.5 | |||
| Positive | 138 | 4.0 | 100 | 8.1 | 541 | 19.5 | |||
| P value | <0.0001 | ||||||||
Abbreviations: HPV, human papillomavirus; IQR, interquartile range.
a “Technical” refers to the last part of high school with an additional year of trade/vocational training.
b Median among nonvirgins.
c HPV types 34, 40, 42, 43, 44, 53, 54, 70, and 74.
Vaccine efficacy
For naïve cohort 1, VE against CIN2+, regardless of HPV type, was 80.8% (95% CI: 52.6%, 93.5%), with 5 cases of CIN2+ in the vaccine arm and 26 cases in the control arm (Table 2). For naïve cohort 2, VE against CIN2+, regardless of HPV type, was 69.0% (95% CI: 40.3%, 84.9%), with 11 cases of CIN2+ in the vaccine arm and 36 cases in the control arm. Although there was an 11.8% difference in VE between naïve cohorts 1 and 2, this difference was not statistically significant (P = 0.1). VE estimates against HPV16/18 type–specific CIN2+ were 100.0% (95% CI: 54.7%, 100.0%) for naïve cohort 1 and 81.6% (95% CI: 25.8%, 97.2%) for naïve cohort 2 (P = 0.08). Finally, VE estimates for HPV16/18 type–specific 12-month viral persistence for naïve cohorts 1 and 2 were 88.6% (95% CI: 77.3%, 94.9%) and 85.5% (95% CI: 75.7%, 91.8%), respectively (P = 0.3).
Table 2.
Vaccine Efficacy Against CIN2+ Regardless of HPV Type, HPV16/18 Type–Specific CIN2+, and HPV16/18 12-Month Persistence for Naïve Cohort 1 and Naïve Cohort 2, Costa Rica HPV16/18 Vaccine Trial, 2004–2005
| Study Group by Endpoint | Naïve Cohort 1a (n = 3,462) |
Naïve Cohort 2b (n = 4,670) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Women | No. of Events | No. of Events per 1,000 Women | 95% CI | Efficacy, % | 95% CI | No. of Women | No. of Events | No. of Events per 1,000 Women | 95% CI | Efficacy, % | 95% CI | |
| CIN2+ regardless of HPV type | ||||||||||||
| Vaccine recipientsc | 1,733 | 5 | 2.9 | 1.1, 6.4 | 2,318 | 11 | 4.7 | 2.5, 8.2 | ||||
| Control groupd | 1,729 | 26 | 15 | 10.1, 21.6 | 80.8 | 52.6, 93.5 | 2,352 | 36 | 15.3 | 10.9, 20.9 | 69 | 40.3, 84.9 |
| HPV16/18 type–specific CIN2+ | ||||||||||||
| Vaccine recipientsc | 1,733 | 0 | 0 | 0.0, 1.7 | 2,318 | 2 | 0.9 | 0.1, 2.8 | ||||
| Control groupd | 1,729 | 8 | 4.6 | 2.2, 8.8 | 100 | 54.7, 100.0 | 2,352 | 11 | 4.7 | 2.5, 8.1 | 81.6 | 25.8, 97.2 |
| HPV16/18 12-month persistence | ||||||||||||
| Vaccine recipientsc | 1,733 | 8 | 4.6 | 2.1, 8.7 | 2,318 | 15 | 6.5 | 3.8, 10.4 | ||||
| Control groupd | 1,729 | 70 | 40.5 | 31.9, 50.6 | 88.6 | 77.3, 94.9 | 2,352 | 105 | 44.6 | 36.8, 53.6 | 85.5 | 75.7, 91.8 |
Abbreviations: CI, confidence interval; CIN2+, cervical intraepithelial neoplasia grade 2 or worse; ELISA, enzyme-linked immunosorbent assay; HPV, human papillomavirus.
a In naïve cohort 1, women were excluded on the basis of 1) HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68/73 DNA positivity and 2) HPV16 or HPV18 seropositivity defined by standard ELISA cutoffs of 8 ELISA units/mL or 7 ELISA units/mL, respectively.
b In naïve cohort 2, women were excluded on the basis of 1) HPV6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, or 59 DNA positivity and 2) HPV16 or HPV18 seropositivity defined by modified ELISA cutoffs of 54 ELISA units/mL or 65 ELISA units/mL, respectively.
c Participants in the vaccine group were vaccinated with Cervarix (GlaxoSmithKine Vaccines, Rixensart, Belgium), a bivalent vaccine containing the recombinant L1 major capsid proteins of HPV16 and HPV18.
d Participants in the control group were vaccinated with a hepatitis A vaccine (modified Havrix, GlaxoSmithKline Vaccines).
For the analysis of VE against CIN2+, regardless of HPV type, 17 additional CIN2+ cases (6 in the HPV arm and 11 in the control arm) were present in naïve cohort 2 compared with naïve cohort 1. Of the 6 women with CIN2+ added to the vaccine arm of naïve cohort 2, 5 had detectable HPV antibodies at the HPV16 and HPV18 ELISA cutoffs of 8 ELISA units/mL and 7 ELISA units/mL, respectively, and 3 (including the 1 seronegative individual) were also HPV DNA positive for either HPV66 or HPV68/73. Of the 11 additional women with CIN2+ added to the control arm of naïve cohort 2, 10 had detectable HPV antibodies at the HPV16 and HPV18 ELISA cutoffs of 8 ELISA units/mL and 7 ELISA units/mL, respectively, and the remaining seronegative individual was HPV DNA positive for either HPV66 or HPV68/73. In addition, there was 1 woman with CIN2+ in the control arm of naïve cohort 1 who was not in naïve cohort 2; she was HPV DNA positive for HPV6 or HPV11. On the basis of these findings, serological cutoffs accounted for 15 of the 17 additional cases of CIN2+ in naïve cohort 2, and HPV DNA positivity criteria accounted for 2. Therefore, it was estimated that serology alone resulted in the greatest reduction of the estimate of VE against CIN2+ regardless of HPV type (approximately 8%), whereas DNA positivity criteria accounted for the remaining 4%.
DISCUSSION
Differences in both serological cutoffs and criteria for HPV DNA positivity together accounted for 11.8% lower VE estimates between naïve cohort 1 (PATRICIA-like criteria) and naïve cohort 2 (FUTURE I/II-like criteria). The lower estimation of VE against CIN2+ regardless of type in naïve cohort 2 resulted from the inclusion of more HPV-exposed women compared with naïve cohort 1. Of the 17 additional cases of CIN2+ in the vaccine and control arms of naïve cohort 2 compared with naïve cohort 1, 15 had detectable HPV antibodies at the more sensitive ELISA cutoffs of 8 ELISA units/mL and 7 ELISA units/mL, and 4 (including 2 seronegative individuals) were HPV DNA positive for either HPV66 or HPV68/73 at enrollment. Given the higher prevalence of HPV66 and HPV68/73 (2.9% and 2.5%, respectively) in the CVT at enrollment compared with HPV6 and HPV11 (1.8% and 0.9%, respectively), exclusion on the basis of HPV66 and HPV68/73 seropositivity may have further resulted in a “cleaner” population of HPV-naïve individuals within naïve cohort 1. In contrast, we observed a smaller difference in VE estimates for HPV16/18 type–specific 12-month persistence (88.6% vs. 85.5%). The explanation for this is unclear; however, 12-month persistence was a more common endpoint and, therefore, provided greater precision in the VE point estimates.
Therefore, on the basis of our findings, methodological differences alone may not account for the entire or even the majority of the 22% difference in estimated VE against CIN2+ reported in the HPV-naïve subcohort analyses of FUTURE I/II and PATRICIA. Another likely explanation for the difference in VE estimates between the 2 trials is a varying degree of cross-protection against oncogenic nonvaccine HPV types for Cervarix and Gardasil (3, 4). Although HPV16 and HPV18 were responsible for the majority of CIN2+ lesions in both trials, a significant proportion of cases were also attributed to nonvaccine types. Because both trials reported nearly identical estimates of VE against CIN2+ attributable to HPV16/18 (100.0% for FUTURE I/II and 99.0% for PATRICIA), the difference in VE estimates for CIN2+ regardless of HPV type may partially be accounted for by differences in CIN2+ cases attributable to oncogenic nonvaccine types (3, 4). Cervarix has been reported to have higher efficacy against persistent HPV31 infections (77.1% vs. 46.2%), as well as higher efficacy against HPV33- and HPV45-associated CIN2+ compared with Gardasil (82.3% vs. 24.0% and 100% vs. −51.9%, respectively (12)). Therefore, Cervarix's higher cross-reactivity against nonvaccine types may have partially contributed to the higher estimate of VE for CIN2+, regardless of type, compared with Gardasil.
Other possible, but less likely, explanations include weaker immunogenicity of Gardasil compared with Cervarix and risk differences between the 2 trial populations. Although Cervarix has been shown to induce significantly higher HPV16/18 antibody levels compared with Gardasil (13, 14), this difference in immunogenicity is unlikely to explain the difference in VE against CIN2+ regardless of type, given that both trials reported nearly 100% VE against CIN2+ attributable to HPV16/18 in the naïve subcohort analyses (3, 4). Additionally, although the FUTURE I/II, PATRICIA, and CVT populations were very similar in their risk profiles, they did vary with respect to baseline HPV16 prevalence (6, 15–17). Baseline HPV16 prevalence rates for FUTURE I/II, CVT, and PATRICIA were 9.1%, 8.3%, and 5.0%, respectively (16, 17). Although the high baseline HPV16 prevalence in FUTURE I/II suggests that this population had a greater risk of HPV exposure than the population in PATRICIA, this is unlikely to explain differences in VE. HPV16 prevalence in the CVT was similar to that in FUTURE II (8.3% vs. 9.1%), yet we have reported in the current study the highest VE estimate against CIN2+ regardless of HPV type (80.8%) among all 3 trials. The higher VE estimate in the CVT compared with estimates in PATRICIA and FUTURE I/II may be partially driven by the fact that a slightly higher proportion of CIN2+ cases were attributable to HPV16/18 in the CVT compared to PATRICIA (31% vs. 26%; no information was provided for FUTURE I/II) (3, 4).
The strength of this analysis is that we used the same testing facilities and identical serological and HPV DNA assays to those used in PATRICIA. Other strengths include the fact that our study was conducted within a well-characterized, single population and, therefore, our findings are internally valid. Although we believe our qualitative results to be valid, our quantitative results may not be directly applicable to the other trials because we could not directly control for all differences between the trials. Examples of these differences include cultural differences in the reporting of sexual behavior, minimum age of enrollment, attack rates, HPV type attribution, and other assay differences (i.e., HPV DNA PCR assays and laboratories used for histopathology). Whether these differences affected our results is unclear; however, in our current analysis we noted a difference in VE estimates against CIN2+ attributable to HPV16/18 between naïve cohorts 1 and 2, which was not observed in the original trials. Perhaps the most important issue we were unable to address was the effect of exclusion on the basis of HPV6 and HPV11 seropositivity. This was due to the fact that HPV6 and HPV11 testing was not performed in the CVT. Exclusion of women who were seropositive for HPV6 or HPV11 in the FUTURE I/II would be expected to result in exclusion of some high-risk women who the PATRICIA analysis would have included, and, therefore, may have partially counteracted the less stringent definition of HPV16/18 seropositivity. However, although exclusion on the basis of HPV6/11 seropositivity may have attenuated some of the differences in the risk of exposure between the 2 studies, HPV6/11 antibodies are not likely to be as closely linked to CIN2+ development as is low-level HPV16/18 seropositivity.
In summary, although the methods used to define the HPV-naïve subcohort in PATRICIA may have resulted in a “cleaner” HPV-naïve population compared with those used in FUTURE I/II, our analysis suggests that differences in serological strategies and HPV DNA criteria may not fully account for the difference in reported VE against CIN2+, regardless of HPV type, between the 2 trials. However, additional studies are warranted.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: National Cancer Institute (NCI), National Institutes of Health (NIH), Bethesda, Maryland (Krystle A. Lang Kuhs, John T. Schiller, Mark Schiffman, Sholom Wacholder, Douglas R. Lowy, Aimée R. Kreimer, Mark E. Sherman, Allan Hildesheim, Mahboobeh Safaeian); Proyecto Epidemiológico Guanacaste, Fundación INCIENSA, San José, Costa Rica (Carolina Porras, Ana Cecilia Rodriguez, Paula Gonzalez, Rolando Herrero); Prevention and Implementation Group, International Agency for Research on Cancer, Lyon, France (Paula Gonzalez, Rolando Herrero); Public Health Foundation of India, New Delhi, India (Arpita Ghosh); Joint Program in Survey Methodology, University of Maryland, College Park, Maryland (Yan Li); GlaxoSmithKline (GSK) Vaccines, Rixensart, Belgium (Sylviane Poncelet); Information Management Services, Inc., Calverton, Maryland (John Schussler); DDL Diagnostic Laboratory, Rijswijk, the Netherlands (Wim Quint, Leen-Jan van Doorn); and Georgetown University, Washington, DC (Mary Sidawy).
This trial is sponsored and funded by the NCI (contract N01-CP-11005), with funding support from the NIH Office of Research on Women's Health. GSK Biologicals SA provided vaccine and support for aspects of the trial associated with regulatory submission needs of the company under a US Food and Drug Administration (FDA) clinical trials agreement (FDA BB-IND 7920) during the 4-year, randomized, blinded phase of our study. Vaccine was provided for our trial by GSK Vaccines under a clinical trials agreement with the NCI. GSK Vaccines also provided support for aspects of the trial associated with regulatory submission needs of the company. The authors received no financial support or other form of compensation related to the development of the manuscript. The trial is registered with clinicaltrials.gov (NCT00128661). The protocol is available at http://proyectoguanacaste.org.
We thank the staff in Costa Rica for their tremendous effort and dedication to this project; the US team from information management services for the development and maintenance of the data system used in the trial; Jean Cyr, Julie Buckland, and Brian Befano for their invaluable contributions; the members of the data and safety monitoring board for protecting the safety and interest of participants in our trial (Drs. Steve Self, Adriana Benavides, Luis Diego Calzada, Ruth Karron, and Ritu Nayar, as well as Nancy Roach); and members of the external scientific HPV working group who have contributed to the success of our efforts over the years (Drs. Joanna Cain, Diane Davey, David DeMets, Francisco Fuster, Ann Gershon, Elizabeth Holly, Henriette Raventós, Wasima Rida, Luis Rosero-Bixby, and Kristen Suthers, as well as Silvia Lara and Sarah Thomas).
Investigators in the Costa Rica Vaccine Trial group include the following: Mario Alfaro, Manuel Barrantes, M. Concepción Bratti, Fernando Cárdenas, Bernal Cortés, Albert Espinoza, Yenory Estrada, Paula González, Diego Guillén, Roland Herrero, Silvia E. Jiménez, Jorge Morales, Luis Villegas, Lidia Ana Morera, Elmer Pérez, Carolina Porras, Ana Cecilia Rodríguez, Libia Rivas (Proyecto Epidemiológico Guanacaste, Fundación INCIENSA, San José, Costa Rica); Enrique Freer, José Bonilla, Alfanso García-Piñeres, Sandra Silva, Ivannia Atmella, Margarita Ramírez (University of Costa Rica, San José, Costa Rica); Allan Hildesheim, Aimée R. Kreimer, Douglas R. Lowy, Nora Macklin, Mark Schiffman, John T. Schiller, Mark Sherman, Diane Solomon, Sholom Wacholder (NCI, Bethesda, Maryland ); Ligia Pinto, Troy Kemp (SAIC, NCI-Frederick, Frederick, Maryland); Claire Eklund, Martha Hutchinson (Women's and Infants’ Hospital, Providence, Rhode Island); Mary Sidawy (Georgetown University, Washington, DC; and Wim Quint and Leen-Jan van Doorn (DDL Diagnostic Laboratory, Rijswijk, the Netherlands).
Conflict of interest: S.P. is an employee of the GSK group of companies. As part of US government–supported research at the NCI/NIH, D.R.L. is an inventor of technology that underlies the L1-based prophylactic viruslike particle (VLP) human papillomavirus (HPV) vaccine and technology that underlies an L2-based candidate prophylactic HPV vaccine. The NIH has licensed the technology for the L1 VLP vaccine to Merck & Co., the manufacturer of Gardasil; to GSK, Inc., the manufacturer of Cervarix; and to Indian Immunologicals Ltd. The L2-based vaccine technology is the subject of a cooperative research and development agreement between the NCI, Johns Hopkins University, and Shantha Biotechnics and has been licensed to Shantha Biotechnics, PanVax, Ltd., Acambis, Inc., and GSK, Inc. US Federal law entitles D.R.L. to a limited share of the royalties the NIH receives for these technologies. J.T.S. is an inventor on US government–owned patents of the HPV VLP vaccine technology. The NCI and Costa Rica investigators are responsible for the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation of the manuscript. GSK Biologicals was provided the opportunity to review a preliminary version of this manuscript for factual accuracy, but the authors are solely responsible for the final content and interpretation.
REFERENCES
- 1.Schiller JT, Castellsagué X, Garland SM. A review of clinical trials of human papillomavirus prophylactic vaccines. Vaccine. 2012;30(suppl 5):F123–F138. doi: 10.1016/j.vaccine.2012.04.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildesheim A, Herrero R, Wacholder S, et al. Effect of human papillomavirus 16/18 L1 viruslike particle vaccine among young women with preexisting infection: a randomized trial. JAMA. 2007;298(7):743–753. doi: 10.1001/jama.298.7.743. [DOI] [PubMed] [Google Scholar]
- 3.Muñoz N, Kjaer SK, Sigurdsson K, et al. Impact of human papillomavirus (HPV)-6/11/16/18 vaccine on all HPV-associated genital diseases in young women. JNatl Cancer Inst. 2010;102(5):325–339. doi: 10.1093/jnci/djp534. [DOI] [PubMed] [Google Scholar]
- 4.Lehtinen M, Paavonen J, Wheeler CM, et al. Overall efficacy of HPV-16/18 AS04-adjuvanted vaccine against grade 3 or greater cervical intraepithelial neoplasia: 4-year end-of-study analysis of the randomised, double-blind PATRICIA Trial. Lancet Oncol. 2012;13(1):89–99. doi: 10.1016/S1470-2045(11)70286-8. [DOI] [PubMed] [Google Scholar]
- 5.Safaeian M, Ghosh A, Porras C, et al. Direct comparison of HPV16 serological assays used to define HPV-naive women in HPV vaccine trials. Cancer Epidemiol Biomarkers Prev. 2012;21(9):1547–1554. doi: 10.1158/1055-9965.EPI-12-0558. [DOI] [PubMed] [Google Scholar]
- 6.Herrero R, Hildesheim A, Rodríguez AC, et al. Rationale and design of a community-based double-blind randomized clinical trial of an HPV 16 and 18 vaccine in Guanacaste, Costa Rica. Vaccine. 2008;26(37):4795–4808. doi: 10.1016/j.vaccine.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrero R, Wacholder S, Rodríguez AC, et al. Prevention of persistent human papillomavirus infection by an HPV16/18 vaccine: a community-based randomized clinical trial in Guanacaste, Costa Rica. Cancer Discov. 2011;1(5):408–419. doi: 10.1158/2159-8290.CD-11-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleter B, van Doorn LJ, ter Schegget J, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153(6):1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleter B, van Doorn LJ, Schrauwen L, et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol. 1999;37(8):2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dessy FJ, Giannini SL, Bougelet CA, et al. Correlation between direct ELISA, single epitope-based inhibition ELISA and pseudovirion-based neutralization assay for measuring anti-HPV-16 and anti-HPV-18 antibody response after vaccination with the AS04-adjuvanted HPV-16/18 cervical cancer vaccine. Hum Vaccin. 2008;4(6):425–434. doi: 10.4161/hv.4.6.6912. [DOI] [PubMed] [Google Scholar]
- 11.Harper DM, Franco EL, Wheeler C, et al. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet. 2004;364(9447):1757–1765. doi: 10.1016/S0140-6736(04)17398-4. [DOI] [PubMed] [Google Scholar]
- 12.Malagón T, Drolet M, Boily MC, et al. Cross-protective efficacy of two human papillomavirus vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(10):781–789. doi: 10.1016/S1473-3099(12)70187-1. [DOI] [PubMed] [Google Scholar]
- 13.Einstein MH, Baron M, Levin MJ, et al. Comparative immunogenicity and safety of human papillomavirus (HPV)-16/18 vaccine and HPV-6/11/16/18 vaccine: follow-up from months 12–24 in a phase III randomized study of healthy women aged 18–45 years. Hum Vaccin. 2011;7(12):1343–1358. doi: 10.4161/hv.7.12.18281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Draper E, Bissett SL, Howell-Jones R, et al. A randomized, observer-blinded immunogenicity trial of Cervarix® and Gardasil® human papillomavirus vaccines in 12–15 year old girls. PLoS One. 2013;8(5):e61825. doi: 10.1371/journal.pone.0061825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garland SM, Hernandez-Avila M, Wheeler CM, et al. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N Engl J Med. 2007;356(19):1928–1943. doi: 10.1056/NEJMoa061760. [DOI] [PubMed] [Google Scholar]
- 16.FUTURE II Study Group. Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N Engl J Med. 2007;356(19):1915–1927. doi: 10.1056/NEJMoa061741. [DOI] [PubMed] [Google Scholar]
- 17.Paavonen J, Jenkins D, Bosch FX, et al. Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: an interim analysis of a phase III double-blind, randomised controlled trial. Lancet. 2007;369(9580):2161–2170. doi: 10.1016/S0140-6736(07)60946-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.