Abstract
HrcA is a transcriptional repressor that regulates stress response genes in many bacteria by binding to the CIRCE operator. We have previously shown that HrcA regulates the promoter for the dnaK heat shock operon in Chlamydia. Here we demonstrate that HrcA represses a second heat shock promoter that controls the expression of groES and groEL, two other major chlamydial heat shock genes. The CIRCE element of C. trachomatis groEL is the most divergent of known bacterial CIRCE elements, and HrcA had a decreased ability to bind to this nonconsensus operator and repress transcription. We demonstrate that the CIRCE element is necessary and sufficient for transcriptional regulation by chlamydial HrcA and that the inverted repeats of CIRCE are the binding sites for HrcA. Addition of a CIRCE element upstream of a non-heat-shock promoter allowed this promoter to be repressed by HrcA, showing in principle that a chlamydial promoter can be genetically modified to be inducible. These results demonstrate that HrcA is the regulator of the major chlamydial heat shock operons, and we infer that the mechanism of the heat shock response in Chlamydia is derepression. However, derepression is likely to involve more than a direct effect of increased temperature as we found that HrcA binding to CIRCE and HrcA-mediated repression were not altered at temperatures that induce the heat shock response.
Chlamydia trachomatis is the cause of the most common reportable infectious disease in the United States (6), as well as a common cause of preventable blindness in the developing world (41). Recent evidence has shown that there is an association between a related species, Chlamydia pneumoniae, and the development of coronary artery disease (11). The hallmark of chlamydial infections is a chronic inflammatory process that leads to tissue damage and scarring. There is a wide body of evidence that implicates the chlamydial heat shock proteins in elicitation of this host inflammatory response. These heat shock proteins are molecular chaperones and proteases that function in protein folding and degradation in response to cellular stress (23), and they are well conserved in chlamydial species (12, 18). The major chlamydial heat shock proteins that have been studied are GroEL (Hsp60), GroES (Hsp10), and DnaK (Hsp70) (34).
It has long been noted that chlamydial infection correlates with antibodies to the chlamydial heat shock proteins (reviewed in reference 22). Serum antibodies to chlamydial GroEL have been associated with trachoma (32) and also with pelvic inflammatory disease, ectopic pregnancy, and infertility in women (1, 5, 33, 54). Antibodies to GroES have been shown to correlate with the severity of genital infection (21). The presence of C. pneumoniae GroEL in atherosclerotic plaques (20) and the correlation of antibodies to GroEL with detection of C. pneumoniae in these plaques (9) support the hypothesis that this species plays a role in atherosclerotic heart disease. Chlamydial GroEL has also been shown to cause a hypersensitivity reaction of the eye (28) and the fallopian tubes (31).
More recently, it has been appreciated that chlamydial heat shock proteins may also induce host cells to produce factors that contribute to chlamydial pathogenesis (reviewed in reference 46). Chlamydial GroEL can interact with cells via Toll-like receptors to induce cellular signaling (40, 53). GroEL activates endothelial cells, smooth muscle cells, and macrophages to produce proinflammatory cytokines and adhesion molecules (19). GroEL also induces expression of matrix metalloproteinase and tumor necrosis factor alpha (20) and promotes cellular oxidization of low-density lipoprotein to make it more atherogenic (16). The other major heat shock protein, DnaK, has been proposed to be a factor for binding of chlamydiae to host cells (44).
Because of the likely role of heat shock proteins in chlamydial pathogenesis, we are interested in the mechanisms that regulate expression of the heat shock genes. Increasing the temperature from 37 to 42°C has been shown to produce a heat shock response in chlamydia-infected cells along with increased transcription of chlamydial dnaK and groE (8). Unlike the mechanism in Escherichia coli, the mechanism does not seem to involve an alternative sigma factor for regulating the transcription of heat shock genes, as the three pathogenic chlamydial species do not contain the heat shock alternative sigma factor σ32 (17, 36, 37, 47). Instead, Chlamydia appears to use a mechanism described for other bacteria (29), in which heat shock genes are transcribed by the major form of RNA polymerase (51) and are regulated by a transcriptional repressor, HrcA, and an operator designated CIRCE. We have shown that chlamydial HrcA can bind to a CIRCE element located upstream of the dnaK promoter and that it can repress transcription of this promoter (56), although we have not directly shown that repression depends on the presence of the CIRCE element. We are also interested in studying a CIRCE-like inverted repeat upstream of the groE promoter (51), whose sequence is significantly different from the well-characterized CIRCE consensus sequence (45), to see if it can function as an operator for HrcA binding and repression.
In this report, we describe our analysis of HrcA-CIRCE interactions in Chlamydia and demonstrate that CIRCE is both necessary and sufficient for HrcA-mediated transcriptional repression in vitro. We present data which show that chlamydial HrcA binds the inverted repeats of the CIRCE element and does not require Chlamydia-specific interactions between HrcA and RNA polymerase for repression. We found that the nonconsensus groE CIRCE showed reduced HrcA binding and HrcA-mediated repression, and below we discuss the implications of these findings for the expression of groEL and groES under non-heat-shock conditions. We also discuss how the insensitivity of HrcA binding and HrcA-mediated repression to higher temperatures suggests that a direct effect of temperature alone is not likely to be the mechanism of derepression in response to heat shock.
MATERIALS AND METHODS
Reagents.
Products were obtained from the following sources and were used according to the manufacturers' specifications: restriction enzymes, calf intestinal alkaline phosphatase, DNase I, and rRNasin, Promega Biotech (Madison, Wis.); T4 polynucleotide kinase, T4 DNA ligase, and pRSET expression vector DNA, Invitrogen (Carlsbad, Calif.); Sequenase kit, U.S. Biochemicals (Cleveland, Ohio); nucleoside triphosphates, 3′-O-methylguanosine 5′-triphosphate, poly(dI-dC), HiTrap chelating column, and HiPrep gel filtration column, Amersham (Arlington Heights, Ill.); Centriplus columns, Millipore (Bedford, Mass.); oligonucleotide primers, Sigma Genosys (The Woodlands, Tex.); 32P-labeled nucleotide triphosphates, ICN (Costa Mesa, Calif.); Pwo DNA polymerase and mini Quick Spin DNA columns, Roche Diagnostics (Indianapolis, Ind.); Bio-Rad protein assay reagent, Bio-Rad (Hercules, Calif.); Klenow fragment of E. coli DNA polymerase I, New England Biolabs (Beverly, Mass.); and E. coli RNA polymerase holoenzyme, Epicentre (Madison, Wis.).
Overexpression and purification of HrcA protein.
His6-HrcA was overexpressed in E. coli BL21(DE3) and purified as previously described (56).
EMSA templates.
CIRCE-containing 110-bp restriction fragments were used as probes for the gel retardation assays. The groE promoter region was amplified by PCR by using C. trachomatis serovar D genomic DNA and primers T215 (5′-ACGTCTAGACTGGAAGAACTAGCTAGCTTGCTATAAA) and T216 (5′-ACGGAATTCAGAACGATAGCAAAGTCCTCGC). A fragment containing one-half of the groE CIRCE element was produced by amplifying a portion of the groE promoter region with primers T157 (5′-GGGAATTCGACTTTGCTATCGTTCTTCCTC) and T158 (5′-GGGGTACCTATTTTTATATTCTATGAGGCCTCGT). PCR products were cloned into the SmaI site of pGEM-7Zf(+) to generate plasmids pMT1175 (groE with CIRCE) and pMT1154 (groE with one-half of CIRCE). The dnaK CIRCE plasmid pMT1134 was constructed as described previously (56). Electrophoretic mobility shift assay (EMSA) restriction fragment templates were digested with XbaI and BamHI, and this was followed by gel purification from a 2% agarose gel.
EMSA.
The probe DNA was labeled with [α-32P]dATP by using the Klenow fragment of E. coli DNA polymerase. Free nucleotides were removed by using a mini Quick Spin DNA column, and probe activity was determined by liquid scintillation counting. The EMSA reaction mixture contained 1× binding buffer (40 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 70 mM KCl, 135 μM EDTA, 100 μM dithiothreitol, 7.5% glycerol), labeled probe at a concentration of 0.3 nM, and various amounts of purified His6-HrcA. Standard reaction mixtures were incubated for 20 min at room temperature. In thermostability studies, the reaction mixtures were incubated for 15 min at room temperature and then shifted to the temperature being examined for 5 min. Samples were immediately loaded onto a 7% polyacrylamide EMSA gel and electrophoresed at 150 V in 0.5× Tris-borate-EDTA (TBE) buffer (35). After electrophoresis, the gels were dried and exposed to phosphorimager plates. The plates were scanned with a Bio-Rad Personal FX scanner, and the data were analyzed with Bio-Rad Quantity One software (Bio-Rad).
Purification of C. trachomatis RNA polymerase.
RNA polymerase was partially purified from C. trachomatis LGV serovar L2 at 20 h postinfection by heparin-agarose chromatography as previously described (49).
Construction of transcription plasmids.
Plasmid pMT1178 contained the promoter region of groE (positions −137 to 5) amplified by PCR from C. trachomatis serovar D genomic DNA by using primers T274 (5′-ACCGAATTCATGCTTAAAGCCTTTTCCTACATCG) and T276 (5′-TCAGAGGAAGAACGATAGCAAAG). Plasmid pMT1177 contained the dnaK promoter (positions −40 to 5) fused to the upstream region of the hctA promoter (positions −65 to −41) amplified from plasmid pMT579 (51) by using primers dnaK2 (5′-AAGTTGGTGTCATTATAGGAAAACC) and T256 (5′-CGTGAATTCTTTAGGATCCTTACCTAGATTCTAGAAAAATTCTTGACCAGAGGCTC). Plasmid pMT1176 contained the hctA promoter (positions −40 to 5) fused to the dnaK CIRCE region (positions −65 to −41) amplified from plasmid pMT1010 (43) by using primers T254 (5′-CGTGAATTCTAGCACTCTTTGCTCGCGAGCGCTAAAAATGGTTGCATGAATT) and T253 (5′-TTTTAATTTTTAATTAGTTTGTTTGTTC). Plasmid pMT1224 contained the dnaK promoter (positions −40 to 5) fused to the groE CIRCE region (positions −65 to −41) amplified from plasmid pMT1161 (56) by using primers T447 (5′-CATGAATTCAATAGCAGTTGATCATGCCAACTGCTAAAATTCTTGACCAGAGGCTCCG) and dnaK2 (5′-AAGTTGGTGTCATTATAGGAAAACC). Each promoter insert was cloned upstream of a promoterless G-less cassette transcription template in plasmid pMT1125 (56). Plasmid pMT1194 was a PacI drop-out of pMT1176 that upon transcription produced a transcript which was 28 nucleotides shorter than the parent plasmid. Construction of plasmids pMT1161 (dnaK) and pMT1158 (omcB) has been described previously (56).
In vitro transcription.
The in vitro transcription reaction mixture contained 50 mM potassium acetate, 8.1 mM magnesium acetate, 50 mM Tris acetate (pH 8.0), 27 mM ammonium acetate, 2 mM dithiothreitol, 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.21 μM [α-32P]CTP (800 Ci/mmol), 100 μM 3′-O-methylguanosine 5′-triphosphate (Na salt), 18 U of rRNasin, 5% glycerol, 15 nM supercoiled DNA template, and various amounts of purified His6-HrcA. The reaction mixture was incubated for 20 min at room temperature. Then 0.5 μl of heparin-agarose-purified C. trachomatis RNA polymerase or 0.5 μl of a 1:10 dilution of E. coli RNA polymerase holoenzyme was added, and the reaction mixture was incubated for an additional 15 min at 37 or 42°C as indicated below. The final reaction volume was 10 μl. The reaction was terminated by addition of 10 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). A 10-μl portion of the sample was electrophoresed on an 8 M urea-6% polyacrylamide gel in 1× TBE buffer. After electrophoresis, the gels were dried and exposed to phosphorimager plates. The plates were scanned with a Bio-Rad Personal FX scanner, and the data were analyzed with Bio-Rad Quantity One software.
DNase I footprinting.
A segment of the dnaK promoter region from position −130 to position 60 relative to the transcription start site was amplified by PCR from C. trachomatis serovar D genomic DNA by using primers T307 (5′-TCGGAATTCTTGGATTGGTGCTCTAAAAATCT) and T308 (5′-TCGATCGATGTAGAGCTTAGTGGCCATAAGTAGAACA). The PCR products were cloned into the SmaI site of pGEM-7Zf(+) to generate plasmid pMT1208. pMT1208 was digested with ApaI and BamHI to generate the probe. The probe DNA was purified from a 1% agarose gel and labeled with [α-32P]dATP by using the Klenow fragment of E. coli DNA polymerase. Free nucleotides were removed by using a mini Quick Spin DNA column, and probe activity was determined by liquid scintillation counting. G and G-A ladders were prepared from end-labeled probes by Maxam-Gilbert sequencing as previously described (25). Labeled probe (1 nM) was incubated with various concentrations of purified His6-HrcA in binding buffer (20 mM HEPES [pH 8.0], 5 mM MgCl, 50 mM potassium glutamate, 1 mM dithiothreitol, 1 ng of sonicated salmon sperm DNA per μl) for 10 min at 37°C. Three microliters of diluted DNase I was then added to each reaction mixture and incubated for an additional 2 min at 37°C. The reactions were then stopped with 1× stop solution (300 mM sodium acetate [pH 7.0], 10 mM EDTA), extracted with phenol-chloroform, and ethanol precipitated. The pellets were resuspended in DNase I gel loading buffer (40% deionized formamide, 5 M urea, 5 mM NaOH, 1 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). A 2-μl sample was electrophoresed on an 8 M urea-8% polyacrylamide gel in 1× TBE buffer. After electrophoresis, the gels were dried and exposed to film.
RESULTS
CIRCE is necessary and sufficient for HrcA-mediated transcriptional repression in vitro.
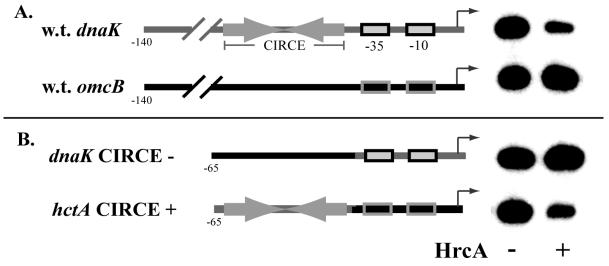
We have shown previously that C. trachomatis dnaK promoter activity is repressed by HrcA (56), and we have inferred that the mechanism of repression depends on binding of HrcA to CIRCE. This reasoning is based on the observation that the dnaK promoter contains a CIRCE element and on our finding that HrcA specifically binds CIRCE (56). However, the direct role of CIRCE in HrcA-mediated in vitro transcriptional repression has not been addressed previously. To examine the role of CIRCE, we performed in vitro transcription reactions with the dnaK promoter, which contains a CIRCE element, and the hctA and omcB promoters, which lack CIRCE. We also created and tested two hybrid promoters, in which we swapped the upstream regions between the dnaK and hctA promoters, so that we could directly determine if CIRCE was sufficient for regulation by HrcA. Figure 1A shows that HrcA repressed transcription of the dnaK promoter almost fivefold but had no effect on the omcB promoter, as we have seen previously (56). HrcA had no effect on transcription of the wild-type hctA promoter (data not shown). Figure 1B shows that a dnaK promoter with the hctA upstream sequence from position −65 to position −42 (thus lacking CIRCE) was not repressed by HrcA, showing that CIRCE is necessary for HrcA-mediated repression. The definitive experiment was performed by using an hctA promoter with the upstream CIRCE sequences of the dnaK promoter, from position to −65 to position −42. This hybrid promoter was repressed nearly fivefold by HrcA, showing that the CIRCE element is sufficient for transcriptional repression by HrcA.
FIG. 1.
In vitro transcription to examine the effects of the CIRCE element and recombinant HrcA: structure of a pair of promoter constructs and results of an in vitro transcription reaction performed with C. trachomatis RNA polymerase in the absence (minus sign) or presence (plus sign) of 1,150 nM HrcA. (A) The wild-type dnaK promoter (upper line), containing a CIRCE element, was transcribed with the omcB promoter (lower line), and these promoters produced 153- and 125-nucleotide transcripts, respectively. w.t., wild type. (B) The dnaK promoter containing the hctA upstream region (upper line) and thus lacking the CIRCE element was transcribed with the hctA promoter containing the dnaK upstream region and its CIRCE element (lower line).
HrcA-mediated transcriptional repression does not require Chlamydia-specific interactions between HrcA and RNA polymerase.
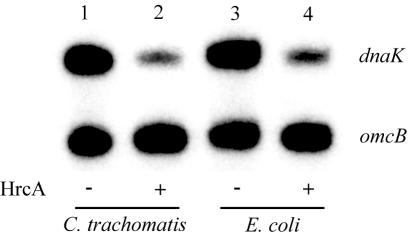
Although HrcA-CIRCE interactions have been studied in many bacterial species (for a review, see reference 29), the precise mechanism by which HrcA represses transcription has not been determined. It has been proposed that HrcA represses transcription by sterically hindering access of RNA polymerase to the core promoter elements (29, 57). This model predicts that specific protein-protein interactions between HrcA and RNA polymerase are not necessary and that HrcA from one bacterium should be able to repress transcription by a heterologous RNA polymerase. To test this hypothesis, we performed an experiment to determine if C. trachomatis HrcA could repress transcription of a CIRCE-containing promoter by E. coli RNA polymerase. The advantages of using E. coli RNA polymerase are that it can transcribe the C. trachomatis dnaK and omcB promoters (43) and that E. coli does not contain an HrcA homolog (4). Transcription of the dnaK promoter by purified E. coli RNA polymerase was decreased fivefold by chlamydial HrcA (Fig. 2, lanes 3 and 4), which was similar to the level of repression obtained with C. trachomatis RNA polymerase (Fig. 2, lanes 1 and 2). These results show that HrcA can repress transcription of E. coli RNA polymerase with an efficiency equivalent to the efficiency of repression seen with C. trachomatis RNA polymerase.
FIG. 2.
In vitro transcription with C. trachomatis and E. coli RNA polymerases. The upper band is the transcript from the dnaK promoter, and the lower band is the transcript from the omcB promoter. Lane 1, C. trachomatis RNA polymerase in the absence of HrcA; lane 2, C. trachomatis RNA polymerase and 1,150 nM HrcA; lane 3, E. coli RNA polymerase in the absence of HrcA; lane 4, E. coli RNA polymerase and 1,150 nM HrcA.
HrcA protects CIRCE from DNase I digestion.
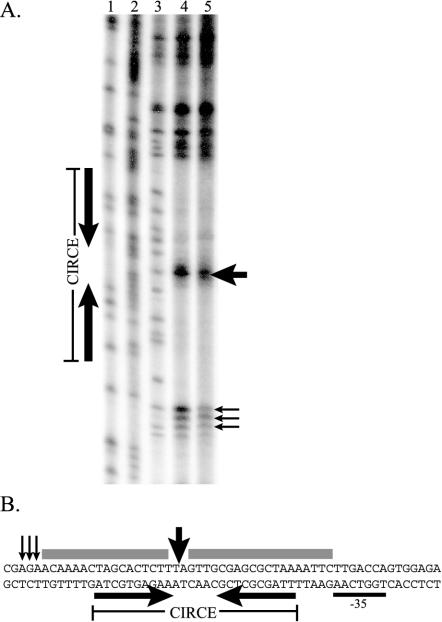
To determine the binding specificity of HrcA and sites of protein-DNA contact, we performed a DNase I footprint analysis of the C. trachomatis dnaK promoter region. As shown in Fig. 3A, addition of HrcA protected the CIRCE inverted repeat from DNase I digestion. The areas of protection covered the entire CIRCE inverted repeat, and there was additional 5- or 6-bp protection upstream of the 5′ inverted repeat. There was an unprotected region between the CIRCE inverted repeats, indicating that HrcA does not bind the spacer region. HrcA also produced DNase-hypersensitive sites in the spacer region and at the ends of both protected regions. Figure 3B shows these protected and hypersensitive sites in the sequence of the dnaK promoter region.
FIG. 3.
DNase I footprint of recombinant HrcA on the dnaK CIRCE region. (A) Each lane contained approximately 10,000 cpm of labeled dnaK probe. Lane 1, G sequencing ladder; lane 2, G-A sequencing ladder; lane 3, no HrcA; lane 4, 144 nM HrcA; lane 5, 288 nM HrcA. (B) Sequence of dnaK CIRCE region. Protected regions are underlined, and DNase-hypersensitive sites are indicated by vertical arrows.
HrcA binds to the C. trachomatis groE promoter in vitro.
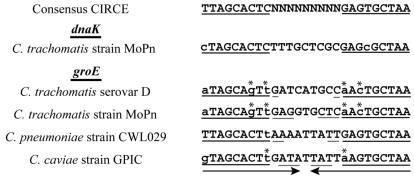
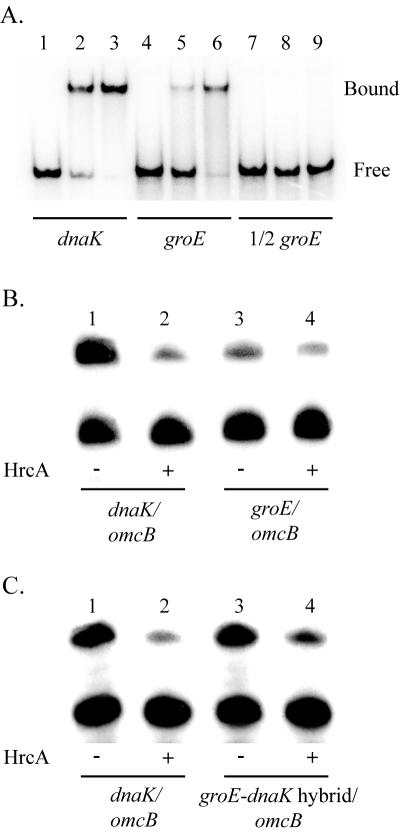
A CIRCE-like inverted repeat upstream of the C. trachomatis groE promoter has been found previously (51). As shown in Fig. 4, the serovar D groE CIRCE-like inverted repeat matches the consensus CIRCE sequence at only 13 of 18 nucleotides, which makes it the least conserved sequence among predicted bacterial CIRCE sequences (13, 45). In fact, the C. trachomatis groE promoter has been reported to lack a CIRCE element (45), probably because of this sequence divergence and unusual upstream location. To examine whether the CIRCE-like sequence in the groE promoter region is functional and can be bound by HrcA, we performed an EMSA analysis using a DNA probe containing the groE inverted repeat element. HrcA bound to the groE CIRCE fragment (Fig. 5A, lanes 4 to 6), but with a lower affinity than it bound to the dnaK CIRCE (lanes 2 and 5). Using additional EMSA experiments, we calculated an apparent KD for groE of 235 nM and an apparent KD for dnaK of 88 nM (data not shown). At high HrcA concentrations, both the groE and dnaK DNA probes were almost exclusively found in the HrcA-bound complex (Fig. 5A, lanes 3 and 6). HrcA did not bind to a DNA probe containing one-half of the groE inverted repeat (lanes 7 to 9), indicating that the complete inverted repeat is required for binding.
FIG. 4.
Alignment of the consensus bacterial CIRCE sequence (29) and the sequences of CIRCE elements from C. trachomatis strain MoPn (51), C. trachomatis serovar D (47), C. pneumoniae strain CWL029 (17), and C. caviae strain GPIC (37). The heavy underlining indicates the CIRCE region; the lowercase letters indicate mismatches with the consensus CIRCE sequence; the light underlining indicates a region of extended complementarity; the arrows indicate the maximum extent of extended complementarity; and the asterisks indicate mismatches with the consensus sequence which are complemented in the inverted repeat.
FIG. 5.
Binding and transcription of the groE promoter by using recombinant HrcA. (A) EMSA with dnaK and groE promoter restriction fragments. The positions of bound and free probe are indicated on the right. Lanes 1 to 3, dnaK probe with no HrcA (lane 1), 100 nM HrcA (lane 2), or 400 nM HrcA (lane 3); lanes 4 to 6, groE probe with no HrcA (lane 4), 100 nM HrcA (lane 5), or 400 nM HrcA (lane 6); lanes 7 to 9, groE probe containing only one arm of the CIRCE inverted repeat with no HrcA (lane 7), 100 nM HrcA (lane 8), or 400 nM HrcA (lane 9). (B) In vitro transcription of wild-type groE promoter. Lanes 1 and 2, transcription of the dnaK promoter (upper band) and the lower omcB promoter (lower band) by using C. trachomatis RNA polymerase with no HrcA (lane 1) or 1,150 nM HrcA (lane 2); lanes 3 and 4, transcription of the groE promoter (upper band) and the omcB promoter (lower band) with no HrcA (lane 3) or 1,150 nM HrcA (lane 4). (C) In vitro transcription of hybrid groE CIRCE-dnaK core promoter. Lanes 1 and 2, transcription of the dnaK promoter (upper band) and the omcB promoter (lower band) by using C. trachomatis RNA polymerase with no HrcA (lane 1) or 1,150 nM HrcA (lane 2); lanes 3 and 4, transcription of the groE/dnaK hybrid promoter (upper band) and the omcB promoter (lower band) with no HrcA (lane 3) or 1,150 nM HrcA (lane 4).
HrcA represses in vitro transcription from the groE promoter.
Having shown that HrcA binds the groE CIRCE, we next asked whether transcription of the groE promoter can be repressed by HrcA. We performed in vitro transcription reactions with dnaK and groE transcription constructs in the presence or absence of HrcA. Addition of HrcA to a dnaK transcription reaction resulted in an almost fivefold reduction in transcription (Fig. 5B, lanes 1 and 2), which is consistent with our previous findings (56). Transcription of the groE promoter in the absence of HrcA was considerably lower than transcription of either the dnaK or omcB promoter (lane 3). Addition of HrcA resulted in a reproducible twofold reduction in transcription from the groE promoter (lane 4). While addition of HrcA resulted in less transcriptional repression of the groE promoter than of the dnaK promoter, the decreased repression was approximately equal to the lower binding of HrcA for the groE CIRCE.
To determine if the decreased repression of the groE promoter was due to the groE CIRCE alone rather than to an effect of the core promoter, we compared HrcA-mediated repression through the groE and dnaK CIRCE elements in the context of the same promoter. We constructed a transcription template with the groE CIRCE placed upstream of the dnaK promoter and compared it to the wild-type dnaK promoter and its native CIRCE element. Basal transcription without HrcA was similar for the hybrid groE/dnaK construct and the wild-type dnaK construct (Fig. 5C, lanes 1 and 3). With the addition of HrcA, however, transcription from the hybrid promoter was repressed twofold (lane 2), compared to the fivefold repression of the wild type dnaK promoter (lane 5). The decreased repression with the groE CIRCE is consistent with the decreased HrcA binding to this nonconsensus CIRCE element by EMSA. We concluded that the level of transcriptional repression by HrcA is dependent on the binding affinity of HrcA for the CIRCE element.
HrcA activity is not affected by heat shock temperatures.
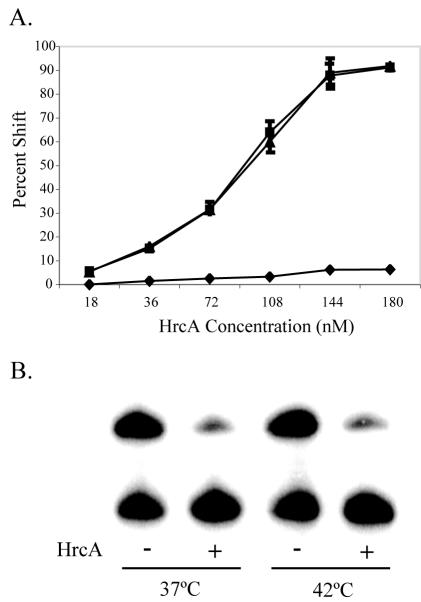
While we have focused so far on HrcA-mediated transcriptional repression, it is important from the perspective of chlamydial biology to also consider how heat shock genes are derepressed during the stress response. An obvious mechanism for derepression in response to heat shock would be that increased temperature has a direct effect on the ability of HrcA to bind CIRCE. To measure the effect of temperature on HrcA binding, we performed a modified EMSA protocol in which HrcA and a dnaK CIRCE fragment were first incubated at room temperature for 15 min so that binding could occur; this was followed by an additional 5 min of incubation at an elevated temperature. We compared the effects of temperature upshifts to 37°C, which represents a normal growth temperature inside human cells, to 42°C, which has been shown to induce the heat shock response with increased transcription of the heat shock genes (8), and to 56°C, a superphysiologic temperature that is predicted to disrupt the HrcA-CIRCE complex. The data from three independent EMSA experiments with a range of concentrations of recombinant HrcA were quantified by phosphorimager analysis and are shown in Fig. 6A. Similar levels of binding of HrcA to CIRCE were obtained with temperature upshifts to 37 and 42°C, as shown by the nearly identical binding curves. In contrast, the upshift to 56°C resulted in very little residual binding of HrcA to CIRCE, indicating that disruption of the HrcA-CIRCE complex had occurred and could be measured with this assay. Based on these in vitro observations, the HrcA-CIRCE complex is thermostable at an increased temperature that is known to induce the heat shock response in C. trachomatis.
FIG. 6.
Binding and transcription with recombinant HrcA at an elevated temperature. (A) EMSA reactions with 0.5 nM 32P-labeled dnaK CIRCE probe and various concentrations of recombinant HrcA were performed in triplicate, and the results were quantified by phosphorimager analysis. The reaction mixtures were preincubated at room temperature for 15 min and then shifted to 37°C (▪), 42°C (▴), or 56°C (♦) for 5 min. The error bars indicate standard deviations from the mean. (B) In vitro transcription with E. coli RNA polymerase. The upper band was from the dnaK promoter, and the lower band was from the omcB promoter. The lanes show (from left to right) transcription at 37°C with no HrcA, transcription at 37°C with 1,150 nM HrcA, transcription at 42°C with no HrcA, and transcription at 42°C with 1,150 nM HrcA.
We also tested the effect of an elevated temperature on transcriptional repression by HrcA. We were not able to perform these experiments with our partially purified C. trachomatis RNA polymerase as there was a significant loss of transcriptional activity at 42°C (data not shown). Instead, we performed these experiments with E. coli RNA polymerase, taking advantage of our ability to measure repression by chlamydial HrcA with this heterologous RNA polymerase, as shown above (Fig. 2). Purified E. coli RNA polymerase is relatively thermostable, as shown by the similar levels of basal transcription at 37 and 42°C for the dnaK and omcB promoters (Fig. 6B, lanes 1 and 3). When we added HrcA, we observed the same fivefold repression of the dnaK promoter at 37 and 42°C (lanes 2 and 4). These DNA-binding and transcription studies demonstrated that an elevated temperature that can induce the heat shock response is not sufficient to alter the ability of chlamydial HrcA to bind CIRCE and repress transcription.
DISCUSSION
In this study, we demonstrated that the CIRCE element is necessary and sufficient for repression of in vitro transcription by HrcA in Chlamydia. Since the chlamydial CIRCE element does not overlap the core promoter sequence (42, 50), we were able to take a defined CIRCE sequence and place it upstream of the hctA promoter (52), a developmentally regulated promoter that is not normally repressed by HrcA. Addition of the CIRCE element allowed this promoter to be repressed by HrcA to the same degree as the wild-type dnaK promoter, suggesting that CIRCE functions independent of the promoter context. To our knowledge, this is the first report of an independent, movable sequence with the demonstrated capacity to regulate a heterologous promoter in Chlamydia. As such, it may be useful for developing an inducible Chlamydia expression system in conjunction with a system for genetic transformation (48) or phage-based genetics (15).
We also demonstrated that HrcA can repress transcription by E. coli RNA polymerase and transcription by C. trachomatis RNA polymerase to the same extent. This finding indicates that HrcA-mediated transcriptional repression does not require Chlamydia-specific interactions between HrcA and RNA polymerase or additional Chlamydia-specific factors and supports a model in which HrcA functions as a repressor by steric hindrance of RNA polymerase binding (39). This result is not completely unexpected since it has been shown that Bacillus subtilis HrcA expressed in E. coli is able to repress transcription of a CIRCE-regulated promoter (26). However, our in vitro system allows us to more precisely define the components necessary for HrcA-mediated transcriptional repression.
DNase footprint analysis revealed that the binding site for chlamydial HrcA symmetrically overlaps the CIRCE inverted repeat. This footprint is similar to the footprint for Staphylococcus aureus obtained by using renatured, wild-type HrcA, with protection of CIRCE extending just beyond the inverted repeat on each side and with an unprotected region within the 9-bp spacer region (7). The other previously described DNase footprint of HrcA, from B. subtilis, showed that there was a larger area of protection, extending well beyond CIRCE, without an unprotected region in the spacer between inverted repeats (38). While the difference in the findings may be attributed to species-specific variation, it is interesting that the B. subtilis footprint was obtained with a mutant HrcA, while the S. aureus and C. trachomatis footprints were obtained by using wild-type HrcA. Only our C. trachomatis footprint was obtained by using wild-type, soluble HrcA protein that had not been denatured during purification.
The finding that the groE promoter is regulated by HrcA supports a model for feedback regulation of the groESL operon through interactions between GroE and HrcA. Several lines of evidence indicate that the GroE chaperonin machinery increases HrcA-mediated repression of heat shock promoters in vitro and in vivo in other bacteria (26, 27, 38). Thus, GroE may negatively regulate expression of its own gene, perhaps as part of a homeostatic mechanism. While the situation in Chlamydia is complicated by the fact that the three pathogenic chlamydial species each encode three GroEL-like proteins, only the major groE operon, which encodes GroEL1, has been shown to have a CIRCE element. Furthermore, only transcription of groEL1, and not transcription of groEL2 or groEL3, is increased in response to heat shock (18).
The CIRCE element of the C. trachomatis groE promoter is unusual compared to the chlamydial dnaK CIRCE and CIRCE elements from other bacteria because it has a divergent sequence and a slightly longer inverted repeat that extends into the spacer region between the repeats. The groE CIRCE sequence matches the consensus CIRCE sequence at 13 of 18 nucleotides, which makes it the most divergent of 70 predicted CIRCE sequences from more than 40 eubacterial species (29, 45). Interestingly, two of the mismatches in the first repeat are complemented by base substitutions in the inverted repeat (Fig. 4), suggesting that there is selective pressure to maintain DNA secondary structure. Figure 4 also shows that similar mismatches and complementary changes are observed in the predicted groE CIRCE elements of the C. trachomatis MoPn strain and Chlamydia caviae. These chlamydial groE CIRCE elements also have a longer inverted repeat that can extend into the spacer region by as much 4 bp, effectively lengthening the stem and reducing or removing the loop from the predicted stem-loop structure of the CIRCE element (30).
We propose that Chlamydia sets the transcription levels of the heat shock genes under nonstress conditions via the sequence of the CIRCE elements. We found that in C. trachomatis serovar D, the groE CIRCE has a divergent sequence that leads to decreased HrcA binding and a corresponding lower level of repression of groE compared to the dnaK promoter. Interestingly, the sequence of the predicted groE CIRCE element in C. pneumoniae (17) almost completely matches the consensus CIRCE sequence (Fig. 4). Thus, we predict that there should be stronger HrcA binding and greater repression leading to lower groE transcript levels in C. pneumoniae under nonstress conditions than the binding and repression observed with C. trachomatis.
In our in vitro system, HrcA activity was not affected by increasing the temperature to 42°C, even though this elevated temperature is known to induce the heat shock response in Chlamydia and to result in increased transcription of groE and dnaK (8). Thus, an increased temperature alone is not sufficient for the chlamydial heat shock response, suggesting that additional factors that are not present in our in vitro system play a role. Studies of the thermostability of HrcA-CIRCE complexes in other bacteria have shown that HrcA dissociates from CIRCE at temperatures above physiologic temperatures (24, 55), although this effect varies with the bacterial species (14) and the presence of additional factors (24, 38). C. trachomatis is an obligate intracellular parasite and, as such, is exposed to a narrower range of growth temperatures than the free-living, gram-positive bacteria in which much of HrcA activity has been characterized previously. An attractive hypothesis to explain the differences between in vivo and in vitro findings involves the GroE regulatory model discussed above (26). Support for this hypothesis comes from the observation that in Streptococcus thermophilus and B. subtilis addition of GroE to a recombinant system increased the activity and thermostability of HrcA (24, 38). We are also mindful that despite the common reference to heat shock proteins, these molecular chaperones are actually stress response proteins, and their expression may be regulated by other forms of cellular stress that chlamydiae may experience during the intracellular chlamydial developmental cycle. In fact, HrcA-mediated transcriptional repression appears to be a general stress response mechanism in other bacteria (27).
The regulation of groESL transcription is clinically relevant because GroEL expression has been associated with two important aspects of chlamydial pathogenesis, the host immune response and the chlamydial persistent state. GroEL is one of the primary antigens in patients with a C. trachomatis infection, and antibodies to chlamydial GroEL are associated with some of the worst outcomes of chlamydial infection. During the persistent state, when there are viable chlamydiae inside the host cells but no infectious progeny (for a review, see reference 3), many chlamydial proteins are down-regulated, but the concentrations of GroEL mRNA and protein are maintained at steady-state or higher levels (2, 10). It remains to be seen whether regulation by HrcA is involved in the differential regulation of GroEL expression during persistence.
Acknowledgments
We thank Hilda Hiu Yin Yu, Chris Schaumburg, Elizabeth Di Russo, and Johnny Akers for their support.
This work was supported by grant AI 44198from the NIH. A.C.W. is supported by predoctoral training grant GMT3207311 from the NIH.
REFERENCES
- 1.Ault, K. A., B. D. Statland, M. M. King, D. I. Dozier, M. L. Joachims, and J. Gunter. 1998. Antibodies to the chlamydial 60 kilodalton heat shock protein in women with tubal factor infertility. Infect. Dis. Obstet. Gynecol. 6:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm of chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Brunham, R. C., R. Peeling, I. Maclean, M. L. Kosseim, and M. Paraskevas. 1992. Chlamydia trachomatis-associated ectopic pregnancy: serologic and histologic correlates. J. Infect. Dis. 165:1076-1081. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Sexually transmitted disease surveillance, 2001. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 8.Engel, J. N., J. Pollack, E. Perara, and D. Ganem. 1990. Heat shock response of murine Chlamydia trachomatis. J. Bacteriol. 172:6959-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong, I. W., B. Chiu, E. Viira, W. Tucker, H. Wood, and R. W. Peeling. 2002. Chlamydial heat-shock protein-60 antibody and correlation with Chlamydia pneumoniae in atherosclerotic plaques. J. Infect. Dis. 186:1469-1473. [DOI] [PubMed] [Google Scholar]
- 10.Gerard, H. C., L. Kohler, P. J. Branigan, H. Zeidler, H. R. Schumacher, and A. P. Hudson. 1998. Viability and gene expression in Chlamydia trachomatis during persistent infection of cultured human monocytes. Med. Microbiol. Immunol. (Berlin) 187:115-120. [DOI] [PubMed] [Google Scholar]
- 11.Grayston, J. T. 2000. What is needed to prove that Chlamydia pneumoniae does, or does not, play an etiologic role in atherosclerosis? J. Infect. Dis. 181(Suppl. 3):S585-S586. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths, E., and R. S. Gupta. 2001. The use of signature sequences in different proteins to determine the relative branching order of bacterial divisions: evidence that Fibrobacter diverged at a similar time to Chlamydia and the Cytophaga-Flavobacterium-Bacteroides division. Microbiology 147:2611-2622. [DOI] [PubMed] [Google Scholar]
- 13.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 14.Hitomi, M., H. Nishimura, Y. Tsujimoto, H. Matsui, and K. Watanabe. 2003. Identification of a helix-turn-helix motif of Bacillus thermoglucosidasius HrcA essential for binding to the CIRCE element and thermostability of the HrcA-CIRCE complex, indicating a role as a thermosensor. J. Bacteriol. 185:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsia, R., H. Ohayon, P. Gounon, A. Dautry-Varsat, and P. M. Bavoil. 2000. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2:761-772. [DOI] [PubMed] [Google Scholar]
- 16.Kalayoglu, M. V., B. Hoerneman, D. LaVerda, S. G. Morrison, R. P. Morrison, and G. I. Byrne. 1999. Cellular oxidation of low-density lipoprotein by Chlamydia pneumoniae. J. Infect. Dis. 180:780-790. [DOI] [PubMed] [Google Scholar]
- 17.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 18.Karunakaran, K. P., Y. Noguchi, T. D. Read, A. Cherkasov, J. Kwee, C. Shen, C. C. Nelson, and R. C. Brunham. 2003. Molecular analysis of the multiple GroEL proteins of chlamydiae. J. Bacteriol. 185:1958-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kol, A., T. Bourcier, A. H. Lichtman, and P. Libby. 1999. Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J. Clin. Investig. 103:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kol, A., G. K. Sukhova, A. H. Lichtman, and P. Libby. 1998. Chlamydial heat shock protein 60 localizes in human atheroma and regulates macrophage tumor necrosis factor-alpha and matrix metalloproteinase expression. Circulation 98:300-307. [DOI] [PubMed] [Google Scholar]
- 21.LaVerda, D., L. N. Albanese, P. E. Ruther, S. G. Morrison, R. Morrison, K. A. Ault, and G. I. Byrne. 2000. Seroreactivity of Chlamydia trachomatis Hsp10 correlates with severity of human genital tract disease. Infect. Immun. 68:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaVerda, D., M. V. Kalayoglu, and G. I. Byrne. 1999. Chlamydial heat shock proteins and disease pathology: new paradigms for old problems? Infect. Dis. Obstet. Gynecol. 7:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 24.Martirani, L., R. Raniello, G. Naclerio, E. Ricca, and M. De Felice. 2001. Identification of the DNA-binding protein, HrcA, of Streptococcus thermophilus. FEMS Microbiol. Lett. 198:177-182. [DOI] [PubMed] [Google Scholar]
- 25.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 26.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mogk, A., A. Volker, S. Engelmann, M. Hecker, W. Schumann, and U. Volker. 1998. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison, R. P., K. Lyng, and H. Caldwell. 1989. Chlamydial disease pathogenesis: ocular hypersensitivity elicited by a genus-specific 57 kD protein. J. Exp. Med. 169:663-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Ohta, T., S. Nettikadan, F. Tokumasu, H. Ideno, Y. Abe, M. Kuroda, H. Hayashi, and K. Takeyasu. 1996. Atomic force microscopy proposes a novel model for stem-loop structure that binds a heat shock protein in the Staphylococcus aureus HSP70 operon. Biochem. Biophys. Res. Commun. 226:730-734. [DOI] [PubMed] [Google Scholar]
- 31.Patton, D. L., Y. T. Sweeney, and C. C. Kuo. 1994. Demonstration of delayed hypersensitivity in Chlamydia trachomatis salpingitis in monkeys: a pathogenic mechanism of tubal damage. J. Infect. Dis. 169:680-683. [DOI] [PubMed] [Google Scholar]
- 32.Peeling, R. W., R. L. Bailey, D. J. Conway, M. J. Holland, A. E. Campbell, O. Jallow, H. C. Whittle, and D. C. Mabey. 1998. Antibody response to the 60-kDa chlamydial heat-shock protein is associated with scarring trachoma. J. Infect. Dis. 177:256-259. [DOI] [PubMed] [Google Scholar]
- 33.Peeling, R. W., J. Kimani, F. Plummer, I. Maclean, M. Cheang, J. Bwayo, and R. C. Brunham. 1997. Antibody to chlamydial hsp60 predicts an increased risk for chlamydial pelvic inflammatory disease. J. Infect. Dis. 175:1153-1158. [DOI] [PubMed] [Google Scholar]
- 34.Peeling, R. W., and D. C. Mabey. 1999. Heat shock protein expression and immunity in chlamydial infections. Infect. Dis. Obstet. Gynecol. 7:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, M. 1996. Electrophoretic mobility shift assay (EMSA), p. 6-11. In K. Docherty (ed.), Gene transcription: DNA binding proteins. John Wiley & Sons, Chichester, West Sussex, United Kingdom.
- 36.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reischl, S., T. Wiegert, and W. Schumann. 2002. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277:32659-32667. [DOI] [PubMed] [Google Scholar]
- 39.Rojo, F. 2001. Mechanisms of transcriptional repression. Curr. Opin. Microbiol. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 40.Sasu, S., D. LaVerda, N. Qureshi, D. T. Golenbock, and D. Beasley. 2001. Chlamydia pneumoniae and chlamydial heat shock protein 60 stimulate proliferation of human vascular smooth muscle cells via Toll-like receptor 4 and p44/p42 mitogen-activated protein kinase activation. Circ. Res. 89:244-250. [DOI] [PubMed] [Google Scholar]
- 41.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 42.Schaumburg, C. S., and M. Tan. 2003. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal −35 promoter element. Nucleic Acids Res. 31:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaumburg, C. S., and M. Tan. 2000. A positive cis-acting DNA element is required for high level transcription in Chlamydia. J. Bacteriol. 182:5167-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmiel, D. H., S. T. Knight, J. E. Raulston, J. Choong, C. H. Davis, and P. B. Wyrick. 1991. Recombinant Escherichia coli clones expressing Chlamydia trachomatis gene products attach to human endometrial epithelial cells. Infect. Immun. 59:4001-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal, R., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Stephens, R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44-51. [DOI] [PubMed] [Google Scholar]
- 47.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 48.Tam, J. E., C. H. Davis, and P. B. Wyrick. 1994. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can. J. Microbiol. 40:583-591. [DOI] [PubMed] [Google Scholar]
- 49.Tan, M., and J. N. Engel. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 178:6975-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan, M., T. Gaal, R. L. Gourse, and J. N. Engel. 1998. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J. Bacteriol. 180:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan, M., B. Wong, and J. N. Engel. 1996. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J. Bacteriol. 178:6983-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao, S., R. Kaul, and W. M. Wenman. 1991. Identification and nucleotide sequence of a developmentally regulated gene encoding a eukaryotic histone H1-like protein from Chlamydia trachomatis. J. Bacteriol. 173:2818-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vabulas, R. M., H. Wagner, and H. Schild. 2002. Heat shock proteins as ligands of Toll-like receptors. Curr. Top. Microbiol. Immunol. 270:169-184. [DOI] [PubMed] [Google Scholar]
- 54.Wagar, E. A., J. Schachter, P. Bavoil, and R. S. Stephens. 1990. Differential human serologic response to two 60,000 molecular weight Chlamydia trachomatis antigens. J. Infect. Dis. 162:922-927. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, K., T. Yamamoto, and Y. Suzuki. 2001. Renaturation of Bacillus thermoglucosidasius HrcA repressor by DNA and thermostability of the HrcA-DNA complex in vitro. J. Bacteriol. 183:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson, A. C., and M. Tan. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, G., and S.-L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]