Summary
TaTEF-7A is a functional regulatory factor gene for grain number and other traits, and its diagnostic markers could be used in marker-assisted selection to improve yield potential in wheat.
Key words: Association analysis, haplotypes, transcript elongation factor, wheat, yield.
Abstract
In this study, TaTEF-7A, a member of the transcript elongation factor gene family, and its flanking sequences were isolated. TaTEF-7A was located on chromosome 7A and was flanked by markers Xwmc83 and XP3156.3. Subcellular localization revealed that TaTEF-7A protein was localized in the nucleus. This gene was expressed in all organs, but the highest expression occurred in young spikes and developing seeds. Overexpression of TaTEF-7A in Arabidopsis thaliana produced pleiotropic effects on vegetative and reproductive development that enhanced grain length, silique number, and silique length. No diversity was found in the coding region of TaTEF-7A, but 16 single nucleotide polymorphisms and Indels were detected in the promoter regions of different cultivars. Markers based on sequence variations in the promoter regions (InDel-629 and InDel-604) were developed, and three haplotypes were identified based on those markers. Haplotype–trait association analysis of the Chinese wheat mini core collection revealed that TaTEF-7A was significantly associated with grain number per spike. Phenotyping of near-isogenic lines (NILs) confirmed that TaTEF-7A increases potential grain yield and yield-related traits. Frequency changes in favoured haplotypes gradually increased in cultivars released in China from the 1940s. Geographic distributions of favoured haplotypes were characterized in six major wheat production regions worldwide. The presence of Hap-7A-3, the favoured haplotype, showed a positive correlation with yield in a global set of breeding lines. These results suggest that TaTEF-7A is a functional regulatory factor for grain number per spike and provide a basis for marker-assisted selection.
Introduction
Transcript elongation is a simple extension of an initiated transcript (Grasser, 2005). In recent years, it has become apparent that synthesis of mRNA is a dynamic and highly regulated step in gene expression, and it requires the concerted action of a variety of elongation factors (Lolas et al., 2010). Transcript elongation factors (TEFs) play a crucial role in regulation, proliferation, and differentiation of cells, and also control various growth processes. The vast majority of studies on the regulation of eukaryotic transcription have focused on the early stages of the transcription cycle. However, studies of Drosophila and zebrafish mutants defective in elongation factors revealed intriguing results about the essential roles of these factors in development. Mutations in the Drosophila gene encoding a member of the ELL family (dELL), which belongs to the TEF group, cause segmentation defects and embryonic lethality, and dELL is required for normal expression of diverse genes during development (Eissenberg et al., 2002). Characterization of the effect of a foggy mutant on the TEFs Pandora/Spt6 and Foggy/Spt5 with an amino acid substitution in zebrafish revealed that the mutation not only affected typical overall anatomy but also influenced the differentiation of cells in specific neuron lineages (Keegan et al., 2002). Recent studies in Arabidopsis and maize also indicated that factors regulating transcript elongation play crucial roles in plant development. Mutations in the Arabidopsis DEFORMED ROOTS AND LEAVES1 (DRL1) gene cause disorganized shoot, inflorescence, flower, and root growth (Nelissen et al., 2003). DRL1 is likely to be involved in regulation of meristem activity and organ growth. Mutations in three genes encoding subunits of the Arabidopsis elongator complex display a pleiotropic phenotype, to some extent resembling the phenotype of the drl1 mutant (Nelissen et al., 2005). Thus, elo mutants show reduced growth of organs that can be attributed to a reduced rate of cell proliferation. Molecular analysis of elo mutants revealed that many genes were abnormally expressed in the mutants compared with the wild types (Nelissen et al., 2005). A study of the maize Etched (et1) mutant revealed the intriguing possibility that control of transcript elongation also plays a crucial role in expression of plastid gens; et1 mutants display delayed greening of seedling leaves. Mature kernels of these plants are severely fissured and cracked. Microscopic examination demonstrated aberrant plastid development in mutant leaves and kernels (Da Costa e Silva et al., 2004).
In rice, a T-DNA insertion mutant in a gene encoding a TEF homologous to yeast elf1 (Os2TEF1) led to a 60–80% reduction in tillering, retarded growth of seminal roots, and sensitivity to salt stress compared with wild-type Basmati 370. Detailed transcriptomic profiling of OsTEF1 revealed that mutation in the TEF differentially regulated expression of >100 genes with known function and finally regulated the tillering process by inducing expression of cytochrome P450 (Paul et al., 2012). These findings show that the TEFs are more pleiotropic than previously thought, and that previous studies failed to identify all functions of the factors.
Bread wheat (Triticum aestivum L.), as one of the three most important cereals, is widely cultivated worldwide, and improvement of wheat yields could prevent global food shortages. As it is an allohexaploid species with an extremely large and complex genome (2n=6x=42) of ~16 000 Mbp (Tilman et al., 2011), gene isolation is extremely difficult. It is well known that grain yield is manifested via a complex relationship among yield component traits such as 1000-kernel weight (TKW), grain number per spike (GN), and effective tiller number (Zhang et al., 2010). Thus, deciphering the molecular mechanisms underlying yield-related genes in wheat is of great interest not only to plant biologists, but also to breeders. Wheat yield, or yield components, is controlled by numerous genes with additive and epistatic effects that are highly interactive with the environment. Quantitative trait locus (QTL) mapping has been widely used to study specific yield-related traits in bread wheat (Zhang et al., 2012). Numerous QTLs have been reported for traits such as TKW, GN, grain weight per spike, and tiller number (Huang et al., 2004; Narasimhamoorthy et al., 2006), but fine mapping of particular genes from QTLs associated with yield is rarely reported (Barrero et al., 2011). Fine mapping is hampered by the large genome and large number of repeated nucleotide sequences. A few genes associated with yield have been isolated by comparative genetics. For example, an orthologue of the rice OsGW2 gene, named TaGW2, was cloned in wheat; this gene significantly affected TKW (Su et al., 2011; Yang et al., 2012). Zhang et al. (2012) characterized TaCKX6-D1 that also increased grain productivity, and further analysis showed that it explained phenotypic variance of 1.3–1.4g per 1000 kernels. The function and mechanism of a glutamine synthetase gene controlling nitrogen use was comprehensively studied by Li et al. (2011) and Quraishi et al. (2011), who found that the best haplotype conferred superior seedling growth, better agronomic performance, and improved N uptake during vegetative growth and grain N concentration. Xue et al. (2011) identified TaMYB13, a transcriptional activator of fructosyltransferase genes, that is involved in β-2, 6-linked fructan synthesis in wheat, and positively associated with grain yield. Currently, there is an increasing need to clone yield-related genes, exploit the more favourable alleles, and develop tailor-made markers for marker-assisted selection (MAS).
In this study, work is presented on isolation of the gene TaTEF-7A in wheat and its functional characterization by expression analysis, association analysis, near-isogenic line (NIL) comparison, and overexpression in Arabidopsis. The findings indicate that TaTEF-7A mainly regulates yield-related traits involved in vegetative growth and reproductive development. Furthermore, the coding and promoter regions were analysed and sequence polymorphisms among wheat cultivars were identified. Association analysis and comparison of NILs showed that TaTEF-7A was associated with grain number. Finally, a functional marker for TaTEF-7A was developed for MAS of high-yielding wheat genotypes.
Materials and methods
Plant materials
A Chinese wheat mini core collection (MCC) was employed for association of yield traits with markers. The MCC contains 262 wheat accessions, comprising 157 landraces, 88 modern cultivars, and 17 introduced lines, and represents 1% of the national wheat collection but >70% of the genetic diversity (Hao et al., 2011). Accessions of the three putative diploid progenitors of hexaploid common wheat (AABBDD), namely Triticum urartu (AA, accession UR203), Aegilops speltoides (SS, closely related to BB, accession Ae49), and Aegilops tauschii (DD, accession Y2282) were used for identification of the genomic origins of TaTEF homoeologues. A set of nullisomic–tetrasomic lines of Chinese Spring was used for chromosomal location of TaTEF genes. A double haploid (DH) mapping population (Hanxuan 10×Lumai 14) was used for linkage mapping. NILs for TaTEF-7A were identified in a BC3F6 of Jinmai 47//4*/Lumai14 population. In addition, a large number of modern wheat cultivars, comprising 384 European, 429 North American, 53 CIMMYT, 82 Russian, and 51 Australian cultivars (Hou et al., 2014), were used to determine the geographic distribution of TaTEF-7A haplotypes.
Phenotypic assessment and statistical analyses
Phenotypic traits of all accessions in the MCC, namely heading date (HD), maturity date (MD), spike length (SL), spikelet number per spike (SN), plant height (PH), grain number per spike (GN), effective tiller number (ETN), 1000-kernel weight (TKW), kernel length (KL), kernel width (KW), and kernel thickness (KT), were collected in four environments—in 2002, 2005, and 2006 at Luoyang, Henan province, and in 2010 at Shunyi, Beijing; they were named 02LY, 05LY, 06LY, and 10SY, respectively. Each accession was planted in a 2 m two-row plot with 30cm between rows, and 40 seeds per row. Ten plants from the middle of each plot were used in investigating the above phenotypic traits. The DH mapping population (Hanxuan 10×Lumai 14) and NILs were planted at Changping, Beijing. Five plants in the middle of each plot were randomly sampled for analysis. Eight agronomic traits were measured, namely PH, SL, TKW, GN, ETN, SN, penultimate internode length, and uppermost internode length.
Mean values of phenotypic traits and standard errors were analysed by SPSS 16.0 software (http://www.brothersoft.com/downloads/spss-16.html). The mean value of each trait in the Chinese wheat MCC was estimated by the best linear unbiased predictor (BLUP) method (Bernardo et al., 1996a , b, c).
DNA and RNA extraction
Genomic DNA was extracted from young leaves of each accession using the cetyltrimethylammonium bromide (CTAB) method (Stewart and Via, 1993). Total RNA was isolated using an RNAprep pure plant kit (Tiangen) according to the manufacturer’s instructions. cDNA was synthesized with M-MLV Reverse Transcriptase (Promega), the cDNA was diluted 10 times, and 2 μl were used for subsequent PCR.
Primers and PCR
Primers were designed by the software Primer Premier Version 5.0 (Premier Biosoft International, Palo Alto, CA, USA), and the primers were synthesized by Sangon (www.sangon.com). The Primer sequences are shown in Supplementary Table S1 available at JXB online. LA-Taq enzyme from TaKaRa (www.takara.com.cn) was used for PCR amplification. PCR were performed in total volumes of 15 μl, including 3 pmol of each primer, 120 μM of each dNTP, 80ng of template DNA or cDNA, 0.75U of La-Taq, and 7.5 μl of 2× buffer [TaKaRa Biotechnology (Dalian) Co. Ltd, Product Code: DRR20AG]. PCRs were performed as follows: 95 °C for 4min; followed by 30–35 cycles of 95 °C for 30 s, annealing (55–62 °C) for 30 s, and extension at 72 °C (30 s to 3min), and 72 °C for 30 s, with a final extension of 72 °C for 10min. The annealing temperatures and extension times depended on the primer sets and lengths of expected PCR products.
Functional marker development
Development of TaTEF-4F/R was based on two selected variations in the promoter region of TaTEF-7A, including a 1bp indel at position –629bp (InDel1-629) and another 1bp indel at position –604bp (InDel1-604). First, genomic-specific primer set TaTEF-3F/R was used to amplify fragments from chromosome 7A in all cultivars. A second PCR was performed as follows: the first PCR solution was diluted 100 times, taking 1 μl as template for the second PCR, primer TaTEF-4F/R was used for the second PCR, with annealing at 54 °C for 30 s, and extension at 72 °C for 30 s.
Chromosomal localization
To determine the chromosomal location, 37 nulli-tetrasomic lines of Chinese Spring provided by the Kansas Wheat Stock Centre (http://www.k-state.edu/wgrc/) were used. Gene-specific primers TaTEF-1F/R, TaTEF-2F/R, TaTEF-3F/R, and TaTEF-6F/R for TaTEF-7A localization were used for PCR. TaTEF-7A was mapped on a mapping population (Hanxuan 10×Lumai 14) using the MAPMAKER/EXP 3.0 (Lander and Botstein, 1989).
Multiple sequence alignment and phylogenetic analysis
Full-length protein sequences of TaTEF-7A from Triticum aestivum (Ta), Brachypodium distachyon (Bd), Oryza sativa (Os), Setaria italica (Si), Zea mays (Zm), Sorghum bicolour (Sb), Arabidopsis thaliana (At), Thellungiella halophila (Th), Brassica rapa (Br), Citrus sinensis (Cs), Physcomitrella patens (Pp), and Saccharomyces cerevisiae (By) were aligned by ClustalX. A phylogenetic tree was created using a Neighbor–Joining method in the MEGA5.05 program (Tamura et al., 2011).
Subcellular location of TaTEF-7A protein by fusion with green fluorescent protein (GFP)
The full-length cDNA clone of TaTEF-7A was fused upstream of the GFP gene and put under control of the constitutive Cauliflower mosiac virus (CaMV) 35S promoter in the pJIT163-GFP expression vector to construct a 35S::TaTEF–GFP fusion protein. Restriction sites, KpnI and XbaI, were added to the 5’ and 3’ ends of the coding region by PCR. The PCR products were digested with restriction endonucleases, ligated with the pJIT163-GFP plasmid, and cut with the corresponding enzymes to create recombinant plasmids expressing the fusion protein. The subcellular location of TaTEF-7A was detected by monitoring transient expression of GFP in wheat protoplast cells as described by Mao et al. (2011)
Expression analysis
Quantitative real-time PCR was performed using SYBR® Premix Ex Taq™ II (Takara) according to the manufacturer’s instructions on a 7300 Real-time PCR System (Applied Biosystems). The relative expression of each gene was calculated according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). The glyceraldehyde-3-phosphate dehydrogenase gene was used as an endogenous reference for real-time PCR, and all analyses were performed with three technical and three biological replicates.
Binary vector construction and transformation of Arabidopsis
The coding sequence of TaTEF-7A, including the XbaI (5′) and BamHI (3’) restriction sites, was first amplified. The PCR products and pbi121 plasmid were digested with XbaI and BamHI, and then cloned into the binary vector pbi121 under control of the CaMV 35S promoter in the sense orientation. The construct was transformed into Arabidopsis (ecotype Col-0) by the floral-dip method according to previously described procedures using Agrobacterium tumefaciens strain GV3101 (Herrera-Estrella et al., 2005). Transgenic plants were selected on solid half-strength Murashige and Skoog (MS) medium containing 50mg l–1 kanamycin (Ahmed et al., 2012).
Transient expression in tobacco
The promoter fragment of TaTEF-7A was fused with the β-glucuronidase (GUS) gene in the pCAMBIA1391Z vector. Restriction sites, BamHI and AvrII, were added to the 5′ and 3′ ends of the coding region by PCR. The PCR products were digested with restriction endonucleases, and then ligated with the plasmid. pBICHP57. The luciferase (LUC) reporter gene (Wu et al., 2009) was cloned in the binary vector pBI121 downstream of the CaMV 35S promoter. Nicotiana benthamiana leaves were co-infiltrated with plasmids and pBICHP57. The GUS activity was normalized by the LUC activity. Plant transformation, and GUS and LUC activity assays were determined according to Wu et al. (2009).
Plant growth and documentation
Arabidopsis thaliana (ecotype Columbia-0) was chosen for transgene analysis, and the T3 generation was grown in a controlled environment chamber at 22 °C, with a 16h/8h photoperiod, light intensity of 120 mmol m−2 s−1, and 70% relative humidity. After sowing, seeds were stratified in darkness at 4 °C for 48h prior to incubation in a plant growth chamber. At least five plants of each line were used for phenotypic characterization. Distances from the stem bases in each line were measured to compare silique lengths. The numbers of siliques on each line were counted. Seeds of each line were photographed and measured using an Olympus BH-2 microscope (Olympus, Japan).
Results
Isolation of TaTEF-7A and phylogenetic analysis of orthologous genes in wheat and other species
Using the rice TEF gene (Os02g04160) as a query, contig08999 was found in the Chinese Spring draft genome sequence (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/Documents/DOC_search_reads.php). A full-length wheat coding sequence (CDS; accession CJ655632) was retrieved from NCBI by blasting the contig sequence against GenBank (Supplementary Fig. S1 available at JXB online). The TaTEF genomic sequence and CDS were amplified by TaTEF-5F/R from Chinese Spring genomic DNA and cDNA, respectively. Their lengths were ~1100bp and 270bp, respectively. cDNA sequences of the three homoeologous genes in the A, B, and D genomes were obtained (Supplementary Figs S2, S3 available at JXB online). Four exons and three introns were detected in this gene (Supplementary Fig. S1 available at JXB online).
TaTEF encodes an 89 amino acid polypeptide that is 84.9% identical to OsTEF protein (Supplementary Fig. S1b available at JXB online). In most species, TEF genes have similar structures with four exons and three introns. A phylogenetic tree of TEF genes was generated based on the predicted polypeptide sequences in monocots, eudicots, and yeast (Supplementary Table S2 available at XB online). All of the proteins shared high similarity of 52.8–95.5%, implying potentially conserved functions among orthologues (Supplementary Fig. S1c available at JXB online). The tree consists of at least three divergent clades with strong bootstrap support; the species include monocots, eudicots, and Saccharomyces cerevisiae.
Diversity mainly occurs in the promoter region of TaTEF on chromosome 7A
A number of single nucleotide polymorphisms (SNPs) and Indels were found in the intron sequences of TaTEF genes from the A, B, and D genomes. This allowed discrimination of TaTEF-7A from the other copies on chromosomes 7B and 7D by amplification of genome-specific primers TaTEF-1F/R, TaTEF-2F/R, and TaTEF-3F/R (Fig. 1). The region of each primer set is shown in Fig. 2. No nucleotide difference was detected in the coding region from 48 cultivars with quite different phenotypes, implying that the mechanism of control of wheat development possibly depends on regions external to the gene. Approximately 700bp upstream of ATG of the 48 accessions were then sequenced. Fourteen nucleotide substitutions and two Indels were detected among the 48 cultivars, and three major haplotypes were formed by 16 representative nucleotides (Fig. 2; Supplementary Table S3 available at JXB online).
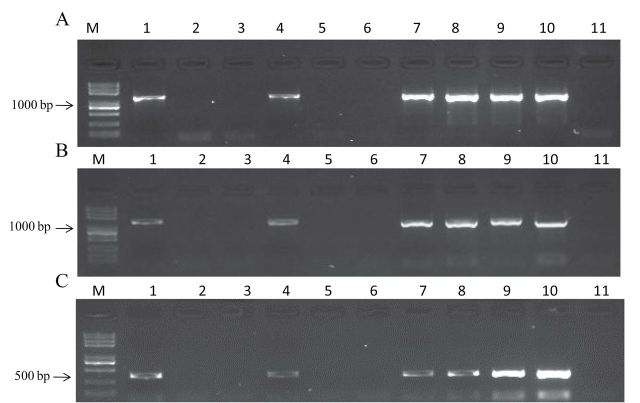
Fig. 1.
Targeted PCR amplification of TaTEF-7A in T. aestivum cv. Chinese Spring, the nullisomic–tetrasomic lines for homoeologous group 7, and three putative diploid progenitors of wheat with genome-specific primer sets. (A) Primer TaTEF-1F/R; (B) primer TaTEF-2F/R; (C) primer TaTEF-3F/R. M, marker; 1, T. urartu; 2, Ae. speltoides; 3, Ae. tauschii; 4, T. aestivum; 5, N7AT7B; 6, N7AT7D; 7, N7BT7A; 8, N7BT7D; 9, N7DT7B; 10, N7DT7A; 11, H2O.
Fig. 2.
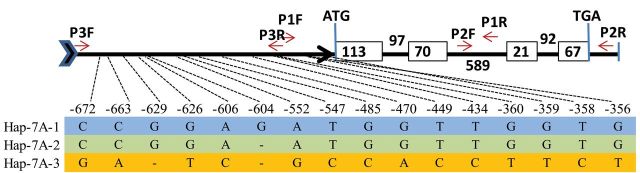
Gene structure and haplotypes in the promoter region of TaTEF-7A. The positions of 14 SNPs and two Indels between cultivars are shown using the TaTEF gene sequence, with the start codon designated as position 0. Haplotypes are highlighted in different colours. Arrow bar, promoter region; open boxes, exons; thin lines, introns; ATG, start codon; TGA, stop codon; red arrows, positions of primers TaTEF-1F/R, TaTEF-2F/R, and TaTEF-1F/R.
The design of primer TaTEF-3F/3R was based on polymorphisms in the promoter region. As shown in Fig. 1C, target fragments were detected only in accessions with the A genome, and were not amplified in Ae. speltoides, Ae. tauschii, NT7A7B, and NT7A7D. This indicated that TaTEF was amplified from chromosome 7A in Triticum spp. Moreover, primer TaTEF-4F/R (Table 1) was used to genotype 150 DHs derived from Hanyuan10×Lumai14. TaTEF was mapped to a region flanked by P3156.3 (7.2 cM distal) and Xwmc83 (6.5 cM proximal) on chromosome 7A (Fig. 3).
Table 1.
Phenotypic analysis of Col-0 and two transgenic plants
| Trait | CK | Line 1 | Line 2 |
|---|---|---|---|
| Silique length (cm) | 1.06±0.05 a(A) | 1.23±0.06 b(B) | 1.13±0.069 c(B) |
| Silique number | 21.0±3.08 a(A) | 31.5±2.87 b(B) | 27.5±2.41 c(C) |
| Grain length (μm) | 441.26±31.37 a(A) | 489.03±7.85 b(B) | 498.99±14.3 6 b(B) |
CK, Col-0.
Upper and lower case letters indicate significance level at P=0.01 and P=0.05, respectively.
Fig. 3.
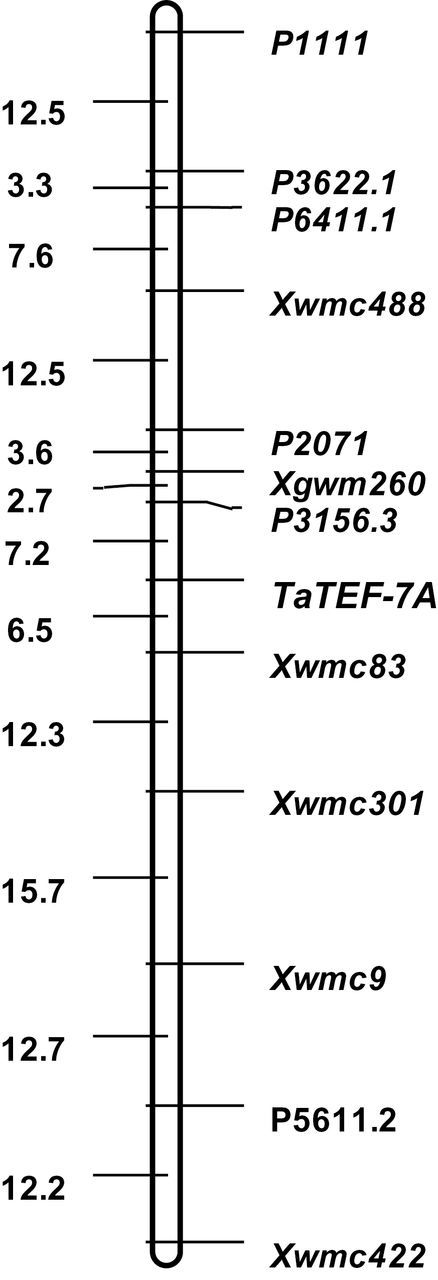
Linkage map of TaTEF-7A and surrounding markers on wheat chromosome 7A.
TaTEF-7A is mainly expressed in young spikes and developing seeds, and the protein is located in the nucleus
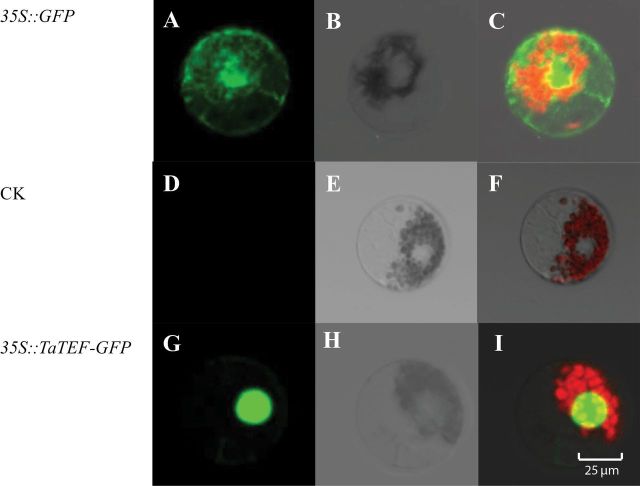
Transient expression of TaTEF-7A fused with GFP in wheat protoplast cells showed that TaTEF7A-GFP accumulated only in the nucleus, whereas GFP alone was present throughout the whole cell (Fig. 4), demonstrating that TaTEF-7A interacted with the cell nucleus. TaTEF-6F/R (Table 1) was designed based on the 5’-untranslated region (UTR) sequence, which was amplified only from the 7A chromosome (Supplementary Fig. S4 available at JXB online). A tissue-specific expression study using quantitative real-time PCR showed that TaTEF-7A was expressed in nearly all tissues tested (Fig. 5). The highest expression levels were detected in young spikes and developing seeds. The transcription level was consistent in spikes at various stages from young to immature seeds and increased gradually during grain filling, indicating that it may have effects on yield traits.
Fig. 4.
Subcellular localization of TaTEF-7A protein in the wheat protoplast. Cell images of green fluorescence in dark field (A, D, G); cell images in bright field (B, E, H); merged images (C, F and I). CK, control.
Fig. 5.

TaTEF-7A expression levels revealed by real-time PCR amplification. E, embryo; SR, seedling root; TN, tillering node; SL, seedling leaf; HR, roots at heading date; HN, nodes at heading date; HS, stems at heading date; FL, flag leaves; YS, young spike 1.5–2.5cm in length; HS, spike 6–7 in cm length; LS, leaf sheath; G4, grain 1–4 DAF; G14, grain 13–14 DAF; G21, grain 20–21 DAF; the expression level of young spikes 1.5–2.5cm in length was assumed to be 1.00 and was regarded as the reference for other tissues.
Overexpression of TaTEF-7A in Arabidopsis alters vegetative and reproductive development
Tissue-specific expression showed that TaTEF-7A was constitutively expressed, but at much higher levels in young spikes and developing seeds. A T-DNA insertion in rice caused 60–80% less tillering, retarded growth of seminal roots, and increased sensitivity to salt stress compared with the wild type (Paul et al., 2012). The question that arises is what traits are affected when TaTEF-7A is overexpressed in Arabidopsis. Transgenic plants harbouring TaTEF-7A constructs driven by the CaMV 35S promoter were generated. Overexpression of TaTEF-7A in Arabidopsis enhanced vegetative growth of both seedlings and booting stage plants (Fig. 6A, B). As shown in Fig. 6C and D, and Table 1, silique and grain lengths were increased in transgenic lines 1 and 2 compared with the control. The expression levels of TaTEF-7A transgenics were also higher (Fig. 6E). The fact that TaTEF-7A affects silique length, silique number, and grain length suggests that this gene should also influence yield traits in wheat.
Fig. 6.
Overexpression of TaTEF-7A in Arabidopsis. Comparisons of two trangenic lines and control plants: (A) growth at 23 days after sowing (DAS); (B) growth at 45 DAS; (C) siliques; (D) seeds; (E) TaTEF-7A expression.
Haplotype association indicates that TaTEF-7A strongly influences yield traits
Compared with Hap-7A-1, Hap-7A-2 has a 1bp deletion and Hap-7A-3 has a 2bp deletion. After amplification, the PCR products were separated using 6% denaturing polyacrylamide gels (Fig. 7). Linkage disequilibrium (LD) was estimated between all pairs of polymorphic sites in the 0.7kb region. Significant LD (r 2=1, P<0.001) was observed across pairwise polymorphisms (Supplementary Fig. S5 available at JXB online).
Fig. 7.
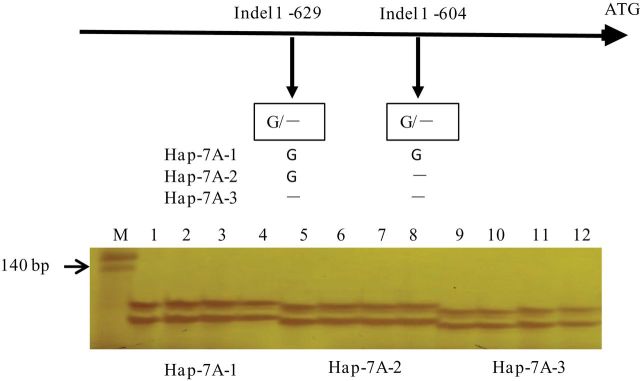
Marker development and polymorphism of different cultivars on 6% denaturing polyacrylamide gels. M, marker; 1, LM14; 2, QCM; 3, WSB; 4, NQ4; 5, NY188; 6, BN6; 7, AM6; 8, PY27; 9, SJZ8; 10, YZ1; 11, S4185; 12, YM18.
The MCC is regarded as a suitable population for detection of major QTLs controlling yield traits (Zhang et al., 2012). In comparing the effects of each haplotype on agronomic traits GN, KL, KW, KT, and TKW in MCC landraces, significant associations were identified, and there was a negative relationship between GN and TKW. However, among modern cultivars, there were significant differences in GN between haplotypes. Interestingly, the phenotypic means for KL, KW, KT, and TKW in Hap-7A-3 accessions were all higher than for other haplotypes in this subset, and GN differences between the three haplotypes were also significant. Thus the average GN of Hap-7A-3 was increased, with TKW enhanced (P<0.05) between landraces and modern cultivars during yield improvement (Table 2). Collectively, these results demonstrated that Hap-7A-3 was positively selected because it had a significantly positive effect on yield.
Table 2.
Comparisons of three haplotypes of TaTEF-7A based on BLUP values of phenotypic traits in MCC from four environments
| Genotype/trait | Hap-7A-1 | Hap-7A-2 | Hap-7A-3 | |||
|---|---|---|---|---|---|---|
| Mean ±SE | Range | Mean± SE | Range | Mean ±SE | Range | |
| Landraces | ||||||
| HD (d) | 196.86±0.42 a | 188.00–209.00 | 197.78±0.98 a | 194.00–202.00 | 192.33±2.96 a | 188.00–198.00 |
| MD (d) | 237.35±0.34 a | 230.00–248.00 | 237.56±0.69 a | 235.00–241.00 | 234.00±1.53 a | 232.00–237.00 |
| SL (cm) | 9.89±0.14 a | 6.34–14.07 | 10.40±0.60 a | 7.40–12.69 | 11.44±0.55 a | 10.40–12.27 |
| SN | 21.65±0.09 a | 18.93–27.64 | 20.94±0.44 a | 19.03–22.78 | 20.78±0.35 a | 20.28–21.46 |
| PH (cm) | 113.54±0.82 a | 58.96–145.23 | 116.15±1.91 a | 109.77–128.73 | 113.96±5.32 a | 108.21–124.58 |
| GN | 49.73±0.46 a | 38.29–72.92 | 45.07±1.84 b | 37.79–52.80 | 43.37±3.55 a | 38.69–50.34 |
| ETN | 10.68±0.10 a | 7.83–14.05 | 10.84±0.40 a | 8.65–12.73 | 10.10±1.21 a | 8.59–12.49 |
| TKW (g) | 32.04±0.38 A | 22.77–46.11 | 35.48±1.26 A | 28.30–39.95 | 46.48±3.83 B | 39.59–52.81 |
| KL (mm) | 6.35±0.04 A | 5.55–7.45 | 6.62±0.15 A | 6.18–7.37 | 7.70±0.26 B | 7.18–8.02 |
| KW (mm) | 3.01±0.01 a | 2.60–3.40 | 3.07±0.04 ab | 2.87–3.27 | 3.22±0.02 b | 3.20–3.26 |
| KT (mm) | 2.75±0.01 a | 2.35–3.16 | 2.85±0.05 ab | 2.63–3.09 | 2.96±0.09 b | 2.85–3.13 |
| Modern cultivars | ||||||
| HD (d) | 194.00±0.47 a | 189.00–208.00 | 195.00±1.52 a | 190.00–201.00 | 192.25±0.65 a | 190.00–198.00 |
| MD (d) | 235.65±0.42 a | 228.00–245.00 | 236.63±1.13 a | 234.00–243.00 | 234.25±0.95 a | 230.00–242.00 |
| SL (cm) | 9.95±0.17 a | 6.88–13.31 | 10.07±0.81 a | 7.77–14.97 | 9.99±0.50 a | 7.67–13.27 |
| SN | 21.38±0.12 a | 19.40–23.88 | 21.34±0.49 a | 19.69–23.85 | 21.49±0.23 a | 20.13–22.56 |
| PH (cm) | 99.52±1.91 a | 60.76–122.50 | 98.80±3.01 a | 85.24–110.30 | 90.53±4.37 a | 67.32–119.04 |
| GN | 50.31±0.68 a | 37.40–63.97 | 48.44±1.89 a | 42.76–58.07 | 55.39±1.87 b | 43.85–65.35 |
| ETN | 9.99±0.18 a | 7.15–13.96 | 10.41±0.38 a | 9.30–12.15 | 9.03±0.24 a | 7.94–10.26 |
| TKW (g) | 39.51±0.69 a | 26.64–50.08 | 39.14±1.54 a | 35.12–47.51 | 42.74±1.03 a | 38.03–47.79 |
| KL (mm) | 6.71±0.06 a | 5.75–7.85 | 6.57±0.10 a | 6.18–7.04 | 6.89±0.08 a | 6.36–7.26 |
| KW (mm) | 3.22±0.02 a | 2.92–3.51 | 3.17±0.05 a | 3.00–3.35 | 3.28±0.03 a | 3.07–3.44 |
| KT (mm) | 2.91±0.02 a | 2.62–3.24 | 2.91±0.05 a | 2.66–3.19 | 2.94±0.04 a | 2.72–3.18 |
Landraces: Hap-7A-1, n=145; Hap-7A-2, n=9; Hap-7A-3, n=3. Modern cultivars: Hap-7A-1, n=68; Hap-7A-2, n=8; Hap-7A-3, n=12
Upper and lower case letters indicate significant differences at P=0.01 and P=0.05, respectively.
Previous studies demonstrated that GN differences were associated with TaELF-7A haplotypes (Table 2). Therefore, differences in GN between TaELF-7A haplotypes under different environmental conditions were examined (Table 3). In landraces, the GN of Hap-7A-1 was higher than those of Hap-7A-2 and Hap-7A-3 in all environments, but a significant difference (P<0.05) occurred only in 02LY. In modern cultivars, in contrast, the GN of Hap-7A-3 was significantly higher than those of Hap-7A-1 and Hap-7A-2 in both 02LY and 10SY. Because Hap-7A-3 had a significantly positive effect on GN, it was considered a potentially superior allele for improvement of grain yield in wheat.
Table 3.
Comparisons of TaTEF-7A haplotypes in MCC for GN in four environments
| Haplotype | 02LY | 05LY | 06LY | 10SY | ||||
|---|---|---|---|---|---|---|---|---|
| No. | Mean ±SE | No. | Mean ±SE | No. | Mean ±SE | No. | Mean ±SE | |
| Landraces | ||||||||
| Hap-7A-1 | 129 | 51.00±0.99 a | 129 | 40.70±0.63 a | 145 | 54.63±0.79 a | 142 | 52.70±0.76 a |
| Hap-7A-2 | 8 | 39.88±4.17 b | 8 | 40.43±3.20 a | 9 | 47.98±2.73 a | 9 | 46.41±3.09 a |
| Hap-7A-3 | 3 | 37.00±2.65 ab | 3 | 37.47±5.76 a | 3 | 45.67±5.53 a | 3 | 46.20±4.70 a |
| Modern cultivars | ||||||||
| Hap-7A-1 | 61 | 50.64±1.47 a | 63 | 46.05±1.11 a | 68 | 53.49±1.16 a | 68 | 52.07±0.99 a(A) |
| Hap-7A-2 | 8 | 49.25±2.86 ab | 8 | 42.05±1.08 a | 8 | 50.08±3.94 a | 8 | 50.90±2.92 a(AB) |
| Hap-7A-3 | 10 | 59.20±2.43 b | 12 | 49.98±2.86 a | 12 | 59.38±4.19 a | 11 | 60.67±2.74 b(B) |
02LY, Luoyang (2002); 05LY, Luoyang (2005); 06LY, Luoyang (2006); 10SY, Shunyi (2010)
Upper and lower case letters indicate significant differences at P=0.01 and P=0.05, respectively.
The effects of TaTEF-7A haplotypes on yield traits were further confirmed in NIL comparisons
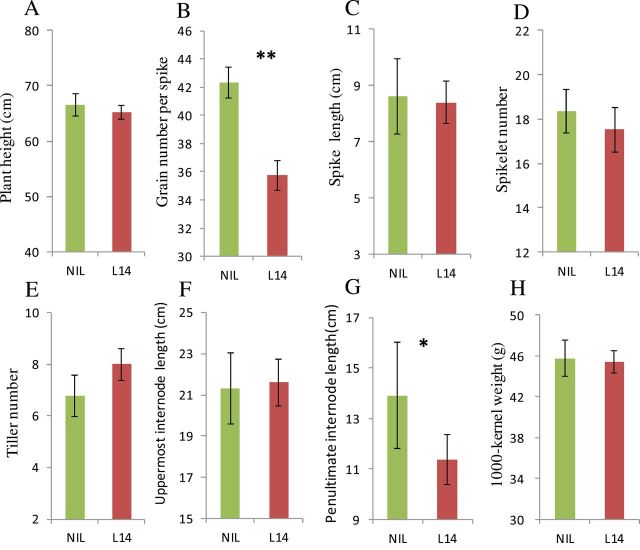
NILs derived from a BC3F6 population of Jinmai 47//4*/Lumai14 were used to evaluate haplotype genetic effects, in which Jinmai 47 represented Hap-7A-3 and Lumai 14 represented Hap-7A-2. Functional analysis of TaTEF-7A in NILs is shown in Fig. 8. It was clear that Hap-7A-3 has higher GN (18.4%) than Hap-7A-2. Moreover, although Hap-7A-3 (NIL) has a longer penultimate internode than Hap-7A-2 (L14) (13.9±2.6cm versus 11.4±1.3cm), there was no significant difference in PH. Hap-7A-3 also exhibited an increase in SN and TKW but a lower TN that did not reach statistical significance. This further confirmed that Hap-7A-3 potentially increases grain yield through regulating grain number per spike.
Fig. 8.
Phenotypic comparison of Lumai 14 (Hap-7A-2) and NIL (Hap-7A-3). (A) Plant height; (B) grain number per spike; (C) spike length; (D) spikelet number; (E) tiller number; (F) uppermost internode length; (G) penultimate internode length; (H) 1000-kernel weight; *P<0.05; **P<0.01.
TaTEF-7A positively regulates yield-related traits
To confirm the expression patterns of TaTEF-7A, young spikes of each haplotype represented by four, one, and four varieties, respectively, were collected. Their relative expression values were measured by quantitative real-time qPCR (Table 4). Hap-7A-3 was the highest overall, and a significant positive correlation between the expression level of TaTEF-7A and GN was observed (r=0.889, P<0.001) The average expression level of TaTEF-7A was significantly higher than that of either TaTEF-7B or TaTEF-7D in a set of NILs. GN was positively correlated with the total expression level of the three TaTEF genes. Furthermore, TaTEF-7A was the highest among the three homoeologues, except in the recurrent parent Lumai 14 (Fig. 9). Association analysis also showed that Hap-7A-3 at TaTEF-7A was significantly related to higher GN, whereas Hap-7A-1 and Hap-7A-2 were associated with lower GN (Table 2). Transient expression analysis in tobacco also showed that the average relative expression of Hap-7A-3 was higher than that for the other two haplotypes (Fig. 10). All of these results further demonstrated that TaTEF-7A positively regulated yield-related traits through its expression level.
Table 4.
Difference in relative expression levels in different wheat varieties
| Cultivar | Haplotype | Relative expression level of TaTEF-7A | GN |
|---|---|---|---|
| YM18 | Hap-7A-3 | 0.53±0.03 | 58.6 |
| WM6 | Hap-7A-3 | 0.39±0.04 | 44.3 |
| JM47 | Hap-7A-3 | 0.36±0.03 | 48.4 |
| S229 | Hap-7A-3 | 0.32±0.02 | 47.8 |
| PY27 | Hap-7A-2 | 0.21±0.02 | 43.6 |
| LM14 | Hap-7A-2 | 0.14±0.01 | 39.5 |
| ZY9507 | Hap-7A-1 | 0.14±0.01 | 40.7 |
| LZ953 | Hap-7A-1 | 0.18±0.02 | 38.4 |
| CS | Hap-7A-1 | 0.11±0.01 | 41.8 |
These cultivars were planted at Shunyi, Beijing.
The expression level of YM18 was assumed to be 1.00.
Fig. 9.
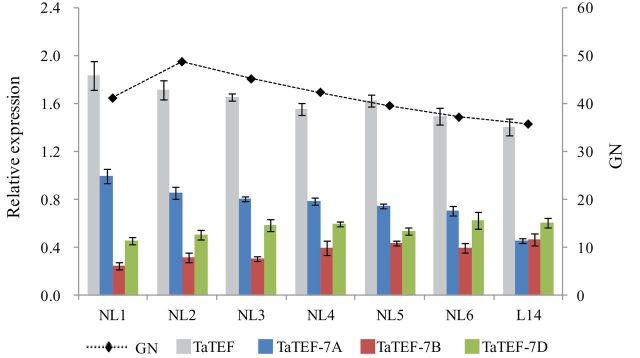
Relative expression levels of TaTEF-7A, TaTEF-7B, and TaTEF-7D in a set of NILs, and their relationship to GN. The expression level of TaTEF-7A was regarded as the reference and assumed to be 1.00.
Fig. 10.
Quantification of the expression of GUS gene driven by each of the three haplotype promoters.
TaTEF-7A co-locates with yield QTLs in wheat
TaTEF-7A was mapped using a DH population derived from Hanxuan 10×Lumai 14, and was flanked by SSR markers P3156.3 (7.2 cM distal) and Xwmc83 (6.5 cM proximal). This region of TaTEF-7A coincides with several previously reported yield-related QTLs, including spikelet number per spike QSpn.nau-7A (Ma et al., 2007; Zhang et al., 2010), flour yield QFy550.b22-7A (Kunert et al., 2007), test weight QTw.crc-7A, and grain yield QYld.crc-7A (Huang et al., 2006). In an immortalized F2 population of 136 lines derived from Nanda 2419×Wangshuibai, QSpn.nau-7A explained 10–12% of the spikelet variation (Ma et al., 2007). Agronomic and quality traits were also detected in this region in a DH population from AC Karma’×87E03-S2B1. QTw.crc-7A for test weight and grain yield per plot explained 10.6% of the phenotypic variance in test weight, and QYld.crc-7A explained 8.1% of the variance in grain yield (Huang et al., 2006). This locus was also related to flour yield in another study (Kunert et al., 2007). An advanced backcross QTL (AB-QTL) strategy utilized to locate QFy550.b22-7A for baking quality traits in two BC2F3 populations of winter wheat explained 11.9% of the genetic variance (Kunert et al., 2007). These results suggest that TaTEF-7A might be related to major yield QTLs mapped to the same region, and that the gene possibly confers complex pleiotropic effects on growth, yield, and quality.
Hap-7A-3 was positively selected in Chinese wheat breeding programmes
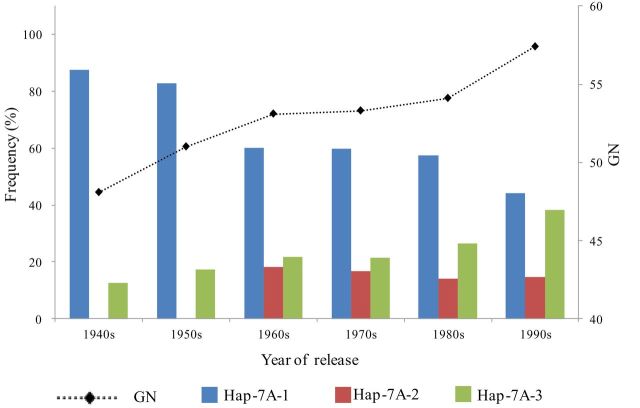
A total of 348 modern cultivars released in China since the 1940s (Hao et al., 2011) were classified into six subgroups (1940s, 1950s, 1960s, 1970s, 1980s, and 1990s) according to decade of release, and were used to determine haplotype frequencies of TaTEF-7A over time. The frequency of Hap-7A-3 increased from 12.5% in the 1940s to 38.2% in the 1990s. Interestingly, the frequencies of this haplotype from the 1940s to 1990s were consistent with yield-related differences in GN. This strongly indicates that Hap-7A-3 was positively selected in Chinese wheat breeding and that its frequency should be closely correlated with increases in GN (Fig. 11).
Geographic distribution of haplotypes of TaTEF-7A in global wheat cultivars
In order to evaluate comprehensively and systematically the distribution of all TaTEF-7A haplotypes in global wheat cultivars, haplotypes were determined in cultivars from North America, Australia, China, CIMMYT, Europe, and Russia. In Chinese landraces, Hap-7A-1 was predominant across all zones (Fig. 12A), whereas the other two haplotypes had lower frequencies in some ecological regions. The favoured haplotype Hap-7A-3 occurred only in two zones (III at a frequency of 8.7% and VII at 11.1%). A population with 348 Chinese modern cultivars was employed to reveal further the haplotype distributions in China. As shown in Fig. 12B, Hap-7A-1 frequency declined in almost all zones except in VII. Interestingly, Hap-7A-3 dramatically increased in different zones, particularly I (14.3%), II (29.9%), and III (29.4%), which have the oldest and strongest breeding programmes in China. It can be inferred that Hap-7A-3 is the most favoured haplotype in breeding, because modern breeding has significantly promoted its frequency in Chinese modern cultivars (Jiang et al., 2011; Su et al., 2011).
Fig. 12.
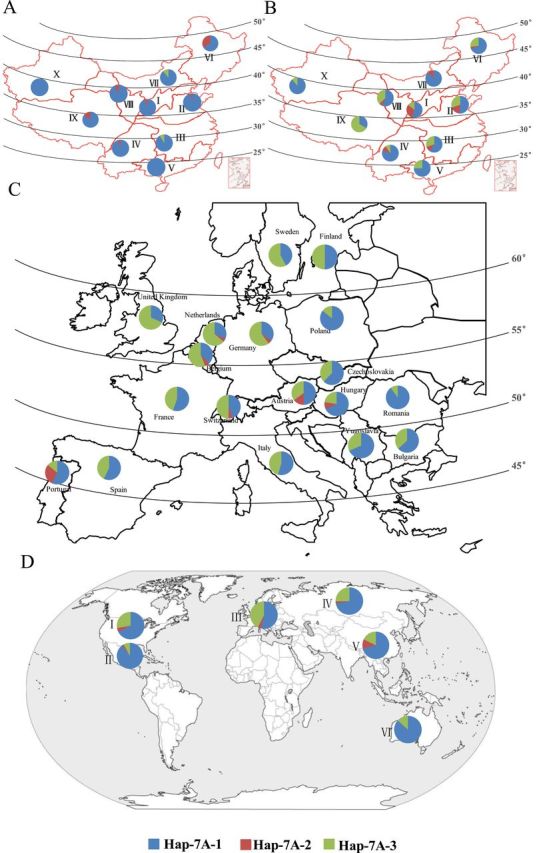
Global distribution of TaTEF-7A haplotypes. Distribution of 157 Chinese landraces (A) and 348 modern cultivars (B) in 10 production zones. I, northern winter wheat region; II, Yellow and Huai River valley winter wheat region; III, low and middle Yangtze River valley winter wheat region; IV, southwestern winter wheat region; V, southern winter wheat region; VI, northeastern spring wheat region; VII, northern spring wheat region; VIII, northwestern spring wheat region; IX, Qinghai–Tibet spring–winter wheat region; X, Xinjiang winter–spring wheat region. (C) Geographic distribution of haplotypes among European wheat cultivars; (D) geographic distribution of haplotypes in cultivars in other regions, I, North America; II, CIMMYT; III, Europe; IV, Former USSR; V, China; VI, Australia.
In addition, haplotype analysis of 384 wheat cultivars released in 18 European countries showed that the average frequency of Hap-7A-3 was 41.4%, ranging from 10% to 68.2%, which was much higher than that in cultivars released in China (Fig. 12C). Hap-7A-3 distribution showed a geographic bias, with a higher frequency in western than in eastern Europe (Fig. 12C). Comparing the global distribution in six major international wheat regions (Fig. 12D), Hap-7A-3 occurred at the highest frequency in varieties released in Russia and North America, but at lower frequencies in CIMMYT and Australian cultivars. This further demonstrated that the favoured haplotype Hap-7A-3 underwent selective pressure but with different degrees in global wheat breeding.
Discussion
Genome rearrangement of the TEF orthologues occurred in grass crop evolution
Poaceae species have diverged >65 million years. Comparative mapping studies indicate that there is high synteny between species at the macro level (Sorrells et al., 2003). An 89 amino acid wheat protein encoding the TaTEF-7A gene was identified using genome-specific primers and Chinese Spring nullisomic–tetrasomic lines. In rice, LOC_Os02g04160, the orthologue of TaTEF-7A, is located on chromosome 2. Collinearity was shown between wheat homoeologous group 6 and rice chromosome 2 (Bolot et al., 2009; Su et al., 2011). To date, there is little information to identify and characterize shared duplications within wheat and between rice and wheat. Interestingly, one model proposed a pathway for evolution of the rice, wheat, sorghum, and maize genomes from a common ancient ancestor (n=5) followed by genome duplication, with breakage and fusion of different chromosomes in the genomes of each of the four species (Salse et al., 2008). Translocations and fusions were found between ancestral chromosomes A2, A4, and A6, which inevitably led to rearrangements of wheat homoeologue groups 6 and 7, and this also disrupted the collinearity between wheat group 6 chromosomes and rice chromosome 2. The amplification products of TaTEF-7A produced by three pairs of genome-specific primers were mapped on chromosome 7A. This confirmed the above opinion and indicated that grass genomes are labile, rapidly evolving entities with complex structural and functional relationships (Quraishi et al., 2011).
Advantages of genetic analysis using natural populations and NILs
Dissecting the genetics underlying complex traits in crops not only provides insights into genetic pathways, but also provides targets for MAS in breeding (Clark, 2010). Recently, candidate gene-based association analysis has been used to trace the origin of agronomically important alleles and to explore the process of domestication of cultivated rice. This approach takes advantage of historical and evolutionary recombination events in natural populations to resolve complex trait variations in individual nucleotides (Lu et al., 2012). Combining linkage analysis and candidate gene association mapping, Zhang et al. (2014) identified a soybean gene related to P efficiency. Moreover, for pleiotropic genes, association mapping also dissects trait correlations at the gene level because polymorphic sites can be independently associated with different traits (Chen and Lubberstedt, 2010). For example, the maize pleiotropic gene Dwarf8, which affects both flowering time and plant height, was shown to contain two SNPs that were independently associated with each trait (Thornsberry et al., 2001). The MCC contains 1% of the basic wheat germplasm collection with an estimated 70% representation of the genetic variation in that collection, making it a suitable population for detection of major QTLs controlling yield traits (Hao et al., 2011; Zhang et al., 2012). In this study, candidate gene association mapping showed that TaTEF-7A was significantly associated with yield traits. The location of this gene was consistent with the location of a previously identified QTL associated with yield and also likely to possess pleiotropic influences on grain yield traits.
NILs are a well known useful and reliable tool to evaluate trait-related allelic effects, but there are few reports of yield traits being validated by NILs in wheat (Lei et al., 2012). In this study, a NIL derived from a backcross of Lumai 14 (recurrent parent, genetic background reconstitution, 93.8%) and Jinmai 47 (donor parent) was used for functional analysis of TaTEF-7A by comparing haplotype Hap-7A-2 with Hap-7A-3. Haplotype Hap-7A-3 had a higher GN and longer penultimate internode (Fig. 8). Hence, this result with NILs was consistent with the association analysis using natural populations and led to the conclusion that TaTEF-7A was the major genetic determinant controlling yield-related traits on chromosome 7A, especially grain number, which was verified by overexpression of the gene in Arabidopsis.
TEF1 affects plant vegetative and reproductive development
Over the past few years, it has become apparent that TEFs regulate various aspects of plant development expressed as changes in flowering, branching, leaf venation patterns, root growth, and seed development (Guo et al., 2002; Grasser et al., 2009; Lolas et al. 2010). In rice, Paul et al. (2010) reported a T-DNA insertion mutant of OsTEF1, an orthologue of TaTEF-7A. Functional loss of this gene led to 60–80% reduced tillering, retarded growth of seminal roots, and sensitivity to salt stress compared with wild-type Basmati 370. Detailed transcriptional profiling of OsTEF1 revealed that mutation in the transcribed elongation factor influenced >100 genes with known function. It finely regulated tillering in rice by induction of expression of MAX1, a member of the cytochrome P450 family. P450s regulates various aspects of biological development, such as cell proliferation (CYP2D6) (Sandee and Miller, 2011), endosperm growth (CYP78A) (Schyman et al., 2010), and grain yield-related parameters (CYP714D1) (Yang et al., 2013). A pathway was proposed for TEF regulation traits through interaction with the Cdc73 component of the Paf1 complex in plants (Kubinski et al., 2006). By its methyltransferase action on histone, the Paf1 complex silences the expression of many genes, such as MAX1 members (Paul et al., 2010). MAX1 acts downstream of HTD1/MAX3 and D3/MAX4 to produce carotenoid-derived branch-inhibiting hormones, such as strigolactone and auxins (Grasser et al., 2009). In this study, overexpression of TaTEF-7A in A thaliana led to pleiotropic effects affecting vegetative development, number and lengths of siliques, and grain length compared with the wild type. Clearly, this gene needs further investigation. It was also shown that TaTEF-7A had a significant influence on wheat yield traits by association analysis; specific haplotypes of this gene were significantly associated with GN, an important component of grain yield. This effect was confirmed in a study of NILs, and the findings indicated that TaTEF-7A probably has pleiotropic effects on plant development, especially on young developing spikes and seed.
Potential application of a functional marker for TaTEF-7A in global wheat breeding for high GN
GN is an important yield trait that continues to attract the attention of wheat breeders. Genes contributing to high GN should be targets for selection in breeding. A significant difference in GN between Hap-7A-3 and other haplotypes indicated that it has been a target of global wheat breeding, especially in China (Figs 11, 12). Since 1950 wheat varieties have changed 4–6 times in China, with ~10% yield increases in each cycle (Zhuang, 2003). Yield increases in China have largely depended on higher TKW and GN (Zhang et al., 2012). As shown in Fig. 11, further increases in the frequency of Hap-7A-3 should lead to higher overall grain yields. Plant breeding through phenotypic selection has made considerable progress in this regard, but it is a time-consuming and relatively inefficient process (Gedye et al., 2012). Recently, MAS has provided a strategy for accelerating progress. Functional markers have been developed for plant height (Wu et al., 2011), vernalization response (Yan et al., 2004), photoperiod response (Beales et al., 2007), TKW (Jiang et al., 2011; Su et al., 2011), disease resistance (Gennaro et al., 2009), and grain quality (Wang et al., 2009). In this study, after functional verification in transgenic Arabidopsis of the yield-related gene TaTEF-7A, molecular markers based on variation in the promoter region of TaTEF-7A were developed to identify haplotype Hap-7A-3. Geographic distributions of Hap-7A-3 indicated that such a marker for increasing GN should have worldwide application.
Fig. 11.
TaTEF-7A haplotype frequencies and GN/spike changes over decades in Chinese modern wheat cultivars released during the 1940s to 1990s.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence characterization of TEF genes in wheat and other plant species.
Figure S2. Gene sequence of TaTEF.
Figure S3. cDNA sequences of TaTEF homoeologues.
Figure S4. PCR amplification of cDNA in nullisomic–tetrasomic lines of homoeologous group 7.
Figure S5. Linkage disequilibrium matrix among pairwise polymorphisms in the promoter region of TaTEF-7A.
Table S1. The 48 cultivars used for identification of haplotypes.
Acknowledgements
We thank R. Singh and A.L. Ordaz Cano, CIMMYT, Mexico, and M. Yang and Z.H. He, ICS-CAAS, Beijing, for gifts of the cultivar collections used in this study. Dr Kai-Jun Zhao, Institute of Crop Science, Chinese Academy of Agricultural Sciences, provided the pBICHP57 vector. We also gratefully acknowledge help from Professor Robert A. McIntosh, University of Sydney, with English editing. This work was supported by grants from the Chinese Ministry of Science and Technology (2014CB138103), the Animal and Plant Transgenic Project (2013ZX08009-001-001-002), the China Agricultural Research System (CARS-03-03B), NSF of Shanxi (2011021031-3), and CAAS-Innovation Team Project.
Glossary
Abbreviations:
- DH
double haploid
- ETN
effective tiller number
- GN
grain number per spike
- GUS
β-glucuronidase
- HD
heading date
- KL
kernel length
- KT
kernel thickness
- KW
kernel width
- LUC
luciferase
- MAS
marker-assisted selection
- MCC
mini core collection
- MD
maturity date
- NIL
near-isogenic line
- PH
plant height
- QTL
quantitative trait locus
- SL
spike length
- SN
spikelet number per spike
- SNP
single nucleotide polymorphism
- TEF
transcript elongation factor
- TKW
1000-kernel weight.
References
- Ahmed SS, Gong ZH, Ji JJ, Yin YX, Xiao HJ, Khan MA, Rehman A, Ahmad I. 2012. Construction of the intermediate vector pVBG2307 by incorporating vital elements of expression vectors pBI121 and pBI221. Genetics and Molecular Research 11, 3091–3104. [DOI] [PubMed] [Google Scholar]
- Barrero RA, Bellgard M, Zhang XY. 2011. Diverse approaches to achieving grain yield in wheat. Functional and Integrative Genomics 11, 37–48. [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. 2007. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics 115, 721–733. [DOI] [PubMed] [Google Scholar]
- Bernardo R. 1996. a Test cross additive and dominance effects in best linear unbiased prediction of maize single-cross performance. Theoretical and Applied Genetics 93, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Bernardo R. 1996. b Marker-based estimate of identity by descent and alikeness in state among maize inbreds. Theoretical and Applied Genetics 93, 262–267. [DOI] [PubMed] [Google Scholar]
- Bernardo R. 1996. c Best linear unbiased prediction of maize single cross performance. Crop Science 36, 50–56. [DOI] [PubMed] [Google Scholar]
- Bolot S, Abrouk M, Masood-Quraishi U, Stein N, Messing J, Feuillet C, Salse J. 2009. The ‘inner circle’ of the cereal genomes. Current Opinion in Plant Biology 12, 119–125. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lubberstedt T. 2010. Molecular basis of trait correlations. Trends in Plant Science 15, 454–461. [DOI] [PubMed] [Google Scholar]
- Clark RM. 2010. Genome-wide association studies coming of age in rice. Nature Genetics 42, 926–927. [DOI] [PubMed] [Google Scholar]
- da Costa e Silva O, Lorbiecke R, Garg P, et al. 2004. The Etched1 gene of Zea mays (L.) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. The Plant Journal 38, 923–939 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. 2002. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proceedings of the National Academy of Sciences, USA 99, 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gedye KR, Gonzalez-Hernandez JL, Owens V, Boe A. 2012. Advances towards a marker-assisted selection breeding program in prairie cordgrass, a biomass crop. International Journal of Plant Genomics 2012, 313545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro A, Koebner RM, Ceoloni C. 2009. A candidate for Lr19, an exotic gene conditioning leaf rust resistance in wheat. Functional and Integrative Genomics 9, 325–334. [DOI] [PubMed] [Google Scholar]
- Grasser KD. 2005. Emerging role for transcript elongation in plant development. Trends in Plant Science 10, 484–490. [DOI] [PubMed] [Google Scholar]
- Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD. 2009. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. Journal of Molecular Biology 386, 598–611. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu JK. 2002. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proceedings of the National Academy of Sciences, USA 99, 7786–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao CY, Wang LF, Ge HM, Dong YC, Zhang XY. 2011. Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One 6, e17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella L, Simpson J, Martinez-Trujillo M. 2005. Transgenic plants: an historical perspective. Methods in Molecular Biology 286, 3–32. [DOI] [PubMed] [Google Scholar]
- Hou J, Jiang QY, Hao CY, Wang YF, Zhang HN, Zhang XY. 2014. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiology 164, 1810–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, Somers DJ, Brown PD. 2006. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theoretical and Applied Genetics 113, 753–766. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Kempf H, Ganal MW, Roder MS. 2004. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theoretical and Applied Genetics 109, 933–943. [DOI] [PubMed] [Google Scholar]
- Jiang QY, Hou J, Hao CY, Wang LF, Ge HM, Dong YC, Zhang XY. 2011. The wheat (T. aestivum) sucrose synthase 2 gene (TaSus2) active in endosperm development is associated with yield traits. Functional and Integrative Genomics 11, 49–61. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D. 2002. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development 129, 1623–1632. [DOI] [PubMed] [Google Scholar]
- Kubinski K, Zielinski R, Hellman U, Mazur E, Szyszka R. 2006. Yeast elf1 factor is phosphorylated and interacts with protein kinase K2. Journal of Biochemistry and Molecular Biology 39, 311–318. [DOI] [PubMed] [Google Scholar]
- Kunert A, Naz AA, Dedeck O, Pillen K, Leon J. 2007. AB-QTL analysis in winter wheat: I. Synthetic hexaploid wheat (T. turgidum ssp. dicoccoides×T. tauschii) as a source of favourable alleles for milling and baking quality traits. Theoretical and Applied Genetics 115, 683–695. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei MP, Li GR, Liu C, Yang ZJ. 2012. Characterization of wheat: Secale africanum introgression lines reveals evolutionary aspects of chromosome 1R in rye. Genome 55, 765–774. [DOI] [PubMed] [Google Scholar]
- Li XP, Zhao XQ, He X, Zhao GY, Li B, Liu DC, Zhang AM, Zhang XY, Tong YP, Li ZS. 2011. Haplotype analysis of the genes encoding glutamine synthetase plastic isoforms and their association with nitrogen-use and yield-related traits in bread wheat. New Phytologist 189, 449–458. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Gronlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD. 2010. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2 . The Plant Journal 61, 686–697. [DOI] [PubMed] [Google Scholar]
- Lu L, Yan WH, Xue WY, Shao D, Xing YZ. 2012. Evolution and association analysis of Ghd7 in rice. PLoS One 7, e34021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZQ, Zhao DM, Zhang C, Zhang ZQ, Xue SL, Lin F, Kong ZX, Tian DG, Luo QY. 2007. Molecular genetic analysis of five spike-related traits in wheat using RIL and immortalized F2 populations. Molecular Genetics and Genomics 277, 31–42. [DOI] [PubMed] [Google Scholar]
- Mao XG, Jia DS, Li A, Zhang HY, Tian SJ, Zhang XK, Jia JZ, Jing RL. 2011. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis . Functional and Integrative Genomics 11, 445–465. [DOI] [PubMed] [Google Scholar]
- Narasimhamoorthy B, Gill BS, Fritz AK, Nelson JC, Brown-Guedira GL. 2006. Advanced backcross QTL analysis of a hard winter wheat×synthetic wheat population. Theoretical and Applied Genetics 112, 787–796. [DOI] [PubMed] [Google Scholar]
- Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, Dyer T, Lust S, Inze D, Van Lijsebettens M. 2003. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. The Plant Cell 15, 639–654.12615938 [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inze D, Van Lijsebettens M. 2005. The elongata mutants identify a functional elongator complex in plants with a role in cell proliferation during organ growth. Proceedings of the National Academy of Sciences, USA 102, 7754–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Awasthi A, Rai AK, Gupta SK, Prasad R, Sharma TR, Dhaliwal HS. 2012. Reduced tillering in Basmati rice T-DNA insertional mutant OsTEF1 associates with differential expression of stress related genes and transcription factors. Functional and Integrative Genomics 12, 291–304. [DOI] [PubMed] [Google Scholar]
- Quraishi UM, Abrouk M, Murat F, et al. 2011. Cross-genome map based dissection of a nitrogen use efficiency ortho-metaQTL in bread wheat unravels concerted cereal genome evolution. The Plant Journal 65, 745–756. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. 2008. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. The Plant Cell 20, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandee D, Miller WL. 2011. High-yield expression of a catalytically active membrane-bound protein: human P450 oxidoreductase. Endocrinology 152, 2904–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schyman P, Usharani D, Wang Y, Shaik S. 2010. Brain chemistry: how does P450 catalyze the O-demethylation reaction of 5-methoxytryptamine to yield serotonin? Journal of Physical Chemistry B 114, 7078–7089. [DOI] [PubMed] [Google Scholar]
- Sorrells ME, La Rota M, Bermudez-Kandianis CE, et al. 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Research 13, 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CJ, Via LE. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–750. [PubMed] [Google Scholar]
- Su ZQ, Hao CY, Wang LF, Dong YC, Zhang XY. 2011. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics 122, 211–223. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ET. 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics 28, 286–289. [DOI] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL. 2011. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences, USA 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, He XY, He ZH, Wang H, Xia XC. 2009. Cloning and phylogenetic analysis of phytoene synthase 1 (Psy1) genes in common wheat and related species. Hereditas 146, 208–256. [DOI] [PubMed] [Google Scholar]
- Wu J, Kong XY, Wan JM, et al. 2011. Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiology 157, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XY, Wang CL, Xie EB, Gao Y, Fan LY, Liu PQ, Zhao KJ. 2009. Molecular cloning and characterization of the promoter for the multiple stress-inducible gene BjCHI1 from Brassica juncea . Planta 229, 1231–1242. [DOI] [PubMed] [Google Scholar]
- Xue GP, Kooiker M, Drenth J, McIntyre CL. 2011. TaMYB13 is a transcriptional activator of fructosyltransferase genes involved in β-2,6-linked fructan synthesis in wheat. The Plant Journal 68, 857–870. [DOI] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. 2004. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theoretical and Applied Genetics 109, 1677–1686. [DOI] [PubMed] [Google Scholar]
- Yang WB, Gao MJ, Yin X, et al. 2013. Control of rice embryo development, shoot apical meristem maintenance, and grain yield by a novel cytochrome p450. Molecular Plant 6, 1945–1960. [DOI] [PubMed] [Google Scholar]
- Yang ZB, Bai ZY, Li XL, Wang P, Wu QX, Yang L, Li LQ, Li XJ. 2012. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theoretical and Applied Genetics 125, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Zhang DL, Hao CY, Wang LF, Zhang XY. 2012. Identifying loci influencing grain number by microsatellite screening in bread wheat (Triticum aestivum L.). Planta 236, 1507–1517. [DOI] [PubMed] [Google Scholar]
- Zhang D, Song HN, Cheng H, Hao DR, Wang H, Kan GZ, Jin HX, Yu DY. 2014. The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genetics 10, e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Liu DC, Guo XL, Yang WL, Sun JZ, Wang DW, Zhang AM. 2010. Genomic distribution of quantitative trait loci for yield and yield-related traits in common wheat. Journal of Integrative Plant Biology 52, 996–1007. [DOI] [PubMed] [Google Scholar]
- Zhuang QS. 2003. Chinese wheat improvemen tand pedigree analysis. Beijing: Agricultural Press; (in Chinese). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.