Abstract
The transcriptional regulation of toxin production in the gram-positive anaerobe Clostridium perfringens involves a two-component signal transduction system that comprises the VirS sensor histidine kinase and its cognate response regulator, VirR. Previous studies showed that VirR binds independently to a pair of imperfect direct repeats, now designated VirR box 1 and VirR box 2, located immediately upstream of the promoter of the pfoA gene, which encodes the cholesterol-dependent cytolysin, perfringolysin O. For this study, we introduced mutated VirR boxes into a C. perfringens pfoA mutant and found that both VirR boxes are essential for transcriptional activation. Furthermore, the spacing between the VirR boxes and the distance between the VirR boxes and the −35 region are shown to be critical for perfringolysin O production. Other VirR boxes that were previously identified from the strain 13 genome sequence were also analyzed, with perfringolysin O production used as a reporter system. The results showed that placement of the different VirR boxes at the same position upstream of the pfoA promoter yields different levels of perfringolysin O activity. In all of these constructs, VirR was still capable of binding to the target DNA, indicating that DNA binding alone is not sufficient for transcriptional activation. Finally, we show that the C. perfringens RNA polymerase binds more efficiently to the pfoA promoter in the presence of VirR, indicating that interactions must occur between these proteins. We propose that these interactions are required for VirR-mediated transcriptional activation.
The gram-positive anaerobe Clostridium perfringens is a causative agent of gas gangrene and food poisoning in humans (32, 33) and of several enterotoxemic diseases of domestic animals (42). It is characterized by its ability to produce many extracellular toxins and enzymes (33), including alpha-toxin (phospholipase C) and theta-toxin (perfringolysin O), which have been shown to act synergistically in gas gangrene (1, 2, 12, 44). The production of these toxins, as well as collagenase (kappa-toxin), sialidase (19), and alpha-clostripain (40), is regulated by a two-component signal transduction system that comprises the VirS sensor histidine kinase and the VirR response regulator. The mutation or inactivation of either the virS or virR gene alters the organism's ability to produce the various toxins and enzymes, with perfringolysin O and alpha-clostripain production being totally dependent on a functional VirS-VirR system (19, 37, 40). In addition to its role in extracellular toxin and enzyme production, the VirS-VirR regulatory network is also involved in the regulation of several housekeeping genes and the hyp7 gene, which encodes a regulatory RNA molecule, VR-RNA (6, 41).
The proposed model of the VirS-VirR cascade involves the detection of an as yet unidentified environmental or growth phase stimulus by the VirS protein, which then undergoes autophosphorylation at His-255. Phosphorylated VirS then acts as a phosphate donor for the phosphorylation of the conserved Asp-57 residue of VirR. Once activated, VirR then modulates the transcription of its target genes either directly or by altering the transcription of other regulatory genes, in particular by the action of VR-RNA (6, 7, 18, 41).
In previous work, we identified the VirR target DNA sequence as being a pair of imperfect direct repeats. These repeats, which are now designated VirR box 1 and VirR box 2, are located within a core 52-bp VirR-binding region situated immediately upstream of the promoter of the perfringolysin O structural gene pfoA (Fig. 1A). By performing in vitro binding studies, we showed that VirR binds to each of these repeats independently, i.e., binding to the VirR boxes is not cooperative (11). The identification of the VirR boxes (11), in conjunction with the sequencing of the C. perfringens strain 13 genome (38), has led to the identification of putative VirR-binding sites upstream of other genes (38), two of which have been demonstrated to be regulated by VirR (40, 41).
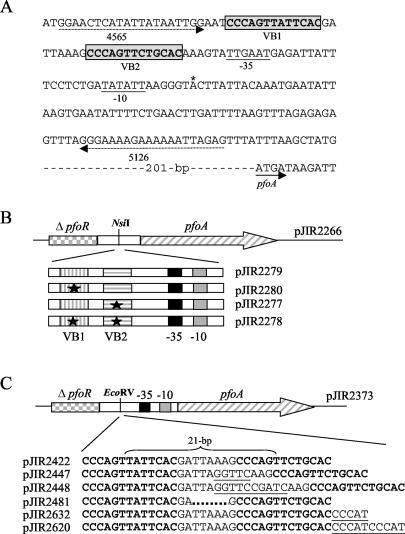
FIG. 1.
Sequence of the VirR binding site upstream of the pfoA gene and analysis of mutated VirR boxes. (A) The VirR boxes, labeled VB1 and VB2, are enclosed in gray boxes, and their sequences are shown in bold. The −35 and −10 boxes of the pfoA promoter are underlined, the transcription start point is indicated by an asterisk, and the start of the pfoA gene is indicated by a solid arrow. The locations of the oligonucleotide primers (4565 and 5126) used to amplify the DNA targets for the gel mobility shift assays are indicated by dashed arrows. (B) Cloning of mutated VirR boxes into pJIR2266. Inserts containing the wild-type VirR boxes or the various VirR box mutations were amplified with primers 14347 and 14348. The resultant PCR products were then cloned into the unique NsiI site in pJIR2266. This vector contains the pfoA gene (hatched arrow) and the 3′ end of the pfoR gene (checked rectangle). Each of the inserts harbored the −10 and −35 boxes, which are represented by gray and black boxes, respectively. The VirR boxes are shown as rectangles with vertical or horizontal stripes. The black stars indicate the mutated boxes, and the resultant plasmids are indicated to the right of each insert. (C) Cloning of VirR box cassettes into pJIR2373. Annealed complementary oligonucleotides containing the wild-type or mutated VirR boxes were inserted into the unique EcoRV site of pJIR2373. This vector contains the pfoA gene (hatched arrow), the −10 box (gray box), the −35 box (black box), and a truncated pfoR gene (checked rectangle). The VirR box sequences are shown in bold, while the inserted nucleotides are underlined. The deletion in the intervening region is indicated by dots. The resultant plasmids are shown to the left of each insert. The 21 bp separating the centers of the VirR boxes are indicated by a brace.
Although VirR has been shown to bind to the VirR boxes in vitro, the in vivo significance or role of each of these boxes is not known. Therefore, the main objective of this study was to examine the function of these binding sites in the native host, using perfringolysin O production as a reporter system. By introducing mutated VirR boxes back into a C. perfringens pfoA mutant, strain JIR4228 (3), we showed that both VirR boxes are required for the production of perfringolysin O. In addition, an alteration of the spacing between the VirR boxes also affects perfringolysin O production, revealing that the helical phasing of the binding sites as well as the distance separating the VirR boxes is important for transcriptional activation. Taken together, these results suggest that protein-protein interactions are essential for the transcriptional activation of the pfoA gene. Evidence for such interactions was obtained by gel mobility shift experiments with VirR and the C. perfringens RNA polymerase (CpRNAP).
MATERIALS AND METHODS
Strains, plasmids, and growth media.
The bacterial strains and plasmids used for this study are listed in Table 1. Escherichia coli strains were cultured at 37°C in 2× YT agar or broth or SOC broth (35) supplemented with ampicillin (100 μg ml−1) or erythromycin (150 μg ml−1). C. perfringens strains were grown at 37°C in Trypticase-peptone-glucose broth (34), brain heart infusion broth (Oxoid), fluid thioglycolate medium (Difco), or nutrient agar (31) supplemented with erythromycin (50 μg ml−1). For the screening of perfringolysin O production, C. perfringens transformants were grown on horse blood agar (19). All agar cultures of C. perfringens were incubated in an atmosphere of 10% (vol/vol) H2 and 10% (vol/vol) CO2 in N2.
TABLE 1.
Relevant strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 gyrA96 relA1 | Life Technologies |
| C43(DE3) | BL21(DE3) carrying an unknown spontaneous mutation | 22 |
| C. perfringens | ||
| JIR325 | Strain 13 Nalr Rifr | 19 |
| JIR4228 | JIR325 ΩTn916 (ΔpfoR pfoA colA luxS) | 3 |
| Plasmids | ||
| pJIR751 | C. perfringens-E. coli shuttle vector, Emr | 5 |
| pJIR1972 | pJIR751(EcoRI/SphI)Ω pTS302(EcoRI/SphI; 3.8 kb) (pfoR+pfoA+) | 4 |
| pTS302 | pUC19Ω(HindIII; C. perfringens; 4.3 kb) (pfoR+pfoA+) | 39 |
| pJIR2098 | pJIR751(SmaI)Ω 3732/14518 PCR product (HindIII/T4 polynucleotide kinase; 1.034 kb)(ΔpfoR) | This study |
| pJIR2266 | pJIR2098(NsiI/Asp718)Ω 14349/5460 PCR product (NsiI/Asp718; 1.9 kb) (ΔpfoR+pfoA+) | This study |
| pJIR2280 | pJIR2266(NsiI)Ω 14347/14348 PCR product (NsiI; 278 bp) (mutation in VirR box 1) | This study |
| pJIR2277 | pJIR2266(NsiI)Ω 14347/14348 PCR product (NsiI; 278 bp) (mutation in VirR box 2) | This study |
| pJIR2278 | pJIR2266(NsiI)Ω 14347/14348 PCR product (NsiI; 278 bp) (mutation in VirR box 1 and VirR box 2) | This study |
| pJIR2279 | pJIR2266(NsiI)Ω 14347/14348 PCR product (NsiI; 278 bp) (wild-type VirR boxes) | This study |
| pJIR2360 | pJIR2266(XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) (ΔpfoR+pfoA+) | This study |
| pJIR2356 | pJIR2280 (XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) (mutation in VirR box 1) | This study |
| pJIR2357 | pJIR2277 (XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) (mutation in VirR box 2) | This study |
| pJIR2358 | pJIR2278 (XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) (mutation in VirR box 1 and VirR box 2) | This study |
| pJIR2359 | pJIR2279 (XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) (wild-type VirR boxes) | This study |
| pJIR2346 | pJIR751(BamHI/SmaI)Ω 9373/17104 PCR product (T4 polynucleotide kinase/BamHI; 395 bp) (ΔpfoR) | This study |
| pJIR2361 | pJIR2346(EcoRV/Asp718)Ω 17116/17117 PCR product (EcoRV/Asp718; 1.866 kb) (ΔpfoR+pfoA+) | This study |
| pJIR2373 | pJIR2361(XbaI/PstI)Ω pJIR2224(XbaI/PstI; 2.0 kb) (Ω fragment+) | This study |
| pJIR2422 | pJIR2373(EcoRV)Ω 17443/17444 annealed complementary primers (EcoRV; 34 bp) (wild-type VirR boxes) | This study |
| pJIR2447 | pJIR2373(EcoRV)Ω 18705/18706 annealed complementary primers (EcoRV; 39 bp) (5-bp insertion between VirR boxes) | This study |
| pJIR2448 | pJIR2373(EcoRV)Ω 18707/18708 annealed complementary primers (EcoRV; 44 bp) (10-bp insertion between VirR boxes) | This study |
| pJIR2481 | pJIR2373(EcoRV)Ω 19211/19212 annealed complementary primers (EcoRV; 29 bp) (5-bp deletion between VirR boxes) | This study |
| pJIR2479 | pJIR2373(EcoRV)Ω 19190/19191 annealed complementary primers (EcoRV; 34 bp) (VirR boxes from upstream of CPE0845) | This study |
| pJIR2480 | pJIR2373(EcoRV)Ω 19206/19207 annealed complementary primers (EcoRV; 34 bp) (VirR boxes from upstream of CPE0920) | This study |
| pJIR2483 | pJIR2373(EcoRV)Ω 19192/19193 annealed complementary primers (EcoRV; 34 bp) (VirR boxes from upstream of CPE0846) | This study |
| pJIR2489 | pJIR2373(EcoRV)Ω 19208/19209 annealed complementary primers (EcoRV; 34 bp) (VirR boxes from upstream of hyp7) | This study |
| pJIR2632 | pJIR2373(EcoRV)Ω 20974/20975 annealed complementary primers (EcoRV; 39 bp) (5-bp insertion between VirR box 2 and −35 box) | This study |
| pJIR2620 | pJIR2373(EcoRV)Ω 20976/20977 annealed complementary primers (EcoRV; 44 bp) (10-bp insertion between VirR box 2 and −35 box) | This study |
Molecular techniques.
Plasmid DNA from E. coli cells was routinely isolated by an alkaline lysis method (23). When it was used for sequencing, DNA was obtained by a modified mini alkaline lysis-polyethylene glycol precipitation procedure outlined in the instructions for a PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems). Competent E. coli cells were prepared and transformed as described previously (15). Electrocompetent C. perfringens cells were prepared and transformed as described previously (36).
PCR amplification was performed with Taq DNA polymerase (Roche) or Pfu DNA polymerase (Promega) and a 0.5 μM concentration of each oligonucleotide primer (Table 2). Denaturation (94°C for 1 min), annealing (50°C for 2 min), and extension (72°C for 3 min) steps were carried out for 30 cycles. PCR products were purified by use of the Wizard PCR Preps DNA purification system (Promega). Nucleotide sequence analysis was performed as previously described (11).
TABLE 2.
Oligonucleotide primers
| Primer or use | Sequence (5′-3′) | Location or usea |
|---|---|---|
| PCR | ||
| 3732 | AATATGAAGTGCTTAGAAAG | pfoR promoter region |
| 14715 | CCAATTATAATATGCATTCCAT | Upstream of VirR boxes |
| 14349 | GAAAATGCATACTTAAAG | Upstream of pfoA |
| 5460 | CTCAGGTACCGAATCTAATACATGTAAACC | Downstream of pfoA |
| 14347 | TTTAAGTATGCATTTTCA | Downstream of VirR boxes |
| 14348 | CTCATGCATATTATAATTGG | Upstream of VirR boxes |
| 9373 | CGCGGATCCTGCTGGTTTTGCTGTAATGT | Within pfoR |
| 17104 | GATATCCCAATTATAATATGCATTCCATTTATG | Introduces EcoRV site upstream of VirR box 1 |
| 17116 | CCGGGTACCTACTTTAGTTTAATTGTA | Introduces Asp718 site at 5′ end of pfoA |
| 17117 | GATATCGTATTGAATGAGATTATTTCCTCTG | Introduces EcoRV site downstream of VirR box 2 |
| Cloning of VirR boxes | ||
| 17444 | CCCAGTTATTCACGATTAAAGCCCAGTTCTGCAC | Wild-type VirR boxes (+) |
| 17443 | GTGCAGAACTGGGCTTTAATCGTGAATAACTGGG | Wild-type VirR boxes (−) |
| 18705 | CCCAGTTATTCACGATTAGGTTCAAGCCCAGTTCTGCAC | 5-bp insertion between VirR boxes (+) |
| 18706 | GTGCAGAACTGGGCTTGAACCTAATCGTGAATAACTGGG | 5-bp insertion between VirR boxes (−) |
| 18707 | CCCAGTTATTCACGATTAGGTTCCGATCAAGCCCAGTTCTGCAC | 10-bp insertion between VirR boxes (+) |
| 18708 | GTGCAGAACTGGGCTTGATCGGAACCTAATCGTGAATAACTGGG | 10-bp insertion between VirR boxes (−) |
| 19211 | CCCAGTTATTCACGAGCCCAGTTCTGCAC | 5-bp deletion between VirR boxes (+) |
| 19212 | GTGCAGAACTGGGCTCGTGAATAACTGGG | 5-bp deletion between VirR boxes (−) |
| 19190 | CCCAGTTTAACATAAAAAATGACCAGTTATGCAC | VirR boxes upstream of CPE0845 (+) |
| 19191 | GTGCATAACTGGTCATTTTTTATGTTAAACTGGG | VirR boxes upstream of CPE0845 (−) |
| 19192 | ACCAGTTATGTATAAATTTTGACCAGTTATGCAA | VirR boxes upstream of CPE0846 (+) |
| 19193 | TTGCATAACTGGTCAAAATTTATACATAACTGGT | VirR boxes upstream of CPE0846 (−) |
| 19206 | CCCAATTATTCATAAAATATTGCCAGTTTTACAC | VirR boxes upstream of CPE0920 (+) |
| 19207 | GTGTAAAACTGGCAATATTTTATGAATAATTGGG | VirR boxes upstream of CPE0920 (−) |
| 19208 | CCCACTTTTACCTGTTTTTTGACCAGTTACGCAC | VirR boxes upstream of hyp7(+) |
| 19209 | GTGCGTAACTGGTCAAAAAACAGGTAAAAGTGGG | VirR boxes upstream of hyp7(−) |
| 20974 | CCCAGTTATTCACGATTAAAGCCCAGTTCTGCACCCCAT | 5-bp insertion between VirR box 2 and −35 box (+) |
| 20975 | ATGGGGTGCAGAACTGGGCTTTAATCGTGAATAACTGGG | 5-bp insertion between VirR box 2 and −35 box (−) |
| 20976 | CCCAGTTATTCACGATTAAAGCCCAGTTCTGCACCCCATCCCAT | 10-bp insertion between VirR box 2 and −35 box (+) |
| 20977 | ATGGGATGGGGTGCAGAACTGGGCTTTAATCGTGAATAACTGGG | 10-bp insertion between VirR box 2 and −35 box (−) |
| Gel mobility shifts | ||
| 4565 | GGAACTCATATTATAATTGG | Upstream of VirR boxes |
| 5126 | CTCTAATTTTTTCTTTTCCC | Downstream of VirR boxes |
| 4566 | TTTAAGTAAACATTTTCATC | Downstream of 5126 |
| 4826 | TTTGCCTTATAATTTATTTC | plc promoter region |
| 4824 | CTTTAGTTGATACCCCAGCCC | plc promoter region |
+, sense primer; −, antisense primer.
Construction of recombinant plasmid vectors.
Standard methods were used for the digestion, modification, ligation, and analysis of plasmid DNAs and restriction fragments (35). To facilitate the cloning of the various VirR box regions, we constructed pJIR2266. This plasmid contains the 3′ end of the upstream pfoR gene and the entire pfoA gene but lacks a 278-bp intergenic region that encompasses the VirR boxes and the pfoA promoter. The pfoR gene region was PCR amplified from wild-type strain JIR325 (Table 1) with oligonucleotides 3732 and 14715 (Table 2), of which the latter incorporated an NsiI site at the 3′ end of the PCR product. The PCR product was then digested with HindIII, filled in by use of T4 polynucleotide kinase, and cloned into pJIR751 (Table 1) to give pJIR2098, which carried the gene in the opposite orientation to that of the lac promoter. The pfoA gene region was PCR amplified from pJIR1972 (Table 1) with oligonucleotides 14349 and 5460 (Table 2). These primers incorporated NsiI and Asp718 sites at the 5′ and 3′ ends of the PCR product, respectively, and enabled the amplified fragment to be inserted into pJIR2098, to give pJIR2266. The missing 278-bp intergenic region containing the wild-type or mutated VirR boxes (16) was then PCR amplified with primers 14347 and 14348 (Table 2), both of which incorporated an NsiI site at the ends of the PCR product, and was inserted into the unique NsiI site of pJIR2266. The intergenic regions were derived from pJIR1546 (wild type), pJIR1804, pJIR1803, and pJIR1821 (mutations in VirR boxes 1, 2, and 1 and 2, respectively) (11). To prevent readthrough from external promoters, we inserted the Ω fragment (17, 27) (Table 1) upstream of the truncated pfoR region of all constructs. All constructs were sequenced to confirm that no mutations had been introduced by PCR.
The plasmid vector pJIR2373, which contains the 3′ end of the upstream pfoR gene and the entire pfoA gene, including the promoter, but lacks the VirR boxes in the intergenic region, was constructed as follows. The 395-bp pfoR gene region was PCR amplified from pTS302 (39) with primers 9373 and 17104 (Table 2), which incorporated BamHI and EcoRV sites at the 5′ and 3′ ends, respectively. The PCR product was treated with T4 polynucleotide kinase and digested with BamHI, and the fragment was cloned into the BamHI and SmaI sites of pJIR751, to give pJIR2346. The 1,866-bp pfoA gene region was PCR amplified with primers 17116 and 17117 (Table 2) to generate a product which contained EcoRV and Asp718 sites at the 5′ and 3′ ends, respectively. This fragment was then inserted into the EcoRV and Asp718 sites of pJIR2346, to give pJIR2361. The vector pJIR2373 was obtained by cloning the Ω fragment from pJIR2224 into the XbaI and PstI sites of pJIR2361 upstream of the truncated pfoR region.
Cloning of VirR boxes into pJIR2373.
Derivatives of the VirR box region were made by annealing two complementary oligonucleotides synthesized with the appropriate VirR box sequences. Each oligonucleotide was resuspended and diluted to a concentration of 100 μM in annealing buffer (30 mM Tris-HCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, pH 7.8) and phosphorylated with T4 polynucleotide kinase. Equimolar amounts of complementary primer pairs were annealed and then ligated into the EcoRV site of alkaline phosphatase-treated pJIR2373. All constructs were sequenced to ensure that only one VirR box cassette, with the correct sequence, had inserted in the desired orientation.
Perfringolysin O assays.
Perfringolysin O activity was determined by measuring the hemolysis of horse erythrocytes. Four-hour C. perfringens cultures and supernatants were obtained as described previously (1). Hemolysin assays were performed by a doubling dilution assay, as described previously (43). The titer was defined as the reciprocal of the last dilution that showed complete hemolysis, which was indicated by a significant decrease in absorbance. The unit of activity was expressed on a logarithmic scale as a log2 value (titer), and consequently each difference in titer of one unit represents a twofold difference in perfringolysin O activity.
Expression and purification of His-tagged VirR.
For protein expression, we used E. coli C43(DE3) cells (22) that had been freshly transformed to ampicillin resistance with pJIR1342 (Table 1). The culture (500 ml) was grown in 2× YT broth supplemented with ampicillin at 37°C with shaking until the turbidity at 600 nm reached approximately 0.5, and it was then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Progen) for 1 h. The His-tagged protein was then purified by the use of Talon resin (Clonetech) as described previously (11). Fractions containing highly purified VirR were pooled and dialyzed overnight in dialysis buffer (50 mM Tris-HCl, 0.3 M NaCl, 0.5 mM EDTA, 50% glycerol, pH 7.5) at 4°C and then were stored at −70°C until use.
Gel mobility shift assays.
Gel mobility shift assays using digoxigenin-11-ddUTP (DIG) (Roche Diagnostics)-labeled target DNAs were performed as described previously (11), with the exception that binding reactions were separated by electrophoresis in a native 0.5× TBE (44.5 mM Tris, 44.5 mM boric acid, 1.0 mM EDTA, pH 8.0)-6% polyacrylamide gel. Gel mobility shift assays using CpRNAP and E. coli RNA polymerase (EcRNAP) were performed as follows. The 278-bp pfoA (−98 to +180) and 394-bp plc (−252 to +142) promoter-containing fragments were amplified by PCRs with the primer pairs 4565-4566 and 4826-4824, respectively. These fragments were end labeled with T4 polynucleotide kinase (U.S. Biochemicals) and [γ-32P]ATP (3,000 Ci/mmol; Amersham). CpRNAP was kindly donated by S. Katayama and was prepared as described previously (16), while EcRNAP was purchased from Epicentre. Binding reactions were carried out in a total volume of 20 μl and contained 0.2 nM labeled DNA mixed with 0.01 U of EcRNAP or 0.01 U of CpRNAP alone or mixed with 1.8 pmol of VirR in the buffer used previously (11) or in RNA polymerase binding buffer [40 mM HEPES (pH 8.0), 100 mM KCl, 1 mM EDTA, 500 μg of bovine serum albumin, 0.033 mg of poly(dI-dC)/ml (Pharmacia Biotech)]. Reactions were incubated for 15 min at room temperature and then were immediately loaded into a 4.5% polyacrylamide gel prepared in TBE buffer. After electrophoresis for 2 h at 13 V/cm, the gel was dried and analyzed by autoradiography.
RESULTS
Both VirR boxes are essential for perfringolysin O production.
In previous studies, the VirR boxes were shown to be directly involved in VirR binding, such that when His-VirR was added to the target DNA, two shifted complexes, complex I (CI) and complex II (CII), were observed in gel mobility shift assays. A 52-bp core region of protection was observed in DNase I footprinting experiments. When the CCA residues of bases 2 to 4 of the VirR boxes were changed to TAG, either individually or together, we found that the alteration of either VirR box resulted in the formation of very little CII, whereas the modification of both VirR boxes almost eliminated VirR binding. These results clearly demonstrated that the VirR boxes were required for VirR binding, with CI representing VirR binding to one VirR box and CII representing VirR binding to both VirR boxes (11).
Since mutation of the VirR boxes had a significant effect on VirR binding, it was important to study the in vivo effects of these mutations on perfringolysin O production. The key question was whether binding to both VirR boxes was required for biological activity. These experiments had to be performed with multicopy plasmids since it is not practical to reproducibly introduce variants of a single gene region-reporter system at the same site on the C. perfringens chromosome. Therefore, for this experiment we cloned the 278-bp VirR box regions (−98 to +180 with respect to the transcription start site) containing the various altered VirR boxes and the −10 and −35 promoter regions into pJIR2266 (Table 1) and introduced the resultant constructs (Fig. 1B) into the C. perfringens pfoA mutant JIR4228. This strain was derived by Tn916 mutagenesis of the wild-type strain, JIR325, and screening for nonhemolytic perfringolysin O mutants. A Southern hybridization analysis of JIR4228 showed that the insertion of Tn916 caused the deletion of a region of the chromosome that encompassed the pfoR, pfoA, and colA genes (3). Since this pfoA deletion mutant was wild type for the virRS operon, it was ideal for use in the following studies, in which a pfoA gene with various VirR boxes was reintroduced into C. perfringens in a shuttle plasmid. In this strain background, the hemolytic activity on horse red blood cells is totally dependent on VirR activation of the plasmid-determined pfoA gene, and therefore a quantitative determination of perfringolysin O activity can be used to assess the biological activity of the interaction between VirR and the modified VirR boxes. In the absence of VirR or VirS, no perfringolysin O activity is detected (19, 37). Note that the upstream pfoR gene, although originally thought to be involved in the regulation of pfoA, is not required for pfoA expression (4).
When introduced into JIR4228, the E. coli-C. perfringens shuttle vector, pJIR751, and the cloning vector, pJIR2266, did not confer any detectable perfringolysin O activity (Table 3). This result was expected, since pJIR751 did not carry the pfoA gene and pJIR2266 had the pfoA gene but not the upstream VirR box-PpfoA region (Fig. 1B). In contrast, the strain harboring the complementation plasmid, pJIR1972, produced perfringolysin O. Similarly, when the wild-type VirR box-PpfoA region was introduced into pJIR2266, the resultant plasmid, pJIR2279, conferred levels of perfringolysin O activity that were similar to that of the positive control (Table 3). This result indicated that the incorporation of the NsiI site, which was introduced to facilitate the cloning of the various VirR box cassettes, had no effect on the VirR-mediated activation of pfoA transcription. Subsequent experiments showed that the introduction of cassettes that contained mutations in VirR box 1 (pJIR2280) or VirR box 2 (pJIR2277) significantly reduced perfringolysin O levels but did not completely eliminate the production of this toxin (Table 3). Finally, the level of perfringolysin O encoded by the plasmid with both VirR boxes mutated (pJIR2278) was the same as when either VirR box was altered. Taken together, these results show that although VirR can still bind to one VirR box when the other box is mutated, this binding is not sufficient to activate transcription of the pfoA gene and produce wild-type levels of perfringolysin O activity. We concluded that both VirR boxes need to be occupied for the efficient activation of perfringolysin O expression.
TABLE 3.
Effect of VirR box mutations on perfringolysin O activity
| Plasmida | Characteristics | PFO titer (log2)b |
|---|---|---|
| pJIR751 | Shuttle vector, negative control | <1.0 |
| pJIR1972 | pfoR+pfoA+, positive control | 5.6 ± 0.7 |
| pJIR2266 | Cloning vector | <1.0 |
| pJIR2279 | Wild-type VirR boxes | 5.7 ± 0.2 |
| pJIR2280 | Mutation in VirR box 1 | 2.3 ± 0.1 |
| pJIR2277 | Mutation in VirR box 2 | 2.1 ± 0.3 |
| pJIR2278 | Mutation in both VirR boxes | 2.1 ± 0.3 |
All plasmids were analyzed in strain JIR4228.
PFO titer refers to the mean perfringolysin O titer (± standard deviation) obtained from duplicate assays using supernatants from three independent cultures of each strain. Note that these titers are reported on a logarithmic, not arithmetic, scale.
Maintenance of correct spacing between VirR boxes is critical for transcriptional activation of pfoA.
The center of each VirR box is separated by 21 bp (Fig. 1C), which is equal to two turns of the DNA helix and implies that the VirR boxes are on the same side of the DNA helix. To examine whether helical phasing was important in transcriptional activation, we altered the VirR box regions such that either a 5-bp (half of a helical turn) or 10-bp (one helical turn) insertion or a 5-bp deletion was made in the spacer region separating the boxes (Fig. 1C). Since the VirR boxes and the intervening sequence only span a short section of DNA, the regions of interest were cloned into a different vector, pJIR2373, as annealed double-stranded oligonucleotides (Table 2). This vector still contained the pfoA gene and a truncated pfoR gene, but it differed from the vector used previously in that only the VirR boxes were absent from the intergenic region (Fig. 1C). The resultant constructs (Fig. 1C) were introduced into JIR4228, and hemolysin assays were performed to assess the effect on perfringolysin O production (Table 4).
TABLE 4.
Effect of altering DNA spacing and other VirR boxes on perfringolysin O activity
| Plasmida | Characteristics | PFO titer (log2)b |
|---|---|---|
| pJIR751 | Shuttle vector, negative control | <1.0 |
| pJIR1972 | pfoR+pfoA+, positive control | 5.9 ± 0.5 |
| pJIR2373 | Cloning vector | 1.4 ± 0.1 |
| pJIR2422 | Wild-type VirR boxes upstream of pfoA | 5.8 ± 0.5 |
| pJIR2447 | Insertion of 5 bp between VirR boxes | 1.3 ± 0.03 |
| pJIR2448 | Insertion of 10 bp between VirR boxes | 2.5 ± 0.1 |
| pJIR2481 | Deletion of 5 bp between VirR boxes | 1.4 ± 0.1 |
| pJIR2632 | Insertion of 5 bp between VirR box 2 and −35 box | 1.1 ± 0.4 |
| pJIR2620 | Insertion of 10 bp between VirR box 2 and −35 box | 1.3 ± 0.4 |
| pJIR2479 | VirR boxes from upstream of CPE0845 | 3.7 ± 0.3 |
| pJIR2483 | VirR boxes from upstream of CPE0846 | 7.6 ± 0.3 |
| pJIR2480 | VirR boxes from upstream of CPE0920 | 4.4 ± 0.1 |
| pJIR2489 | VirR boxes from upstream of hyp7 | 6.7 ± 0.2 |
All plasmids were analyzed in strain JIR4228.
PFO titer refers to the mean perfringolysin O titer (± standard deviation) obtained from duplicate assays using supernatants from three independent cultures of each strain. Note that these titers are reported on a logarithmic, not arithmetic, scale.
The results showed that a low level of perfringolysin O activity was observed with the strain carrying the cloning vector, pJIR2373, either as a result of residual VirR binding to regions outside the VirR box region or as a result of the capacity of RNA polymerase to bind weakly to the promoter region, which is located on a multicopy plasmid. In contrast, the vector plasmid with the inserted wild-type VirR boxes (pJIR2422) showed the same level of perfringolysin O activity as the positive control. The insertion (pJIR2447) or deletion (pJIR2481) of 5 bp in the intervening sequence between the VirR boxes resulted in a perfringolysin O titer that was similar to that of the cloning vector, indicating that these modifications were detrimental to transcriptional activation, and subsequently, to perfringolysin O production. However, the insertion of 10 bp between the VirR boxes (pJIR2448) significantly reduced perfringolysin O production, but did not reduce it to basal levels. These results indicated that both helical phasing and the spacing between the VirR boxes are critical for pfoA activation.
Maintenance of correct spacing between VirR box 2 and the promoter is critical for transcriptional activation of pfoA.
The VirR boxes are located 6 bp upstream of the −35 box of the pfoA promoter. To test the effect of altering the distance between the VirR boxes and the promoter, we introduced 5-bp and 10-bp insertions immediately downstream of VirR box 2 (Fig. 1C). The resultant constructs (Table 1) were introduced into JIR4228, and culture supernatants were assayed for perfringolysin O activity as described above. The results showed that the addition of either 5 bp (pJIR2632) or 10 bp (pJIR2620) at this position reduced perfringolysin O activity to basal levels (Table 4). These data suggest that the correct spacing between the VirR binding site and the promoter is also critical for biological activity.
VirR still binds to VirR box regions with altered helical phasing or DNA spacing.
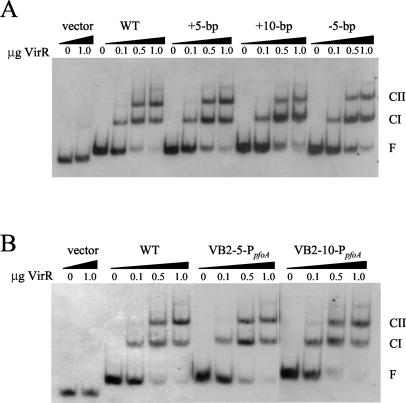
The reduction in perfringolysin O activity observed with the DNA spacing mutants may have been due either to VirR being unable to bind to the target binding sites or to the inability of the bound VirR proteins to activate the transcription of the pfoA gene. To distinguish between these possibilities, we performed gel mobility shift experiments using increasing amounts of purified VirR. For these assays, the different DNA targets were PCR amplified with primers 4565 and 5126 (Fig. 1A), purified, and subsequently labeled with DIG at the 3′ termini by the use of terminal transferase. The results showed that VirR could bind to the VirR boxes in a concentration-dependent manner to produce CI and CII, even when the VirR boxes were on opposite sides of the helix (Fig. 2A) or were further away from the −35 box of the promoter (Fig. 2B). Therefore, we conclude that the binding of VirR to the VirR boxes is not sufficient for in vivo transcriptional activation.
FIG. 2.
Gel mobility shift analysis of altered VirR box regions. Each 183-bp DIG-labeled DNA target was incubated in the presence of various amounts of purified VirR, as indicated above each lane. Note that 1 μg of VirR represents a final concentration of 1.55 μM. The free DNA (F), CI, and CII bands are labeled. The wedges above the lanes are used to distinguish the separate DNA targets. (A) WT, +5-bp, +10-bp, and −5-bp indicate wild-type VirR boxes and VirR boxes containing a 5- or 10-bp insertion or a 5-bp deletion, respectively. (B) WT, VB2-5-PpfoA, and VB2-10-PpfoA indicate wild-type VirR boxes and DNA targets containing a 5- or 10-bp insertion between VirR box 2 and the promoter, respectively.
VirR boxes found upstream of other VirR-regulated genes differentially activate gene expression.
The genome sequence of C. perfringens strain 13 was recently published, and VirR boxes were identified upstream of four additional open reading frames. Two of these open reading frames, CPE0845 and CPE0920, were found to encode hypothetical proteins, while CPE0846 and hyp7 encode the protease alpha-clostripain (40) and the regulatory RNA molecule, VR-RNA (6, 41), respectively. The expression of both CPE0846 and hyp7 has been demonstrated to be VirR dependent, and it is assumed that the expression of CPE0845 and CPE0920 is regulated by VirR in a similar manner (6, 40, 41).
An alignment of the various VirR boxes showed that the sequences of these boxes were similar, but not identical (38) (Fig. 3A). To determine whether these differences had an effect on gene expression, we synthesized the various VirR box regions, cloned them into pJIR2373 upstream of the pfoA gene, and used the resultant constructs to transform JIR4228 to erythromycin resistance. The perfringolysin O titers resulting from the various VirR boxes were different (Table 4). The VirR boxes upstream of CPE0845 (pJIR2479) and CPE0920 (pJIR2480) conferred less perfringolysin O activity than the equivalent pfoA VirR box region (pJIR2422). In contrast, higher levels of perfringolysin O activity were produced by derivatives carrying the VirR boxes from CPE0846 (pJIR2483) and hyp7 (pJIR2489). An analysis of the VirR box sequences did not reveal any obvious features that would determine the relative strength of the VirR-dependent transcriptional activation process.
FIG. 3.
(A) Alignment of the various VirR boxes. The VirR boxes are labeled and enclosed in gray boxes. The open reading frames found downstream of the VirR boxes are indicated to the left of the VirR box sequences. The conserved nucleotides in each VirR box are shown in bold, while the consensus sequence is shown below the alignment. Modified from reference 38. (B) Gel mobility shift analysis of VirR boxes upstream of different open reading frames in the C. perfringens genome. The DIG-labeled DNA targets containing the various VirR boxes were incubated with various amounts of VirR, as indicated above each lane. Note that 1 μg represents a final VirR concentration of 1.55 μM. The wedges above the lanes indicate the different DNA targets. The free DNA (F), CI, and CII bands are labeled.
To see if these differences in gene expression were related to the ability of VirR to bind to the various upstream sites, we performed gel mobility shift assays using DNA targets containing the different VirR boxes, as described above. The results demonstrated that VirR was able to bind to each of the VirR boxes, yielding CI and CII profiles that were comparable to that of the pfoA control (Fig. 3B). Under the conditions used, some variations in the amounts of DNA that were bound were observed (Fig. 3B), but these differences did not reflect the different levels of perfringolysin O activity that were obtained in the hemolysin assays. These results suggested that factors other than the binding of VirR to the VirR boxes, such as interactions with RNA polymerase, are also important for the transcriptional activation of downstream genes.
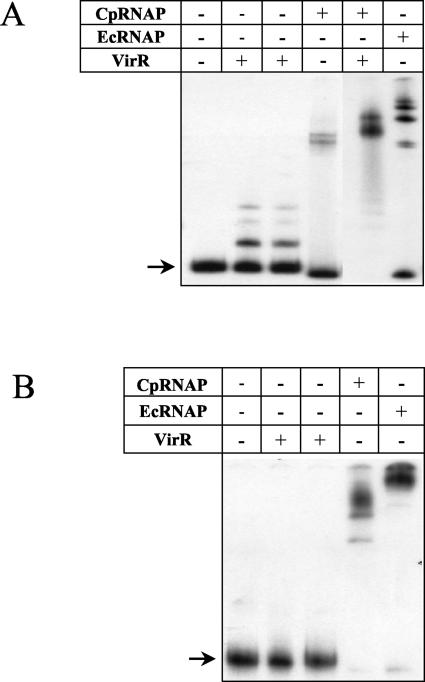
CpRNAP binds more efficiently to the pfoA promoter in the presence of VirR.
Evidence that VirR plays a role in the binding of RNA polymerase to the pfoA promoter was obtained from gel mobility shift experiments carried out with VirR and EcRNAP or CpRNAP (Fig. 4A). The results showed that when it was incubated in either VirR binding buffer (11) or RNA polymerase binding buffer, VirR was able to bind to a 278-bp fragment containing the VirR boxes and the pfoA promoter. Since less VirR was used in these experiments, some target DNA remained unbound. Both EcRNAP and CpRNAP were able to bind to the pfoA promoter region to produce shifted DNA-protein complexes of different sizes (Fig. 4A). However, while EcRNAP shifted the majority of the target DNA, CpRNAP could only bind and shift a minor portion of the target DNA. In contrast, when the same amount of VirR was mixed with CpRNAP and added to the binding reaction, all of the target DNA was bound, producing a shifted complex of a different size (Fig. 4A).
FIG. 4.
Gel mobility shift analysis of the pfoA (A) and plc (B) promoter regions. [γ-32P]ATP-labeled DNA fragments were incubated with various combinations of 1.8 pmol of VirR, 0.01 U of CpRNAP, and 0.01 U of EcRNAP, as indicated above each lane. All reactions were incubated in RNA polymerase buffer (see Materials and Methods), with the exception of the first binding reaction containing VirR alone. This reaction was incubated in a previously described buffer (11). Unbound target DNAs are indicated by arrows.
As a control, similar experiments were performed with the 394-bp plc promoter region, which does not contain any VirR boxes. The plc gene encodes alpha-toxin and has been found to be indirectly regulated by the VirS-VirR system via VR-RNA (6, 41). Previous in vitro binding experiments showed that VirR does not bind to this promoter region (11), as was observed in this study (Fig. 4B). CpRNAP was able to bind and shift all of the plc promoter target in the absence of VirR (Fig. 4B), demonstrating that CpRNAP alone is capable of binding to target DNA to give a complete shift. These results therefore indicate that although CpRNAP is capable of binding independently to the plc promoter, it requires VirR bound at the VirR boxes to bind efficiently to the pfoA promoter.
DISCUSSION
Previous studies (11) showed that the mutation of conserved sequences in either VirR box 1 or 2 altered the ability of the resultant boxes to act as a recognition sequence for the binding of the VirR response regulator. When VirR box 1 was mutated, DNase I footprinting showed that VirR was still able to bind to VirR box 2, and vice versa. We have now introduced these mutated VirR boxes back into the native C. perfringens host on a low-copy-number plasmid to assess the in vivo effect of the mutations on transcriptional activation. For these studies, we used a pfoA null mutant to develop a perfringolysin O reporter system that utilized the native gene target in the native host and therefore was ideal for use in studies aimed at the quantitative analysis of the effect of the upstream region on gene expression. With this system, we have now shown that both VirR boxes are required for VirR-mediated transcriptional activation and that mutation of either VirR box individually not only affects DNA binding (11) but also drastically reduces biological activity in C. perfringens.
When both VirR boxes were mutated, binding to the DNA target was almost eliminated (11). However, the perfringolysin O activity conferred by the plasmid carrying these mutations was very similar to that observed for mutations in only one binding site. Based on these results, we conclude that although VirR can bind to the VirR boxes independently, both VirR boxes must be bound for transcriptional activation to occur. We postulate that although binding at the VirR boxes is clearly not cooperative, the VirR proteins bound at VirR boxes 1 and 2 need to subsequently interact in a cooperative manner to both bind RNA polymerase and activate transcription. Evidence for this interaction was obtained in gel mobility shift assays, in which VirR bound to the VirR boxes was required for the efficient and specific binding of CpRNAP to the pfoA promoter. These results are in agreement with those of other studies that have shown that response regulators can interact with components of RNA polymerase (20, 26, 45, 46).
In all of the upstream regions of genes directly regulated by VirR, the VirR boxes are in the same relative positions with respect to the promoter and are on the same face of the helix. To examine whether the helical phasing or the DNA spacing played an important role in transcriptional activation by VirR, we altered the spacer region by a 5-bp insertion or deletion or a 10-bp insertion. Using a slightly different reporter plasmid but the same reporter strain and assay system, we showed that when the VirR boxes were on the opposite sides of the helix, perfringolysin O production was reduced to a basal level. When a 10-bp sequence was inserted into the intervening region between the VirR boxes, the resultant perfringolysin O activity was higher than that of the mutants containing the 5-bp insertion or deletion but was significantly lower than that of the wild type. We concluded that in the VirR system, proper helical phasing of the binding sites and the spacing between the VirR boxes both play a crucial part in transcriptional activation.
VirR was still able to bind efficiently to the VirR boxes in these spacer constructs, which provided further evidence that the binding of the VirR boxes is not cooperative, since VirR was still able to bind and form CII when the boxes were on opposite sides of the DNA helix. The binding data, in combination with the assay results, suggested that binding alone was not sufficient for transcriptional activation. We propose that the insertion or deletion of 5 bp from the intervening region would have placed the VirR boxes on opposite faces of the DNA helix and that when bound at these positions, the VirR proteins could not interact either with each other or with RNA polymerase, the result of which was the reduction of perfringolysin O activity to a basal level. Similarly, these proposed protein-protein interactions would have been affected by the insertion of 10 bp in the spacer region. Although in this construct the binding sites would be on the same face of the helix, we propose that the addition of 10 extra nucleotides would have altered the ability of the bound VirR molecules to form stable contacts either with each other or with RNA polymerase. It is also possible that the introduction of the extra nucleotides in the spacer region had an effect on the DNA conformation. DNase I footprinting experiments that were performed previously showed the appearance of hypersensitive sites when VirR was added to the binding reactions (11). In general, hypersensitive sites are indicative of localized distortions of the helix due to DNA bending (25). It is therefore possible that in the wild type, the binding of VirR at both of the VirR boxes induces localized DNA bending so that the bound proteins are then able to carry out protein-protein interactions. This hypothesis is consistent with a previous DNase I footprinting analysis of the VirR box mutants (11) by which the hypersensitive site profiles of the VirR box mutants were shown to be different from those of the wild-type construct. Taken together, the results of the DNase I footprinting and in vivo assays suggest that changes in the DNA conformation in the intervening region may be required to facilitate interactions between the bound VirR proteins or between those proteins and RNA polymerase.
The importance of the spatial organization of the VirR box region was also demonstrated when 5 or 10 bp was inserted between VirR box 2 and the six bases preceding the −35 box. The insertion of these sequences significantly reduced transcriptional activation by VirR, even when a full helical turn was inserted to restore the VirR boxes to their original phasing with respect to the −35 box. Given that VirR was found to be required for proper CpRNAP binding to the pfoA promoter region, we postulate that the maintenance of the correct spacing between the VirR boxes and the −35 box is crucial for interactions that may potentially occur between bound VirR and RNA polymerase.
Transcriptional initiation involves the recruitment of RNA polymerase and its binding to the promoter to form a closed complex, followed by isomerization, whereby the DNA strands are locally melted to facilitate the conversion of the closed complex to an open complex. Finally, to obtain full-length transcripts, the RNA polymerase must escape from the promoter region (13, 14, 30). Activators can exert an effect at each of these steps (13, 14), but most response regulators appear to be involved in the recruitment of RNA polymerase to the promoter. It has been postulated that protein-protein interactions between the activator and RNA polymerase stabilize the binding of the enzyme to the promoter (14, 30). For example, in the BvgA (8) and PhoB (21) systems, RNA polymerase is not able to bind to promoter DNA in the absence of the response regulator. Alternatively, activators such as NtrC (24) and NifA (10) stimulate the isomerization of bound RNA polymerase from a closed complex to an open complex (29). It is unlikely that VirR would act in the same fashion as NtrC and NifA, as these proteins bind well upstream of the promoter (9, 46). In contrast, the VirR boxes are located immediately upstream of the −35 box of the pfoA promoter region. Since we have shown that the binding of CpRNAP to the pfoA promoter increased upon the addition of VirR, it is more likely that the mode of action of VirR does not involve isomerization but a direct interaction to promote RNA polymerase binding to the promoter.
We have also shown that VirR is able to recognize and bind to VirR boxes other than those in the pfoA promoter region. When other VirR boxes that have been detected in the C. perfringens genome (38) were inserted upstream of pfoA, in lieu of the pfoA VirR boxes, we found that they conferred different levels of perfringolysin O activity in the reporter system. The results suggested that differences in the in vitro binding abilities were not a likely cause of the different levels of transcriptional activation that were observed. Differences in the VirR box regions may affect DNA bending, the interaction of RNA polymerase with the promoter, the putative contact between the bound VirR molecules, or interactions between VirR and RNA polymerase. All of these factors have been found to influence the promoter activities of other genes (25, 28).
In conclusion, based on the results presented in this paper and those obtained previously, we propose that direct VirR-mediated transcriptional activation in C. perfringens requires independent VirR binding to both VirR boxes. These binding sites must be located on the same face of the DNA helix, must be correctly spaced, and must be the correct distance from the −35 box of the promoter. Once bound to the different VirR boxes, the VirR proteins cooperatively lead to the recruitment of RNA polymerase to the promoter and subsequent transcriptional activation. Although further studies that involve the detailed analysis of the interaction of bound VirR molecules with RNA polymerase are required to determine the precise nature of these interactions, they are clearly critical components of this biologically important regulatory process.
Acknowledgments
This research was supported by grants from the Australian National Health and Medical Research Council.
We thank Sheena McGowan for helpful discussions and Sei-ichi Katayama for kindly providing C. perfringens RNA polymerase.
REFERENCES
- 1.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 2.Awad, M. M., D. M. Ellemor, R. L. Boyd, J. J. Emmins, and J. I. Rood. 2001. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect. Immun. 69:7904-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awad, M. M., and J. I. Rood. 1997. Isolation of α-toxin, θ-toxin and κ-toxin mutants of Clostridium perfringens by Tn916 mutagenesis. Microb. Pathog. 22:275-284. [DOI] [PubMed] [Google Scholar]
- 4.Awad, M. M., and J. I. Rood. 2002. Perfringolysin O expression in Clostridium perfringens is independent of the upstream pfoR gene. J. Bacteriol. 184:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:223-235. [DOI] [PubMed] [Google Scholar]
- 6.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 7.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher, P. E., and S. Stibitz. 1995. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J. Bacteriol. 177:6486-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck, M., W. Cannon, and J. Woodcock. 1987. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol. Microbiol. 1:243-249. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, W., and M. Buck. 1992. Central domain of the positive control protein NifA and its role in transcriptional activation. J. Mol. Biol. 225:271-286. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellemor, D. M., R. N. Baird, M. M. Awad, R. L. Boyd, J. I. Rood, and J. J. Emmins. 1999. Use of genetically manipulated strains of Clostridium perfringens reveals both alpha-toxin and theta-toxin are required for vascular leukostasis to occur in experimental gas gangrene. Infect. Immun. 67:4902-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiselmann, J. 1997. The role of DNA conformation in transcriptional initiation and activation in Escherichia coli. Biol. Chem. 378:599-607. [PubMed] [Google Scholar]
- 14.Hochschild, A., and S. L. Dove. 1998. Protein-protein contacts that activate and repress prokaryotic transcription. Cell 92:597-600. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 16.Katayama, S., O. Matsushita, C.-M. Jung, J. Minami, and A. Okabe. 1999. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 18:3442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koga, T., K. Ishimoto, and S. Lory. 1993. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect. Immun. 61:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyristis, M. 1996. Identification and molecular analysis of the virR/virS regulatory locus from Clostridium perfringens. Ph.D. thesis. Monash University, Melbourne, Australia.
- 19.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 20.Makino, K., M. Amemura, T. Kawamoto, S. Kimura, H. Shinagawa, A. Nakata, and M. Suzuki. 1996. DNA binding of PhoB and its interaction with RNA polymerase. J. Mol. Biol. 259:15-26. [DOI] [PubMed] [Google Scholar]
- 21.Makino, K., M. Amemura, S.-K. Kim, A. Nakata, and H. Shinagawa. 1993. Role of the σ70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 7:149-160. [DOI] [PubMed] [Google Scholar]
- 22.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 23.Morelle, G. 1989. A plasmid extraction procedure on a miniprep scale. Focus 11:7-8. [Google Scholar]
- 24.Ninfa, A. J., L. J. Reitzer, and B. Magasanik. 1987. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell 50:1039-1046. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Martín, J., F. Rojo, and V. De Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pratt, L. A., and T. J. Silhavy. 1994. OmpR mutants specifically defective for transcriptional activation. J. Mol. Biol. 243:579-594. [DOI] [PubMed] [Google Scholar]
- 27.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 28.Pruss, G. J., and K. Drlica. 1989. DNA supercoiling and prokaryotic transcription. Cell 56:521-523. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 30.Rhodius, V. A., and S. J. W. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 31.Rood, J. I. 1983. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can. J. Microbiol. 29:1241-1246. [DOI] [PubMed] [Google Scholar]
- 32.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 33.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rood, J. I., E. A. Maher, E. B. Somer, E. Campos, and C. L. Duncan. 1978. Isolation and characterization of multiple antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob. Agents Chemother. 13:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, second ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Scott, P. T., and J. I. Rood. 1989. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene 82:327-333. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu, T., A. Okabe, J. Minami, and H. Hayashi. 1991. An upstream regulatory sequence stimulates expression of the perfringolysin O gene of Clostridium perfringens. Infect. Immun. 59:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, T., K. Shima, K. Yoshino, K. Yonezawa, T. Shimizu, and H. Hayashi. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 184:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu, T., H. Yaguchi, K. Ohtani, S. Banu, and H. Hayashi. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43:257-265. [DOI] [PubMed] [Google Scholar]
- 42.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens, D. L., J. Mitten, and C. Henry. 1987. Effects of α and θ toxins from Clostridium perfringens on human polymorphonuclear leukocytes. J. Infect. Dis. 156:324-333. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, D. L., R. K. Tweten, M. M. Awad, J. I. Rood, and A. E. Bryant. 1997. Clostridial gas gangrene: evidence that α and θ toxins differentially modulate the immune response and induce acute tissue necrosis. J. Infect. Dis. 176:189-195. [DOI] [PubMed] [Google Scholar]
- 45.Stibitz, S. 1998. Mutations affecting the α subunit of Bordetella pertussis RNA polymerase suppress growth inhibition conferred by short C-terminal deletions of the response regulator BvgA. J. Bacteriol. 180:2484-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su, W., S. Porter, S. Kustu, and H. Echols. 1990. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA. 87:5504-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]