Abstract
Thalassospira bacteria are widespread and have been isolated from various marine environments. Less is known about their genetic diversity and biogeography, as well as their role in marine environments, many of them cannot be discriminated merely using the 16S rRNA gene. To address these issues, in this report, the phylogenetic analysis of 58 strains from seawater and deep sea sediments were carried out using the multilocus sequence analysis (MLSA) based on acsA, aroE, gyrB, mutL, rpoD and trpB genes, and the DNA-DNA hybridization (DDH) and average nucleotide identity (ANI) based on genome sequences. The MLSA analysis demonstrated that the 58 strains were clearly separated into 15 lineages, corresponding to seven validly described species and eight potential novel species. The DDH and ANI values further confirmed the validity of the MLSA analysis and eight potential novel species. The MLSA interspecies gap of the genus Thalassospira was determined to be 96.16–97.12% sequence identity on the basis of the combined analyses of the DDH and MLSA, while the ANIm interspecies gap was 95.76–97.20% based on the in silico DDH analysis. Meanwhile, phylogenetic analyses showed that the Thalassospira bacteria exhibited distribution pattern to a certain degree according to geographic regions. Moreover, they clustered together according to the habitats depth. For short, the phylogenetic analyses and biogeography of the Thalassospira bacteria were systematically investigated for the first time. These results will be helpful to explore further their ecological role and adaptive evolution in marine environments.
Introduction
Thalassospira are a genus consisting of Gram-negative, motile, vibrio- or spiral-shaped, halotolerant and chemoheterotrophic bacteria belonging to the family Rhodospirillaceae within the class Alphaproteobacteria and was created by López-López et al in 2002 [1]. In the following years, more and more isolates of Thalassospira were obtained from various marine environments and identified using the polyphasic taxonomy. Currently, the Thalassospira genus has accommodated eight species: T. lucentensis [1], T. profundimaris [2], T. xiamenensis [2], T. tepidiphila [3], T. xianhensis [4], T. alkalitolerans and T. mesophila [5], T. povalilytica [6], except a non-validly published species named T. permensis [7].
We found that bacteria of Thalassospira were widespread in the marine environments and occupied a variety of ecological niches, such as surface and deep seawater, deep sediment, halobios etc, covering the Pacific Ocean, the Atlantic Ocean, the Indian Ocean and even the Arctic Ocean [8]–[11]. They aroused extensive attention because of their potential in eliminating marine oil pollution, especially in polycyclic aromatic hydrocarbons (PAHs) degradation [3], [4], [7], [12]. Recently, Thalassospira were found to closely related with the phosphorus cycling in marine environments, especially in the oligotrophic open ocean environments [13], [14]. Additionally, some strains of this genus can produce thalassospiramides [15], [16], beta-galactosidase [17] and biosurfactants [18]. Accumulation of these bacteria with different functions and originations urges analysis on their diversity, evolution and geographic distribution.
In recent years, a large number of bacteria within the genus Thalassospira were isolated from various marine samples enriched with different hydrocarbons, including hexadecane, BTEX (benzene, toluene, ethylbenzene and xylene), and crude oil in our laboratory. Nevertheless, most of the Thalassospira bacteria shared relatively high identities to the 16S rRNA gene (>98%) with each other, with only one exception of T. lucentensis DSM 14000T, thus they could not be classified based upon the 16S rRNA gene. Therefore, an effective method is imperative to discriminate these closely related strains.
Since 2005, MLSA has rapidly become a powerful tool and has been performed frequently for the taxonomy and phylogenetic analysis of the closely related strains [19]–[21]. In general, the several housekeeping genes are widely applied in MLSA schemes, such as acsA, gyrB, mutL, rpoD, etc [22]–[25], because they encode core metabolic enzymes and own by all members of the organisms. Furthermore, the concatenated housekeeping genes could overcome conflicting signals from horizontal gene transfer and recombination. In addition, DNA fingerprinting [26], [27], DNA-DNA hybridization (DDH) [21], [23] and average nucleotide identity (ANI) [28], [29] are dependable methods for identification of the closely related strains.
In this study, we employed MLSA using six housekeeping genes, acsA, aroE, gyrB, mutL, rpoD and trpB, combined with the 16S rRNA gene, DDH and ANI values from draft genome sequences, to explore the diversity, and geographic distribution of the Thalassospira bacteria originating from diverse marine environments.
Materials and Methods
Ethics Statement
The bacterial strains in this study were isolated from the various areas beyond the international sea area or the exclusive economic zone of China. The type strains were obtained from the public culture collections. So no specific permissions were required for these collections. Moreover, the sample did not involve endangered or protected species.
Bacterial strains
A total of 58 Thalassospira strains were analyzed in this study. Specifically, 52 Thalassospira strains were isolated in our laboratory and deposited at the Marine Culture Collection of China (MCCC), including two type strains, T. xiamenensis M-5T and T. profundimaris WPO0211T. T. tepidiphila 1-1BT was generous gift from Dr. Kazuya Watanabe [3]; T. lucentensis DSM 14000T, T. xianhensis CGMCC 1.6849T, T. alkalitolerans JCM 18968T, T. mesophila JCM 18969T and T. permensis NBRC 106175T (non-valid) were purchased from DSMZ, CGMCC, JCM and NBRC, respectively. The detail information of all strains was presented in Table 1. They were isolated from 32 different locations across the Pacific Ocean, the Atlantic Ocean, the Indian Ocean, the Yellow Sea, the East China Sea and the Mediterranean Sea. The sampling sites were shown in Figure S1. At the time of this study, type strain T. povalilytica Zumi 95T was not reported and therefore not included in our study.
Table 1. The 58 Thalassospira bacteria in this study.
| MCCC NO. | Original No. | Closest type strain of 16S rRNA gene (Identity %) | Geographic origin | Source | Depth (m) |
| 1A00207 | WP0211T | T. profundimaris | The Pacific Ocean | Sediment | 4,480 |
| 1A00209 | M-5T | T. xiamenensis | The Taiwan strait | Superficial seawater | 0 |
| 1A00350 | R8-17 | T. profundimaris (99.7) | The Indian Ocean | Deep seawater | 668 |
| 1A00370 | R8-8 | T. profundimaris (99.4) | The Indian Ocean | Deep seawater | 668 |
| 1A00383 | QMT2T | T. lucentensis | The Mediterranean Sea | Upper seawater | 100 |
| 1A00385 | R4-5 | T. profundimaris (99.4) | The Indian Ocean | Deep seawater | 2,268 |
| 1A00624 | SMB34T | T. permensis | Berezniki, Perm region, Russia | Soil | 0 |
| 1A00753 | MBE#61T | T. alkalitolerans | The coastal area, Japan | Sunken bamboo | 0 |
| 1A00756 | MBE#74T | T. mesophila | The coastal area, Japan | Sunken bamboo | 0 |
| 1A01013 | W3-1 | T. xiamenensis (99.4) | The Pacific Ocean | Sediment | 2,682 |
| 1A01017 | DBT-2 | T. xiamenensis (99.4) | The Pacific Ocean | Sediment | 2,682 |
| 1A01041 | PTG4-18 | T. xiamenensis (99.5) | The Indian Ocean | Sediment | 2,946 |
| 1A01051 | MARC2PI | T. xiamenensis (99.5) | The Atlantic Ocean | Sediment | 3,542 |
| 1A01057 | MARC4CW | T. profundimaris (99.7) | The Atlantic Ocean | Sediment | 3,962 |
| 1A01072 | MARC2COD | T. xiamenensis (99.5) | The Atlantic Ocean | Sediment | 3,542 |
| 1A01103 | PB8B | T. profundimaris (99.6) | The Indian Ocean | Deep seawater | 1,800 |
| 1A01109 | PB9B | T. profundimaris (99.4) | The Indian Ocean | Deep seawater | 1,800 |
| 1A01140 | MARMC3G | T. xiamenensis (99.5) | The Atlantic Ocean | Sediment | 4,046 |
| 1A01148 | MARC2CO7 | T. profundimaris (99.7) | The Atlantic Ocean | Sediment | 3,542 |
| 1A01166 | 35 | T. profundimaris (99.4) | The Indian Ocean | Sediment | 2851 |
| 1A01167 | 78 | T. profundimaris (99.4) | The Indian Ocean | Sediment | 2,851 |
| 1A01172 | MARC2PPNC | T. profundimaris (99.4) | The Atlantic Ocean | Sediment | 3,542 |
| 1A01275 | MC2-14 | T. profundimaris (99.7) | The Atlantic Ocean | Deep seawate | 3,542 |
| 1A01288 | S31-2-1 | T. profundimaris (98.3) | The Indian Ocean | Superficial seawater | 0 |
| 1A01300 | S27-11 | T. xiamenensis (99.5) | The Indian Ocean | Superficial seawater | 0 |
| 1A01318 | S25-3-2 | T. profundimaris (98.3) | The Indian Ocean | Superficial seawater | 0 |
| 1A01330 | S29-3-A | T. xiamenensis (99.5) | The Indian Ocean | Superficial seawater | 0 |
| 1A01423 | S25-4 | T. profundimaris (98.3) | The Indian Ocean | Superficial seawater | 0 |
| 1A01448 | S31-7 | T. xiamenensis (99.5) | The Indian Ocean | Superficial seawater | 0 |
| 1A01449 | S31-6 | T. profundimaris (98.3) | The Indian Ocean | Superficial seawater | 0 |
| 1A02030 | PR54-5 | T. profundimaris (99.5) | The Indian Ocean | Deep seawater | 4,146 |
| 1A02031 | 2CR55-14 | T. profundimaris (99.5) | The Indian Ocean | Deep seawater | 3,946 |
| 1A02039 | PR57-5 | T. profundimaris (99.7) | The Indian Ocean | Deep seawater | 3,546 |
| 1A02040 | PR57-2 | T. profundimaris (99.7) | The Indian Ocean | Deep seawater | 3,546 |
| 1A02041 | 2CR55-15 | T. profundimaris (99.6) | The Indian Ocean | Deep seawater | 3,946 |
| 1A02042 | 2CR-54-5 | T. profundimaris (99.6) | The Indian Ocean | Deep seawater | 4,146 |
| 1A02059 | NIC1013S-2 | T. profundimaris (99.4) | The Indian Ocean | Deep seawater | 2,455 |
| 1A02060 | RC911-4 | T. profundimaris (99.4) | The Indian Ocean | Deep seawater | 668 |
| 1A02093 | PC99-15 | T. profundimaris (99.7) | The Indian Ocean | Deep seawater | 1,068 |
| 1A02094 | MC2-9 | T. xiamenensis (99.5) | The Atlantic Ocean | Overlaid seawater | 3,542 |
| 1A02096 | PC92-18 | T. profundimaris (99.7) | The Indian Ocean | Deep seawater | 2,468 |
| 1A02616 | P-4T | T. xianhensis | Xianhe town, China | Soil | 0 |
| 1A02753 | IB2 | T. xiamenensis (99.4) | Yellow Sea, China | Upper seawater | 10 |
| 1A02758 | IB13 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 10 |
| 1A02767 | ID7 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 5 |
| 1A02785 | IH1 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 5 |
| 1A02803 | IK1 | T. profundimaris (100) | Yellow Sea, China | Upper seawater | 40 |
| 1A02843 | IP8 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 30 |
| 1A02866 | IU14 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 30 |
| 1A02873 | IV17 | T. xiamenensis (99.2) | Yellow Sea, China | Upper seawater | 40 |
| 1A02878 | IX2 | T. xiamenensis (99.4) | Yellow Sea, China | Upper seawater | 50 |
| 1A02898 | JB7 | T. profundimaris (100) | Yellow Sea, China | Upper seawater | 2.5 |
| 1A02921 | JG3 | T. xiamenensis (99.4) | East China Sea | Upper seawater | 30 |
| 1A02935 | JK1 | T. xiamenensis (99.2) | East China Sea | Upper seawater | 30 |
| 1A03005 | L6 | T. xiamenensis (99.5) | The Atlantic Ocean | Sediment | 3,962 |
| 1A03052 | AS-I2-11 | T. xiamenensis (99.5) | The Indian Ocean | Sediment | 1,420 |
| 1A03093 | AS-M6-11 | T. xiamenensis (99.5) | The Atlantic Ocean | Sediment | 2,987 |
| 1A03514 | 1-1BT | T. tepidiphila | Kamaishi Bay, Japan | Upper seawater | 15 |
Cultivation and DNA preparation
All strains were incubated on 216 L agar medium (CH3COONa 1.0 g, tryptone 10.0 g, yeast extract 2.0 g, sodium citrate 0.5 g, NH4NO3 0.2 g, agar 15 g, 1 L sea water, pH 7.5) [30] at 28°C for 48 h. Genomic DNA was extracted using the SBS extraction kit (SBS Genetech Co., Ltd. in Shanghai, China) following the manufacturer's instructions.
PCR amplification and sequencing of 16S rRNA and six housekeeping genes
The 16S rRNA gene was amplified using universal primers 27F and 1492R, while six housekeeping genes were amplified, respectively, with specific primers designed in light of the genome sequences of the six type strains using the Primer Premier 5.0 (Table S1). These genes were amplified under nearly identical conditions. The 50 µL reaction mixtures contained 37 µL of double distilled water, 5 µL of 10×Ex Taq buffer (Mg2+ Plus), 4 µL of dNTP mixture (10 mM), 1 µL of each primer (20 µM), 1 µL of genomic DNA template (10–30 ng/μL), 1 µL of Ex Taq™ DNA polymerase (TaKaRa, 5 U/μL). The reaction mixture was subjected to the following parameters in a Biometra T-Professional thermocycler (Biometra; Goettingen, Germany): initial denaturation at 94°C for 5 min; then 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 1.0–1.5 min of extension at 72°C; and a final extension at 72°C for 10 min. More detail information was listed in Table S1. After amplification, these PCR amplicons were separated by electrophoresis on a 1% agarose gel and then purified using the PCR purification kit (Axygen Scientific, Inc., USA) according to the manufacturer's instructions. Finally, the purified amplicons were sequenced with the ABI3730xl platform (Shanghai Majorbio Bio- Pharm Technology Co., Ltd., China) using the corresponding sequencing primers (Table S1).
The sequences of the 16S rRNA gene and six housekeeping genes were assembled and modified using DNAMAN version 5.0. All sequences were submitted to the GenBank database. The accession numbers were assigned and listed in Table S2.
Analysis of nucleotide diversity
The determined sequences of the 16S rRNA gene and six housekeeping genes were compared to the National Center of Biotechnology Information (NCBI) database using BLAST. For each housekeeping gene, different alleles were assigned successively different numbers, and a unique combination of 6 allele numbers (allelic profile) unambiguously was defined the sequence type (ST) of a bacterium. When several strains shared the same allelic profiles, they unquestionably possessed the same ST. A total of allelic profiles were utilized for subsequent analysis. Meantime, the genetic distances and sequence similarities of the single gene were calculated using Kimura's 2-parameter model [31] with the MEGA version 5.0 [32]. The numbers of polymorphic sites and the mean G+C (guanine-cytosine) content in each gene were obtained by the MEGA version 5.0. The nucleotide diversity (π) per site for each gene was performed using the data analysis in molecular biology and evolution (DAMBE) version 5.0 [33]. In addition, the selective pressure on six housekeeping genes were evaluated with the ratios of Ka/Ks (Ka: the number of non-synonymous substitutions per non-synonymous site, Ks: the number of synonymous substitutions per synonymous site) using the MEGA version 5.0.
Population genetic analyses
The assessment of substitution saturation for each gene was determined using the DAMBE version 5.0. In order to measure the possibility of reticulate evolution, split decomposition analysis was performed for all single gene and the concatenated genes using the neighbor-net algorithm in SplitsTree version 4.0 by default settings [34]. The possible existence of the recombination in all genes was estimated using the pairwise homoplasy index (PHI) test as implemented in SplitsTree version 4.0. Meantime, the level of Linkage disequilibrium, as measured by the standardized index of association (IS A), were investigated using the LInkage ANalysis (LIAN) version 3.6 (http://guanine.evolbio.mpg.de/cgi-bin/lian/lian.cgi.pl/query) [35]. In addition, radiation times were estimated with each other based on the rate of synonymous substitution for single housekeeping genes and the concatenated housekeeping genes. These rates were normalized with the known radiation time between Escherichia coli and Salmonella enterica (ca. 1 million years) [36].
Phylogenetic analysis based on the 16S rRNA gene, individual housekeeping gene, the six concatenated housekeeping genes and DDH
The phylogenetic analysis was performed using MEGA version 5.0. The sequences of the 16S rRNA gene and six housekeeping genes were aligned using ClustalW option implemented in MEGA. A series of phylogenetic trees based on the 16S rRNA gene and single housekeeping gene were constructed respectively using the neighbour-joining algorithm by Kimura's 2-parameter model with the MEGA version 5.0. A phylogenetic tree based on the concatenated gene (in the following order: acsA, aroE, gyrB, mutL, rpoD and trpB) was also constructed. Rhodospirillum rubrum ATCC 11170T was used as the outgroup in all phylogenetic analysis. Bootstrap confidence analysis was carried out with 1000 replications for evaluating the robustness of the tree topologies. Additionally, phylogenies from distance matrix by neighbour-joining method the clustering tree based on the DDH was constructed with the web server (http://genome.csdb.cn/cgi-bin/emboss/fneighbor).
Correlation analysis between similarities of the MLSA, DDH and ANI
The genome sequences of sixteen representative Thalassospira strains selected based on the phylogenetic analysis were determined by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China), using Solexa paired-end (500 bp library) sequencing technology. A total of 500 M bp clean data for each strain was generated to reach about 100-fold depth of coverage with an Illumina/Solexa Genome Analyzer IIx (Illumina, SanDiego, CA). The clean data was assembled by SOAPdenovo2 [37]. Two of them (T. profundimaris WP0211T and T. xiamenensis M-5T) had been reported by our lab [38], [39]. DDH values were estimated using the genome-to-genome distance calculator website service (GGDC 2.0) [40], [41]. ANI values of these strains were calculated using the software JSpecies (V1.2.1) [29]. Correlation analysis between similarities of the MLSA and DDH was simulated using the R version 3.0.1.
Results
Individual gene analyses
The sequences of the 16S RNA gene and the six housekeeping genes of 58 strains were determined. The characteristics of gene(s) were listed in Table 2.
Table 2. Characteristics of the 16S rRNA gene, single housekeeping gene and the concatenated genes from 58 strains.
| Locus | Length (bp) | No. of Alleles | No. (%) of Polymorphic sites | π | Mean G+C content (mol %) | K2P distance range | K2P distance mean | Ka/Ks |
| 16S rDNA | 1,444–1,449 | 20 | 64 (4.42–4.43) | 0.012 | 55.1 | 0.000–0.027 | 0.009 | NA |
| acsA | 858 | 24 | 371 (43.2) | 0.180 | 53.4 | 0.000–0.247 | 0.170 | 0.035 |
| aroE | 663 | 25 | 354 (53.4) | 0.192 | 54.7 | 0.000–0.345 | 0.190 | 0.099 |
| gyrB | 918 | 25 | 368 (40.1) | 0.135 | 56.1 | 0.000–0.218 | 0.124 | 0.052 |
| mutL | 726 | 25 | 298 (40.0) | 0.148 | 60.8 | 0.000–0.215 | 0.140 | 0.039 |
| rpoD | 510 | 25 | 179 (35.1) | 0.119 | 53.1 | 0.000–0.192 | 0.111 | 0.060 |
| trpB | 738 | 28 | 280 (38.0) | 0.140 | 57.2 | 0.000–0.221 | 0.134 | 0.049 |
| MLSA | 4,413 | 31 | 1,850 (41.9) | 0.148 | 56.0 | 0.000–0.220 | 0.138 | NA |
Nucleotide diversity (π): the average number of nucleotide differences per site between two randomly-selected strains.
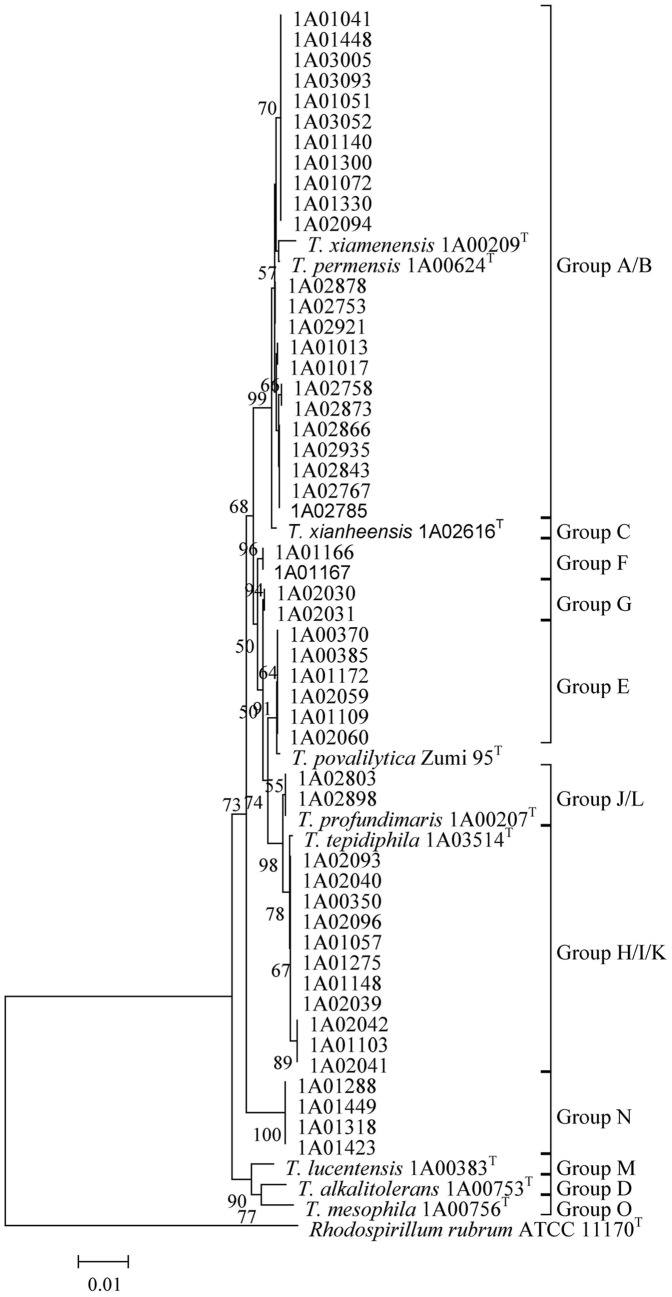
The length of the 16S RNA gene was 1,444 bp, except for three strains named MCCC 1A00383, MCCC 1A00753 and MCCC 1A00753 (1,449 bp). The number of alleles and polymorphic sites of the 16S RNA gene were 20 and 64, respectively; while the proportion of polymorphic sites and nucleotide diversity were 4.42% (4.43%) and 0.012, respectively. The mean G+C content was 55.1 mol%. The range of genetic distance of the 16S RNA gene was 0.000–0.027 (mean 0.008), corresponding to 97.3–100% identity. Measure substitution saturation of the 16S rRNA gene with DAMBE indicated that little saturation existed. The highly conserved 16S rRNA gene was unsuitable for distinguishing 58 Thalassospira strains. Despite the low resolution of the 16S rRNA gene, 58 strains were still divided into 11 groups labeled with the group name A to O (Figure 1), the same group names used as in the MLSA phylogenetic tree. In brief, the largest Group A/B contained 25 strains with two type strains, T. xiamenensis M-5T and T. permensis NBRC106175T (non-valid). The second Group H/I/K consisted of 12 isolates including the type strains T. tepidiphila 1-1BT. The Group J/L consisted of 3 isolates including the type strain T. profundimaris WPO0211T. Three type strains, T. lucentensis DSM 14000T, T. alkalitolerans JCM 18968T and T. mesophila JCM 18969T, located on the root of the phylogenetic tree of the 16S rRNA gene and each represented a group (Group M, D and O). The group C was represented by the type strain T. xianhensis P-4T. The strains of group E formed a clade with recently described type strain T. povalilytica Zumi 95T. The rest of three groups (F, G and N) represented potential novel taxa based on the analyses below. However, low bootstrap values at many nodes of the 16S rRNA gene phylogenetic tree implied that topology structure of tree was unstable and the taxonomic affiliation of all strains was inaccurate. Therefore, the 16S rRNA gene was inappropriate for the phylogenetic analysis of the genus Thalassospira.
Figure 1. Phylogenetic tree based on the 16S rRNA gene of 59 Thalassospira bacteria.
The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. Rhodospirillum rubrum ATCC 11170T (NR_074249) was used as the outgroup.
As the 16S rRNA gene was lack of discriminatory power for the closely related strains of the genus Thalassospira, six housekeeping genes (acsA, aroE, gyrB, mutL, rpoD and trpB) were utilized for assessing the diversity. The length of six housekeeping genes was from 510 bp for the rpoD genes to 918 bp for the gyrB gene. The allele numbers of the five housekeeping genes was similar except the trpB gene. The aroE gene exhibited the highest proportion of polymorphic sites (53.4%), followed by acsA (43.2%) and gyrB (40.1%). Only 179 polymorphic sites (35.1%) were identified in the rpoD gene. It was worthwhile to note that the mean G+C content of six housekeeping gene had significantly different. Similarly, the nucleotide diversity (π, the average number of nucleotide differences per site between two randomly-selected isolates) ranged from 0.119 (rpoD) to 0.192 (aroE) (Table 2), which showed the different evolution rates of six housekeeping genes in the MLSA scheme. Among the six housekeeping genes, the aroE gene exhibited the highest resolution based on the highest mean genetic distance (0.190), whereas rpoD displayed the lowest resolution with the lowest mean genetic distance (0.111). Six housekeeping genes showed low Ka/Ks ratios of 0.035–0.099, suggesting that they are under negative selection pressure. These results demonstrated that the six housekeeping genes were better than the 16S rRNA gene for differentiating Thalassospira bacteria; moreover, the aroE gene possessed the highest resolution power among all the tested housekeeping genes.
In this study, 31 unique STs were identified on basis of the six concatenated genes, suggesting high genotypic diversity (Table S3). Of these, 16 STs (27.59%) corresponded to single strain and 14 STs (60.34%) corresponded to two to four isolates. Whereas, the 28th ST (12.07%) was the best represented ST and found in seven different bacteria. Prior to the phylogenetic analysis, the examination of substitution saturation and recombination for six housekeeping genes were subjected using the above-mentioned software, respectively. The measures of the saturation test for single gene with DAMBE version 5.0 shown that the index of ISS was smaller than the index of ISS.C in each gene, suggesting no signal of substitution saturation was not found (Table S4). At the same time, the analysis of split decomposition indicated some degree of reticulation (i.e. conflicting phylogenetic signals) for two genes, acsA and rpoD (Figure S2–S7). The PHI test also demonstrated the presence of recombination for the acsA gene (p<0.05) and the rpoD gene (p<0.05). Nevertheless, the parallelogram network of the two genes was not very obvious, suggesting that intragenic recombination was not remarkable. Besides, the intergenic recombination was estimated using the LIAN version 3.6. The IA and standardized index of association (IS A) were 5.32 and 0.8634, respectively. Therefore, the null hypothesis of linkage equilibrium (IS A = 0) by both parametric and Monte Carlo methods (100 resamplings) was rejected, indicating linkage disequilibrium among the housekeeping genes. As a result, the MLSA analysis was reliable using these housekeeping genes.
In comparison with the phylogenetic tree of the 16S rRNA gene, six phylogenetic trees based on single housekeeping gene sequences had high bootstrap values and presented roughly congruent topology (Figure S8–S13). In detail, the 58 strains were also separated into 15 groups named Group A to O in six phylogenetic trees. Some differences were observed in the topology at the deepest branching points of six phylogenetic trees. For example, the Group D was close to the Group M in the phylogenetic tree of the mutL gene; in contrast, it had close relationships to the Group A, B and C in other trees. Interestingly, the strains of Group A and B were mixed in the phylogenetic trees of the rpoD gene, this location conflict implies that the acquisition of the ropD gene of strain MCCC 1A1300 or MCCC 1A1330 was probably by the lateral gene transfer (LGT) event. Other slight differences were also observed in the six phylogenetic trees, as shown fully in the Figure S8–S13.
The MLSA analysis
The concatenated genes of six protein-coding genes contained 4,413 bp, 31 alleles and 1,850 polymorphic sites with a mean G+C content of 56.0 mol%. The genetic distance of 58 tested strains ranged from 0.000 to 0.220 with a mean 0.138.
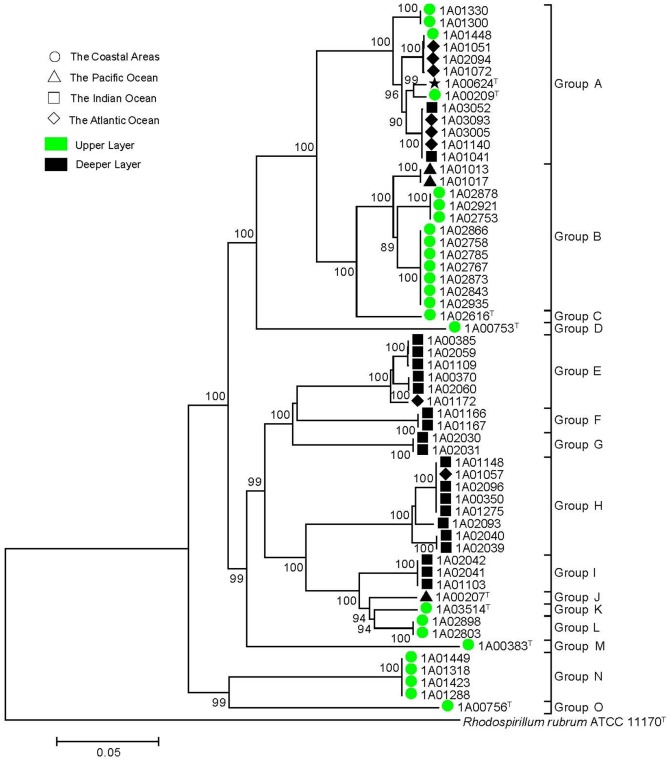
The concatenated genes tree showed a similar topology to the individual gene tree including 15 Groups from A to O with very high bootstrap support (Figure 2). Specifically, the Group A was the largest and had five branches including 13 strains with two type strains T. xiamenensis M-5T and T. permensis NBRC106175T (non-valid). The second largest Group B was separated into three different branches with 12 strains. The Group E contained six strains with three branches including five genetic types. The Group H was split into three branches corresponding to three genetic types with eight strains. The six type strains, T. xianhensis P-4T, T. alkalitolerans JCM 18968T, T. profundimaris WP0211T, T. tepidiphila 1-1BT, T. lucentensis DSM 14000T, and T. mesophila JCM 18969T, formed respectively a separate group marked as Group C, D, J, K, M and O. The rest of five minority groups (Group F, G, I, L and N) and Group B, E and H, cannot be allocated to any previously described species. Therefore, they are new taxon, representing eight potential novel species.
Figure 2. Phylogenetic tree of the MLSA based on six concatenated housekeeping genes of the genus Thalassospira.
The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup. Meantime, the isolated areas and water depth of the Thalassospira bacteria were divided artificially and marked by different symbols or colors.
Correlation analysis between similarities of the MLSA and DDH, ANI, and the evaluation of radiation times
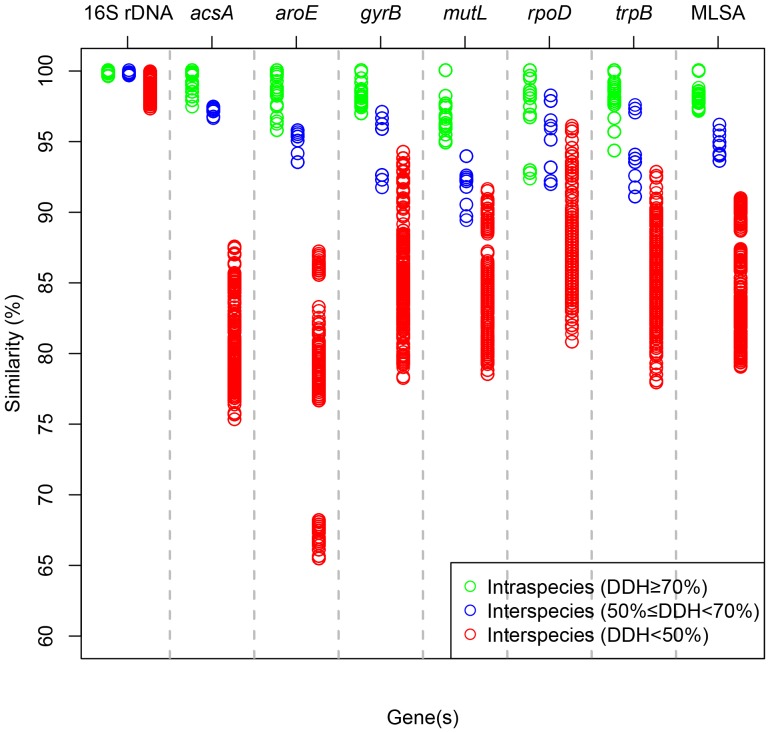
In order to further confirm the accuracy of the MLSA analysis, the DDH values of the 16 representative strains of the 13 groups except the two newly described species (Group D and Group O) were calculated in silico from draft genomes (Table S5). Considering the 70% DDH values as a gold standard for the species boundary in prokaryote taxonomy, 16 strains of the Thalassospira genus were classified into the 13 species corresponding to the 13 group of the MLSA analysis. And then, the intraspecies and interspecies similarities of the individual gene and the concatenated genes among 58 strains were determined based on the species given by DDH values and shown in Figure 3. Disappointingly, identity values of the intraspecies and the interspecies partially overlapped in the most individual genes except the mutL gene. The overlap of identity values was encountered by the strains between Group B and C, and among the strains of Group J, K and L, which shared the high interspecies identity values, with the low intraspecies identity values existing in the strains within Group A, B or H. In contrast, identity values of the concatenated genes exhibited a distinct demarcation (96.16–97.12%) between intraspecies and interspecies. Consequently, compared with the single gene, the MLSA has a great advantage in distinguishing the Thalassospira bacteria.
Figure 3. Intraspecies and interspecies identity of 16S rRNA gene and housekeeping gene(s) of the genus Thalassospira bacteria.
The identity values of each other were displayed with three color circles (intraspecies: green, interspecies: blue/red).
The correlation between the identity of the concatenated genes and DDH in 16 strains was determined by a nonlinear simulate analysis method. As can be seen in Figure S14, the identity values of the six concatenated housekeeping genes was highly correlated (R2 = 0.9661) with the DDH values. Based on the simulative logarithmic equation (y = 11.87*lnx+46.58), DDH value of 70% was equivalent to 97% identity of the MLSA, suggesting that 97% identity of the MLSA as interspecies gap can be used for the cut-off of the species for the Thalassospira bacteria. In addition, as shown in Table S6, the ANIm values of interspecies ranged from 83.34% to 95.76%, whereas the values of intraspecies were from 97.20% to 97.93%. These ANIm values were in accordance with standard ANI criteria for species identity (95–96%) [29].
The radiation times of each other in 58 strains of the genus Thalassospira were estimated using the rate of synonymous substitution for six housekeeping genes. On interspecies level, the radiation time between the strain MCCC 1A02803 (representing the Group L) and the strain MCCC 1A03514 (representing the Group K) was the lowest, about 22.21 million years. By contrast, these two strains had highly related genome, with 64.3% DDH values. This result indicates that the differentiation of two young species started at 22.21 million years ago. On intraspecies level, the highest radiation time between the strain MCCC 1A00624 and MCCC 1A01300 from the same group was 19.30 million years, the DDH value of the two strains was 80.7%. Certainly, these two strains would be divided into different species only several millions years ago. A common ancestor of the strains in the genus Thalassospira might occur 161.48 million years ago (Figure S15).
Biogeography analysis
In this study, the geographical locations of the 57 strains of the genus Thalassospira except of T. permensis NBRC106175T covered a wide range of marine environments. According to the origin of these strains, four large areas, the coastal area (circle), the Pacific Ocean (triangle), the Indian Ocean (square) and the Atlantic Ocean (diamond), were marked using four symbols in MLSA phylogenetic tree, respectively. As shown in Figure 2, strains from the coastal area and the Indian Ocean clustered respectively together to some extent; for instance strains of Group B, L and N were from coastal area, Group E, F, G, H and I were from Indian Ocean; whereas the strains from the Pacific Ocean and the Atlantic Ocean scattered in several groups in the MLSA phylogenetic tree.
In addition, to explore the affection of seawater depth, the habitats were artificially partitioned into the upper layer (0–100 m water depth) and the deeper layer (668–4,480 m water depth) and marked in green and black, respectively, in the MLSA phylogenetic tree (Figure 2). Intriguingly, the most strains tended to cluster together to a certain degree according to the habitats depth. In detail, the strains of the Group B (not including MCCC 1A01013 and MCCC 1A01017) and the Group C were all isolated from the upper layer (marked in Green) and gathered together, while the strains of the Group E, F, G, H, I and J came from the deeper layer (marked in Black) got together. These results indicate that the water depth is one of the key environmental factors for affecting the differentiation of the Thalassospira bacteria.
Discussion
Bacteria of the genus Thalassospira frequently occur in various marine environments,such as seawater, sediment, halobios from every oceans and seas, and frequently retrieved from the bacterial communities enriched using PAHs or crude oil. Recently, they were found to involve the phosphorus cycling in marine environments [13], [14]. The accumulations of isolates of this genus urge the systematic phylogeny analyses. In this report, the genetic diversity and geographic distribution of the Thalassospira bacteria were analyzed for the first time using many approaches including the MLSA with six protein-coding genes, DDH and ANI.
In this study, we demonstrated that MLSA was a powerful method for the discrimination, classification and phylogenetic analysis of the Thalassospira bacteria, which cannot be distinguished by the 16S rRNA gene. The similar situations were frequently encountered in other genera, such as Bacillus, Vibrio, Pseudomonas, Enterobacter and Aeromonas etc. [20], [24], [25], [42], [43]. Recently, the single housekeeping gene (gyrB, rpoB, pyrE, aroE and so on) replaced the 16S rRNA gene and was widely used as a phylogentic marker for differentiating the closely related strains [21], [24], [43], [44]. Due to the horizontal gene transfer [45] and genetic recombination [25], single housekeeping gene may distort the phylogenetic tree. In this report, for example, the strain MCCC 1A02616 occurred in the horizontal gene transfer in the rpoD gene; while the MLSA based on several housekeeping genes are more reliable. In this study, the 58 strains of the genus Thalassospira were split into 15 groups using the MLSA; each of the seven groups (Group A, C, D, J, K, M and O) represented by a described type strain, and eight groups (Group B, E, F, G, H, I, L and N) represented a potential novel species awaiting for further classification of polyphasic taxonomy (Six strains of the Group E probably belong to recently described species T. povalilytica). In addition, DDH value of 70% was used as the gold standard for bacterial species delineation [46]. In this report, we chose 16 representative strains from different groups for DDH and ANI calculation. As a result, the DDH values verified the results of MLSA analysis, and in addition defined the interspecies gap for the genus Thalassospira, with an identity range from 96.16% to 97.12% based on six housekeeping genes. The interspecies gap of ANI value ranged from 95.76% to 97.20%. In the recent studies, the classification and the genetic diversity of the closely related strains in the intragenus were explored by the MLSA combined with the DDH [21], [42], [47], [48]. Therefore, the MLSA is an effective, straightforward, reproducible and comparable tool to explore the taxonomy of the bacteria, to study the diversity of the strains from various environments, to understand evolution of species.
The isolated origins of all culturable strains in our lab laboratory and the NCBI database indicate that they should be the obligate marine bacteria though the non-validly published species, T. permensis SMB34T, was isolated from primitive technogeneous soil formed on salt-mine spoils at the Verchnekamsk deposit of potassium magnesium salts (Berezniki, Perm region, Russia). The geographic distribution patterns showed that the bacteria from the coastal area clustered together to some extent, as well as those from the Indian Ocean. However, the strains from the Pacific Ocean and the Atlantic Ocean scattered in the MLSA phylogenetic tree (Fig. 3). Many similar studies have been reported in recent years. The bacteria of Bacillus pumilus group from marine and terrestrial environments clustered respectively based on the gyrB gene phylogenetic tree [24]. Likewise, Khan et al. discovered that the oceanic Pesudomonas aeruginosa strains trended distinct to cluster from those of other P. aeruginosa strains [44]. In addition, The Vibrio parahaemolyticus bacteria from different epidemiological sources in Thailand revealed a degree of clustering in phylogenetic analysis [49]. The inherent mechanism of the intriguing clustering in different origin isolates remains unclear, and then explores further in future investigations.
Obviously in the tree (Fig. 3), two ecotypes were separated from each other according to the water depths. Thus, the surface ecotype and deep-sea ecotype naturally formed in the vast marine system. Our results were consistent with the previous reports. For example, the marine bacterium Alteromonas macleodii are found to be generally clustered with the depth in the water column from which the isolate originated [50]. Klochko et al found that peculiarities of A. macleodii strains reflected their deep/surface habitation rather than geographical distribution [51]. Similarly, Tarasov et al exposed significant differences between deep and shallow-water hydrothermal vent communities [52].
In summary, we reported the diversity and biogeography of Thalassospira bacteria from diverse marine environments for the first time, based on the MLSA, DDH and ANI. Fifteen distinct lineages corresponding to seven known and eight potential novel taxonomic groups were revealed, and the interspecies gap of MLSA for the genus Thalassospira is 96.16–97.12%. Interestingly, the Thalassospira bacteria exhibited surface and deep ecotypes according to the water depths. These results help to understand their adaptive evolution in marine environments. Further polyphasic characterization and comparative genomic analysis are just under study to understand their role in marine environments.
Supporting Information
The map of geographical distribution of the 58 strains from various marine environments. Each red dot represents a strain, some dots overlapped.
(DOCX)
Split decomposition analysis of the acsA gene.
(DOCX)
Split decomposition analysis of the aroE gene.
(DOCX)
Split decomposition analysis of the gyrB gene.
(DOCX)
Split decomposition analysis of the mutL gene.
(DOCX)
Split decomposition analysis of the rpoD gene.
(DOCX)
Split decomposition analysis of the trpB gene.
(DOCX)
Phylogenetic tree based on the acsA gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the aroE gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the gyrB gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the mutL gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the rpoD gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the trpB gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
The correlation of the DDH and MLSA identity of the 16 Thalassospira bacteria.
(DOCX)
The clustering tree of the DDH values and Radiation time of strains in the three clusters.
(DOCX)
Characteristics of the primers used in this study.
(DOCX)
GenBank accession numbers of all strains used in this study.
(DOCX)
Allelic profiles of all strains used in this study.
(DOCX)
Nucleotide diversity, mearsure substitution saturation and the PHI test for single gene.
(DOCX)
The identity matrix of DDH and MLSA in the 16 Thalassospira bacteria.
(DOCX)
The identity matrix of ANIm in the 16 Thalassospira bacteria.
(DOCX)
Acknowledgments
We thank Dr. Kazuya Watanabe for generous gift of Thalassospira tepidiphila 1-1BT.
Funding Statement
This work was financially supported by COMRA program (No. DY125-15-R-01) (http://www.comra.org/), Public Welfare Project of SOA (201005032) (http://www.soa.gov.cn/), National Natural Science Foundation of China (41176154/41276005) (http://www.nsfc.gov.cn/) and National Infrastructure of Microbial Resources (No. NIMR-2013-9 and NIMR-2014-9) (http://www.nstic.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lopez-Lopez A, Pujalte MJ, Benlloch S, Mata-Roig M, Rossello-Mora R, et al. (2002) Thalassospira lucentensis gen. nov., sp. nov., a new marine member of the alpha-Proteobacteria. Int J Syst Evol Microbiol 52: 1277–1283. [DOI] [PubMed] [Google Scholar]
- 2. Liu C, Wu Y, Li L, Ma Y, Shao Z (2007) Thalassospira xiamenensis sp. nov. and Thalassospira profundimaris sp. nov. Int J Syst Evol Microbiol 57: 316–320. [DOI] [PubMed] [Google Scholar]
- 3. Kodama Y, Stiknowati LI, Ueki A, Ueki K, Watanabe K (2008) Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int J Syst Evol Microbiol 58: 711–715. [DOI] [PubMed] [Google Scholar]
- 4. Zhao B, Wang H, Li R, Mao X (2010) Thalassospira xianhensis sp. nov., a polycyclic aromatic hydrocarbon-degrading marine bacterium. Int J Syst Evol Microbiol 60: 1125–1129. [DOI] [PubMed] [Google Scholar]
- 5. Tsubouchi T, Ohta Y, Haga T, Usui K, Shimane Y, et al. (2013) Thalassospira alkalitolerans sp. nov. and Thalassospira mesophila sp. nov., isolated from a decaying bamboo sunken in the marine environment, and emended description of the genus Thalassospira . Int J Syst Evol Microbiol 64: 107–115. [DOI] [PubMed] [Google Scholar]
- 6. Nogi Y, Yoshizumi M, Miyazaki M (2014) Thalassospira povalilytica sp. nov., a polyvinyl-alcohol-degrading marine bacterium. Int J Syst Evol Microbiol 64: 1149–1153. [DOI] [PubMed] [Google Scholar]
- 7. Plotnikova EG, Anan'ina LN, Krausova VI, Ariskina EV, Prisyazhnaya NV, et al. (2011) Thalassospira permensis sp. nov., a new terrestrial halotolerant bacterium isolated from a naphthalene-utilizing microbial consortium. Mikrobiologiia 80: 691–699. [PubMed] [Google Scholar]
- 8. Cui Z, Lai Q, Dong C, Shao Z (2008) Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ Microbiol 10: 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang B, Lai Q, Cui Z, Tan T, Shao Z (2008) A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ Microbiol 10: 1948–1963. [DOI] [PubMed] [Google Scholar]
- 10. Liu Z, Liu J (2013) Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. MicrobiologyOpen 2: 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L, Wang W, Lai Q, Shao Z (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12: 1230–1242. [DOI] [PubMed] [Google Scholar]
- 12. Bassey D, Grigson S (2011) Degradation of benzyldimethyl hexadecylammonium chloride by Bacillus niabensis and Thalassospira sp. isolated from marine sediments. Toxicol Environ Chem 93: 44–56. [Google Scholar]
- 13. Hutz A, Schubert K, Overmann J (2011) Thalassospira sp. isolated from the oligotrophic eastern Mediterranean Sea exhibits chemotaxis toward inorganic phosphate during starvation. Appl Environ Microbiol 77: 4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas S, Burdett H, Temperton B, Wick R, Snelling D, et al. (2010) Evidence for phosphonate usage in the coral holobiont. ISME J 4: 459–461. [DOI] [PubMed] [Google Scholar]
- 15. Um S, Pyee Y, Kim EH, Lee SK, Shin J, et al. (2013) Thalassospiramide G, a new gamma-amino-acid-bearing peptide from the marine bacterium Thalassospira sp. Mar Drugs 11: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ross AC, Xu Y, Lu L, Kersten RD, Shao Z, et al. (2013) Biosynthetic multitasking facilitates thalassospiramide structural diversity in marine bacteria. J Am Chem Soc 135: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosh M, Pulicherla KK, Rekha VP, Raja PK, Sambasiva Rao KR (2012) Cold active beta-galactosidase from Thalassospira sp. 3SC-21 to use in milk lactose hydrolysis: a novel source from deep waters of Bay-of-Bengal. World J Microbiol Biotechnol 28: 2859–2869. [DOI] [PubMed] [Google Scholar]
- 18. Rizzo C, Michaud L, Hormann B, Gerce B, Syldatk C, et al. (2013) Bacteria associated with sabellids (Polychaeta: Annelida) as a novel source of surface active compounds. Mar Pollut Bull 70: 125–133. [DOI] [PubMed] [Google Scholar]
- 19. Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, et al. (2005) Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol 3: 733–739. [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, et al. (2011) Multilocus phylogenetic analysis of the genus Aeromonas . Syst Appl Microbiol 34: 189–199. [DOI] [PubMed] [Google Scholar]
- 21. Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol 35: 7–18. [DOI] [PubMed] [Google Scholar]
- 22. Winkelmann N, Jaekel U, Meyer C, Serrano W, Rachel R, et al. (2010) Determination of the diversity of Rhodopirellula isolates from European seas by multilocus sequence analysis. Appl Environ Microbiol 76: 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamamoto S, Bouvet PJ, Harayama S (1999) Phylogenetic structures of the genus Acinetobacter based on gyrB sequences: comparison with the grouping by DNA-DNA hybridization. Int J Syst Bacteriol 49: 87–95. [DOI] [PubMed] [Google Scholar]
- 24. Liu Y, Lai Q, Dong C, Sun F, Wang L, et al. (2013) Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PloS one 8: e80097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alvarez-Perez S, de Vega C, Herrera CM (2013) Multilocus sequence analysis of nectar pseudomonads reveals high genetic diversity and contrasting recombination patterns. PLoS One 8: e75797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert M, Zhang J, et al. (2004) BOX-pCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus . Syst Appl Microbiol 27: 84–92. [DOI] [PubMed] [Google Scholar]
- 27. Kim W, Hong YP, Yoo JH, Lee WB, Choi CS, et al. (2002) Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol Lett 207: 21–27. [DOI] [PubMed] [Google Scholar]
- 28. Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lai Q, Yuan J, Gu L, Shao Z (2009) Marispirillum indicum gen. nov., sp. nov., isolated from a deep-sea environment. Int J Syst Evol Microbiol 59: 1278–1281. [DOI] [PubMed] [Google Scholar]
- 31. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia X (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol 30: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 35. Haubold B, Hudson RR (2000) LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- 36. Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS (2000) Parallel evolution of virulence in pathogenic Escherichia coli . Nature 406: 64–67. [DOI] [PubMed] [Google Scholar]
- 37. Luo R, Liu B, Xie Y, Li Z, Huang W, et al. (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai Q, Shao Z (2012) Genome sequence of Thalassospira profundimaris type strain WP0211. J Bacteriol 194: 6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai Q, Shao Z (2012) Genome sequence of Thalassospira xiamenensis type strain M-5. J Bacteriol 194: 6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meier-Kolthoff J, Auch A, Klenk H-P, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meier-Kolthoff J, Göker M, Spröer C, Klenk H-P (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195: 413–418. [DOI] [PubMed] [Google Scholar]
- 42. Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, et al. (2005) Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71: 5107–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P (2013) Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter . Syst Appl Microbiol 36: 309–319. [DOI] [PubMed] [Google Scholar]
- 44. Khan NH, Ahsan M, Yoshizawa S, Hosoya S, Yokota A, et al. (2008) Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa isolates from the ocean. Appl Environ Microbiol 74: 6194–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim BJ, Hong SH, Kook YH, Kim BJ (2013) Molecular evidence of lateral gene transfer in rpoB gene of Mycobacterium yongonense strains via multilocus sequence analysis. PLoS One 8: e51846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. [DOI] [PubMed] [Google Scholar]
- 47. Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX (2012) Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS One 7: e44936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang M, Lv Y, Xiao J, Wu H, Zheng H, et al. (2012) Edwardsiella comparative phylogenomics reveal the new intra/inter-species taxonomic relationships, virulence evolution and niche adaptation mechanisms. PLoS One 7: e36987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Theethakaew C, Feil EJ, Castillo-Ramirez S, Aanensen DM, Suthienkul O, et al. (2013) Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier, and environmental sources in Thailand, determined by multilocus sequence analysis. Appl Environ Microbiol 79: 2358–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ivars-Martinez E, D'Auria G, Rodriguez-Valera F, Sanchez-Porro C, Ventosa A, et al. (2008) Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol Ecol 17: 4092–4106. [DOI] [PubMed] [Google Scholar]
- 51. Klochko V, Zelena L, S IV, Ostapchuk A (2012) Peculiarities of Alteromonas macleodii strains reflects their deep/surface habitation rather than geographical distribution. J Gen Appl Microbiol 58: 129–135. [DOI] [PubMed] [Google Scholar]
- 52. Tarasov V, Gebruk A, Mironov A, Moskalev L (2005) Deep-sea and shallow-water hydrothermal vent communities: two different phenomena? Chem Geol 224: 5–39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The map of geographical distribution of the 58 strains from various marine environments. Each red dot represents a strain, some dots overlapped.
(DOCX)
Split decomposition analysis of the acsA gene.
(DOCX)
Split decomposition analysis of the aroE gene.
(DOCX)
Split decomposition analysis of the gyrB gene.
(DOCX)
Split decomposition analysis of the mutL gene.
(DOCX)
Split decomposition analysis of the rpoD gene.
(DOCX)
Split decomposition analysis of the trpB gene.
(DOCX)
Phylogenetic tree based on the acsA gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the aroE gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the gyrB gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the mutL gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the rpoD gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
Phylogenetic tree based on the trpB gene. The tree was constructed using the neighbor-joining method with MEGA 5.0. Bootstrap values over 50% (1000 replications) were shown at each node. Bar, % estimated substitution. The bacteria of Rhodospirillum rubrum ATCC 11170T (NC_007643) was used as the outgroup.
(DOCX)
The correlation of the DDH and MLSA identity of the 16 Thalassospira bacteria.
(DOCX)
The clustering tree of the DDH values and Radiation time of strains in the three clusters.
(DOCX)
Characteristics of the primers used in this study.
(DOCX)
GenBank accession numbers of all strains used in this study.
(DOCX)
Allelic profiles of all strains used in this study.
(DOCX)
Nucleotide diversity, mearsure substitution saturation and the PHI test for single gene.
(DOCX)
The identity matrix of DDH and MLSA in the 16 Thalassospira bacteria.
(DOCX)
The identity matrix of ANIm in the 16 Thalassospira bacteria.
(DOCX)