Abstract
The capsule (cps) locus of Streptococcus pneumoniae is flanked by the pbp2x and pbp1a genes, coding for penicillin-binding proteins, enzymes involved in cell wall synthesis that are targets for β-lactams. This linkage suggested to us that selection for β-lactam resistance might coselect for capsular transformants. The recombination event would then involve PBP genes, as well as the cps operon, and would change both the serotype and the resistance profile of the strain. We transformed β-lactam-susceptible strain TIGR4 by using whole genomic DNA extracted from multidrug-resistant strain GA71, a serotype 19F variant of pneumococcal clone Spain23F-1, and selected β-lactam-resistant transformants. Smooth colonies appearing on selective plates were subcultured, serotyped by the Quellung reaction, and genotyped to confirm the presence of the GA71 pbp2x-cps19-pbp1a locus in the TIGR4 genetic background by restriction fragment length polymorphism analysis of the whole locus and its flanking regions. The results showed that a new serotype, combined with resistance to β-lactams, could emerge in a susceptible strain via a single transformation event. Quantitative analysis showed that transfer of the cps locus had occurred at an elevated rate in β-lactam-selected transformants. This suggests that in natural settings selection by host immunity and selection by antibiotics may be interrelated because of “hitchhiking” effects due to linkage of resistance determinants and the capsule locus.
Seventy-five years ago, in a milestone experiment, Griffith demonstrated the astonishing ability of Streptococcus pneumoniae to adapt under the pressure of the host immune system, by acquiring what we now know to be DNA encoding biosynthetic enzymes for a polysaccharide capsule, which protects the organism against phagocytosis during infection (16). In the same year that Griffith's experiments were published, Alexander Fleming discovered penicillin—an event that reshaped the history of both humans and pneumococci (13). Before the antibiotic era, the population of pneumococci isolated from humans was mainly dominated by strains with capsular polysaccharides of serotypes 1, 2, and 3 (12, 16). Within the last 25 years, we have witnessed the emergence of penicillin- and multidrug-resistant S. pneumoniae clones, a number of which have spread worldwide. Certain serotypes, this time 6B, 9V, 14 19A, 19F, and 23F, have also dominated this new population, and many of the worldwide clones have appeared with different capsular types (28, 30, 37). The most successful clone in terms of geographical dispersion and prevalence is the multidrug-resistant Spain23F-1 pandemic clone, nine serotype variants of which have been identified so far, including all six of those listed above, in addition to serotypes 3, 9N, and 11 (3, 10, 31; http://www.mlst.net). Such variants are thought to arise through natural transformation involving recombinational replacements, within and around the capsular biosynthesis (cps) locus, of DNA fragments sometimes as large as 25 kb (4-6). Epidemic, multidrug-resistant strains dominate clinical S. pneumoniae isolates in many regions of the world today (28).
In all of the pneumococcal strains analyzed so far, cps genes are clustered and located between the dexB and aliA genes (14, 24; http://www.sanger.ac.uk/Projects/S_pneumoniae). Genes for two penicillin-binding proteins (PBPs) are located on either side of this region, pbp2x upstream of dexB and pbp1a downstream of aliA, each at a remove of ≥10 kb (23, 40, 41). The PBPs are enzymes involved in cell wall synthesis that are targets for β-lactam drugs (20). For example, the cps locus of penicillin-susceptible strain TIGR4 is a 15.2-kb region encoding 13 open reading frames, flanked by the pbp2x gene 10.9 kb upstream and by the pbp1a gene 11.6 kb downstream, and genes coding for all other PBPs are located at least 450 kbp away from the cps locus (40).
There are six PBPs in a pneumococcal cell (PBP1a, -1b, -2a, -2b, -2x, and -3), of which PBP2x and PBP2b have been confirmed to be essential for cell growth (20, 21, 25). The pbp genes in S. pneumoniae have a mosaic structure and can undergo inter- and intraspecies recombination (5, 8, 19). Strains resistant to β-lactams have some of these enzymes modified to express reduced affinity for a β-lactam (20). In general, the resistance profile of the particular isolates results from an interaction between PBPs of various degrees of modification (9, 22), and probably between these and a modified muropeptide branching enzyme, encoded by the murM gene (11, 39).
Resistance to β-lactam drugs used in the treatment of pneumococcal infections requires, in all of the cases studied so far, alterations in at least PBP2x or -2b (22). Modified, low-affinity PBP2x is sufficient for the cell to become nonsusceptible to cephalosporins (e.g., cefotaxime) and is also necessary for penicillin resistance to emerge. Resistance to penicillin requires modified, low-affinity forms of at least PBP1a in addition to modifications in PBP2x (2, 38). The presence of modified forms of PBP2b, PBP2a, and MurM seems to increase further the level of resistance to β-lactams (2, 9, 22, 39).
The existence of a selectable phenotype, resistance to β-lactams, requiring replacements in pbp2x or in both pbp1a and pbp2x, which flank the cps locus, suggested that it might be possible to select single-step capsular transformants in vitro under selective pressure of cefotaxime or penicillin. The recombination event would then involve one or both PBP-encoding genes, as well as the capsule locus, and would change both the serotype and the resistance profile of the strain. In case of selection for resistance to penicillin, a single event like this would require replacement of 42 kb or almost 2% of the TIGR4 strain genome, twice the size of any naturally transformed fragment of S. pneumoniae homologous DNA previously reported (4). Although some transposons of comparable size have been reported to integrate into the S. pneumoniae genome following transformation, their mechanism of integration into the host genome is different (36, 42).
(These results were presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 2003 [abstr. C1-1113].)
MATERIALS AND METHODS
S. pneumoniae strains.
The penicillin-susceptible TIGR4 strain of serotype 4 used in this study was obtained from the American Type Culture Collection (40). Penicillin-resistant serotype 19F strain GA71 was kindly donated by Brian Spratt (6). Bacterial strains were routinely grown at 37°C in 5% CO2 on tryptic soy agar II supplemented with 5% defibrinated sheep blood or in Todd-Hewitt broth supplemented with 0.5% yeast extract (Becton Dickinson, Sparks, Md.), stored in Todd-Hewitt broth supplemented with 0.5% yeast extract supplemented with 10% glycerol, and frozen at −70°C. MICs of cefotaxime and penicillin G were evaluated with E-test strips (AB Biodisk, Solna, Sweden).
Genetic transformation.
Growth media and culture methods for genetic transformation have been used as previously described (33). The transformation of TIGR4 was induced by competence-stimulating peptide variant 2, and the donor DNA concentration was 1 μg/ml of the transformation culture. The genomic DNA of GA71 was prepared with the QIAGEN Genomic DNA system (QIAGEN, Valencia, Calif.). Samples containing approximately 1 μg of chromosomal DNA were evaluated for fragment size distribution by pulsed-field gel electrophoresis with the CHEF-DRIII system (Bio-Rad, Hercules, Calif.) (35). Transformants were selected on blood agar base medium 2 (Becton Dickinson) supplemented with 8% defibrinated sheep blood and various concentrations of penicillin G, trimethoprim (ICN Biochemicals Inc., Aurora, Ohio), and cefotaxime (Sigma-Aldrich, St. Louis, Mo.) alone or in combinations.
Serotyping of isolates.
Polysaccharide capsule types were determined on the basis of Quellung tests with factor-specific typing sera (Statens Serum Institute, Copenhagen, Denmark) (27).
RFLP.
Restriction fragment length polymorphism (RFLP) analysis of pbp2x, pbp2b, and pbp1a and of the cps locus cpsA-cpsB fragment was performed as previously described (15, 26), as a genotypic screen for the presence of donor PBPs and capsule in transformants. The DNA templates used for this screening were obtained as previously described (43). To evaluate the sizes of DNA fragments replaced during the transformation and to identify recombination crossover points, RFLP analysis of the pbp2x-pbp1a region in the donor, the recipient, and five transformants was performed. DNA templates were prepared with the QIAGEN Genomic DNA system, and products were amplified with the TripleMaster PCR system (Eppendorf, Hamburg, Germany) under the cycling conditions recommended by manufacturer. The following pairs of primers were used: TTM13 (5′ATGAAGGCAATATCTGGGGTGACTAC-3′) and pbp2xr2 (15) to amplify fragments from the position 10,075 bp upstream to that 32 bp downstream of pbp2x in the TIGR4 genome (and corresponding fragments from other isolates), pbp2xf (15) and TTM07 (41) to amplify the fragment from base 226 of pbp2x to base 1504 of dexB, 1430 and 1402 (24) to amplify the fragment from base 1475 of dexB to base 48 of aliA, TTM08 (41) and pbp1af (15) to amplify the fragment from position 21 of aliA to the position 159 bp downstream of pbp1a, and pbp1ar (15) and TTM15 (5′ATCGCGGTTGTAAGCATCTGTAAT-3′) to amplify the fragment from the position 89 bp upstream to that 5,865 bp downstream of pbp1a. Products of primer pair TTM13-pbp2xr2 (pbp2x locus and upstream) were digested with the DdeI enzyme, and those of primer pair 1430-1402 (dexB-cps-aliA locus) were digested with the RsaI enzyme. Products of the pbp2xf-TTM07 and TTM08-pbp1af primer pairs (pbp2x-dexB and aliA-pbp1a loci, respectively) were digested with AluI, DdeI, HaeIII, HinfI, MseI, and RsaI, and those of primer pair pbp1ar-TTM15 (pbp1a locus and downstream) were digested with DdeI, HaeIII, HinfI, and RsaI (all restriction enzymes supplied by New England Biolabs, Beverly, Mass.). Fingerprints generated for transformants were compared with those of strains TIGR4 and GA71 with DNAStar software. The regions of possible crossover sites were identified between (i) the last restriction site identified in recipient strain TIGR4 but not in donor strain GA71 or in the transformant and (ii) the first site next to this one that was present in the recipient and in the transformant but not in the donor.
RESULTS AND DISCUSSION
We transformed the β-lactam-susceptible, serotype 4 TIGR4 strain with whole genomic DNA extracted from GA71, a penicillin- and cefotaxime-resistant, serotype 19F variant of the pandemic Spain23F-1 clone. Transformants were selected for the ability to grow on solid media supplemented with penicillin (0.04 mg/liter) or cefotaxime (0.1 mg/liter) (see Table 1 for the penicillin and cefotaxime MICs for the strains described in this study). New serotype 19F polysaccharide capsule variants of TIGR4 were selected among resistant colonies on the basis of the smooth, viscous morphology of the large colonies typical of GA71 but not observed for TIGR4. The presence of the cps19F locus was detected by RFLP analysis of the cpsA-cpsB region in all resistant transformants with colony morphology typical of serotype 19F, and the presence of the new 19F capsular polysaccharide was confirmed by the Quellung reaction (26, 27).
TABLE 1.
Penicillin and cefotaxime MICs for the isolates analyzed in this study
| Strain | Serotype | Drug to which transformant resistance was selecteda | MIC (mg/liter)
|
Reference | |
|---|---|---|---|---|---|
| Penicillin | Cefotaxime | ||||
| TIGR4b | 4 | 0.032 | 0.047 | 4 | |
| GA71c | 19F | 1.5 | 1.0 | 19 | |
| TIGR19Fa | 19F | Penicillin | 0.125 | 1.0 | This study |
| TIGR19Fb | 19F | Penicillin | 0.125 | 1.0 | This study |
| TIGR19Fc | 19F | Cefotaxime | 0.125 | 1.0 | This study |
| TIGR19Fd | 19F | Penicillin | 0.125 | 1.0 | This study |
| TIGR19Fe | 19F | Cefotaxime | 0.125 | 1.0 | This study |
The β-lactam concentration used to select the transformants listed was 0.04 mg/liter for penicillin and 0.1 mg/liter for cefotaxime.
Recipient strain in transformation experiments.
Donor strain.
In serotype 19F transformants selected for further analysis (Table 1), resistance phenotypes matched PBP profile changes identified in RFLP analysis of pbp alleles. For all of them, cefotaxime MICs were equal to those observed for GA71 (1 mg/liter in all of these isolates), suggesting that all of the determinants necessary for emergence of the full cefotaxime resistance phenotype in strain TIGR4 were transformed. On the other hand, while penicillin MICs increased in all of the transformants, they remained more than 10 times lower than that for the GA71 strain, suggesting that not all of the donor's resistance determinants were involved in the transformation. RFLP analysis of pbp1a, pbp2b, and pbp2x showed that in all of these transformants recombinational replacements led to changes in the pbp1a and pbp2x genes but not in pbp2b. The possible role of other β-lactam resistance determinants has not been investigated.
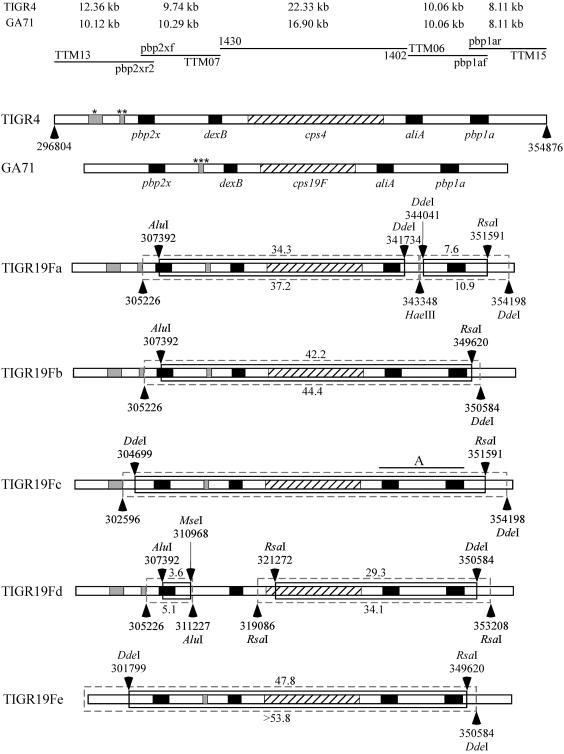
The complete genomic sequence of TIGR4 (40) and part of the sequence of the Sanger serotype 23F S. pneumoniae strain, which is a member of the same pandemic Spain23F-1 clone as GA71 but differs in serotype, were available at the time of this study (http://www.sanger.ac.uk/Projects/S_pneumoniae). This sequence information was used to measure the sizes of replaced fragments and to track crossover points by RFLP analysis of large PCR products. Figure 1 shows the results of mapping of the 58.1-kb fragment of the TIGR4 genomic DNA, covering the whole pbp2x-cps-pbp1a locus and its flanking regions, the corresponding 51-kb fragment of GA71, and the corresponding fragments of five serotype 19F β-lactam-resistant transformants of TIGR4 generated in independent transformation experiments (isolates are described in Table 1). The sizes of the replaced fragments were calculated for each transformant on the basis of the RFLP patterns, and results are presented in Fig. 1. For each of the five transformants, mapping results were consistent with the hypothesis that a single transformation event led to recombinational replacement of the cps locus and at least one of the flanking pbp genes. In all of the transformants analyzed, regions in which recombinational crossover points were predicted to have occurred were localized within the regions of expected high homology between donor and recipient strains. The largest single replacement was identified in isolate TIGR19Fe. Fingerprints of the whole pbp2x-cps-pbp1a region of TIGR19Fe exactly matched those of the donor strain, and possible crossover points were identified upstream of pbp2x and downstream of pbp1a, indicating that in a single event both PBP genes and the capsule locus might have been replaced and at least a 47.8-kb fragment of the TIGR4 genome has been replaced with a GA71 fragment of more than 43.0 kb. The size of the replacement, covering more than 2% of the pneumococcal genome, highlights the considerable ability of S. pneumoniae to adapt under the selective pressure of antimicrobial use and host immune responses. Pulsed-field gel electrophoresis of the donor DNA used for transformation showed a high concentration of fragments in the range of 35 to 250 kb. Thus, it seems unlikely that cotransformation of small DNA fragments was the main mechanism for the replacements observed or that the sizes of replacements were limited by the length of the donor DNA.
FIG. 1.
Graphic representations of the recombinational replacements within and outside the pbp2x-cps-pbp1a region identified by RFLP analysis in five isolates of TIGR4 transformed independently to penicillin nonsusceptibility and serotype 19F with chromosomal DNA of strain GA71. Horizontal lines at the top represent products of PCR amplified with the oligonucleotides indicated above (forward primer) and below (reverse primer) the line. See Materials and Methods for detailed descriptions of the primers used. The sizes of the amplicons generated for the recipient (TIGR4) and donor (GA71) strains with a particular pair of primers are shown above the lines representing PCR products. Horizontal bars represent regions of genomic DNA in the strains analyzed (indicated on the left). The cross-hatched regions represent cps loci, and the black rectangles depict genes present in the regions flanking the cps locus in all natural isolates of S. pneumoniae. Gray regions indicate indels located outside the dexB-aliA locus. The possible sites of the indels were identified on the basis of the Spain23F-1 clone (serotype 23F isolate genome sequences were made available by the Sanger Institute [http://www.sanger.ac.uk/Projects/S_pneumoniae]). The top bar presents the genetic organization of the 58.1-kbp fragment of the TIGR4 strain from position 296804 to position 354876 of its published genome sequence (GenBank accession number NC003028) (40). The single star marks a 1,709-bp indel at position 300888 to position 302596, and the double star marks a 553-bp indel at position 304674 to position 305226, both upstream of pbp2x in TIGR4. The second horizontal bar represents the organization of the same region in isolate GA71, with a size of 51.0 kbp, calculated on the basis of the PCR products generated in this study. The triple star marks the 559-bp indel present in the Sanger Spain23F-1 clone strain at position 3809 bp downstream of pbp2x, also identified in GA71. Arrows above and below bars point at the restriction sites identified as the limits of possible crossover, as described in Materials and Methods. Lack of an enzyme name next to the restriction site genome position number indicates that the crossover site was eliminated by the indel. Inner, solid boxes represent the minimal replacements identified by RFLP analysis in capsule and PBP transformants of TIGR4. Estimated minimal TIGR4 genomic DNA fragments involved in recombinational replacements are depicted above boxes. Outer, dashed boxes represent maximal replacements identified the same way, and the estimated sizes of maximal replacements are depicted below boxes. Fingerprints generated for isolates TIGR19Fa, TIGR19Fc, and TIGR19Fd indicate the presence of two recombinational replacements. The RFLP technique used did not allow us to narrow the sizes of replacements in TIGR19Fc. Region A, extending from a position 611 bp upstream of aliA to one 159 bp downstream of pbp1a, marks the possible site of the additional recombinational crossover(s) in this transformant. Because of the presence of these additional crossovers, the sizes of the replacements were not calculated.
Analysis of the RFLP patterns showed that a single transformation event could lead to simultaneous exchange of the capsule and PBPs in S. pneumoniae. Thus, loss of β-lactam susceptibility and the simultaneous emergence of a new capsule type in pneumococci are feasible. However, RFLP analysis showed also that in a majority of the transformants examined, more than one recombinational event took place.
The results obtained thus far did not quantitatively address the hypothesis that capsular transformants would “hitchhike” during selection for resistance to β-lactams, i.e., that selection for resistance would increase the frequency of capsular transformants. To test this hypothesis rigorously, it was necessary to show that capsular transformants were a greater proportion among strains selected for β-lactam resistance than among transformants selected for an unlinked resistance determinant. Table 2 shows that serotype 19F cells reached frequencies 50 to 400 times higher among strains transformed with GA71 genomic DNA and selected for β-lactam resistance than among strains transformed with the same DNA and selected for resistance to trimethoprim, which is mediated by alterations in dhfr, a gene located 0.9 Mb away from the cps locus in TIGR4 (40).
TABLE 2.
Frequency of recombinational replacements in cells of penicillin-susceptible S. pneumoniae strain TIGR4 transformed with chromosomal DNA of penicillin-resistant serotype 19F strain GA71 on the basis of the number of colonies selected for resistance to different concentrations of antimicrobial agents
| Antimicrobial agent used to select transformants (concn [mg/ml]) | Frequency of resistant transformants among all cells in transformation culturea | Frequency of 19F serotype cells among resistant transformants |
|---|---|---|
| Trimethoprim (25) | 3.6 × 10−3 | 2.1 × 10−5 |
| Penicillin (0.1) | 1.1 × 10−5 | 2.7 × 10−3 |
| Penicillin (0.3) | 2.5 × 10−7 | NDb |
| Trimethoprim (25) + penicillin (0.1) | 1.8 × 10−8 | ND |
| Cefotaxime (0.03) | 7.3 × 10−4 | 1.1 × 10−3 |
| Cefotaxime (0.3) | 1.3 × 10−5 | 8.5 × 10−3 |
| Trimethoprim (25) + cefotaxime (0.3) | 2.0 × 10−9 | ND |
The cell density in transformation cultures varied at the time of selective-medium inoculation from 1.1 × 108 to 1.5 × 108 CFU/ml.
ND, not detected.
In principle, at least two mechanisms could result in an abundance of transformants bearing one replacement (cps) among cells bearing a second replacement (PBP loci). The mechanism we originally hypothesized was a single transformation bearing both replacements, which are closely linked on the genome. An alternate hypothesis is that some cells are more transformable than others; if this were the case, any two markers would be present together more often than if their transformation were independent, because the presence of either would indicate a highly transformable recipient cell. Porter and Guild (32) showed that there are two “transformable units” per cell in pneumococci, so that the null expectation, if transformations occur independently and all cells are competent, is that the frequency of dual transformants bearing unlinked markers would be one-half of the product of the frequencies of single transformants. Our results closely match that expectation. We found that transformants resistant to both trimethoprim (25 mg/liter) and cefotaxime (0.3 mg/liter) occurred at a frequency of 2.0 × 10−9, 0.43 times the product of the single transformation frequencies. Similarly, dual transformants resistant to penicillin (0.1 mg/liter) and trimethoprim (25 mg/liter) occurred at a frequency of 1.8 × 10−8, 0.45 times the product of the single transformation frequencies. Since the hypothesis that the small minority of cells was highly transformable under the protocol used would predict ratios substantially larger than 0.5, we rejected this hypothesis.
On the basis of these quantitative considerations, we concluded that selection for β-lactam resistance results in selection for capsular switches owing to genetic “hitchhiking” effects. For the β-lactams used in this study, the strongest effect (indicated by the proportion of serotype 19F cells among resistant transformants [last column of Table 2]) was observed among transformants selected for the high concentration of cefotaxime, probably because low-affinity forms of PBP2x and PBP1a, but not PBP2b, are necessary for the emergence of this particular phenotype (29). A weaker effect was observed for penicillin, for which resistance also requires low-affinity forms of two PBPs, but not necessarily the two flanking the cps locus. The weakest effect was observed among transformants selected for the low concentration of cefotaxime, where low-affinity PBP2x is the only replacement necessary for the resistance to emerge. This indicates that the hitchhiking effect was more apparent in penicillin-susceptible TIGR4 when capsule transformants were selected for the presence of two flanking resistance markers at once.
Table 2 also shows a decrease in the number of transformants selected for resistance to increasing concentrations of penicillin or cefotaxime, probably representing the complexity of interactions between transformed PBPs. The complexity is present not only because various PBPs may undergo transformation but also because the size of the replacement within each particular gene affects the level of resistance conferred. Whereas a single amino acid substitution within dhfr is sufficient for a pneumococcal isolate to become resistant to trimethoprim, usually changes within three different regions of any pbp gene are necessary to synthesize a modified, low-affinity form of the PBP (1, 9).
Our results showed that a new serotype, combined with β-lactam resistance, could emerge in a susceptible strain via a single transformation event. The possibility that a single transformation event can lead to both a capsular polysaccharide type and PBP exchange in S. pneumoniae has been previously anticipated (4, 18) since regions of homology that putatively served as recombinational crossover points for replacement of capsule biosynthetic genes have been identified within the cps locus and in pbp1a of the serotype variants of the pandemic Spain9V-3 clone (4). However, all of these replacements took place among isolates already resistant to penicillin, and to our knowledge there are no published data documenting simultaneous transformation of penicillin-susceptible S. pneumoniae to a new serotype and to penicillin nonsusceptibility. Integrons and plasmids carrying multiple determinants encoding both virulence and antimicrobial resistance have been described in a number of bacterial species (17, 34). In S. pneumoniae, the major mechanism of horizontal gene transfer is transformation, and the organization of the pbp2x-cps-pbp1a region in S. pneumoniae may provide an analogous opportunity for cotransfer and associated linkage selection of determinants of resistance and pathogenicity. It is also well documented that both the cps operon and the PBP-encoding genes are highly variable and show a mosaic structure, changing frequently and independently from other nearby regions (4, 6, 19).
Our finding suggests that selection by host immunity and selection by antibiotics may be interrelated because of hitchhiking effects due to linkage of resistance determinants and the capsule locus; in natural settings, it suggests that creation of new strains, with different capsular types, may be enhanced by selection by β-lactams. It has long been observed that penicillin resistance is found only in a limited number of pneumococcal serotypes, but no compelling mechanism for this restriction has been proposed. Our observations suggest one hypothesis: that strains acquiring penicillin resistance will often do so by acquiring a new polysaccharide capsule type from the donor strain, thereby preserving the association between capsule and penicillin resistance. Such a hypothesis generates the prediction that the pbp2x and pbp1a genes of penicillin-resistant strains will be more closely related to corresponding alleles from other penicillin-resistant strains of the same serotype than from other penicillin-resistant strains of other serotypes. The ability of the pbp2x-cps-pbp1a region to move as a block also suggests that selection by the host immune system (for novel capsule types) could indirectly select for resistance (or susceptibility) to penicillin.
Recently published data show that introduction of the heptavalent conjugate vaccine has led not only to a reduction in the proportion of vaccine serotypes among clinical isolates of pneumococci but also to a reduction in the proportion of resistant strains, since the vaccine serotypes are most prevalent among resistant strains (7, 44). Concern has arisen that this decline in resistance may be short-lived, because antimicrobial resistance may arise in non-vaccine-type pneumococci as the latter become more common, or resistant clones may acquire non-vaccine-type capsules. Our findings suggest that once resistance to penicillin becomes established in non-vaccine types, selection by both vaccine-induced immunity and continuing antimicrobial use may work together to promote the spread of both resistance and non-vaccine-type capsule genes in the pneumococcal population.
Acknowledgments
We thank Brian G. Spratt for the GA71 strain, Susan K. Hollingshead for critical discussion of the results, and Howard Gold and Lata Venkataraman for access to the pulsed-field apparatus.
This work was supported by National Institutes of Health grant 1R01AI48935 and by a New Investigator Award to M.L. from the Ellison Medical Foundation.
REFERENCES
- 1.Adrian, P. V., and K. P. Klugman. 1997. Mutations in the dihydrofolate reductase gene of trimethoprim-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2406-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcus, V. A., K. Ghanekar, M. Yeo, T. J. Coffey, and C. G. Dowson. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol. Lett. 126:299-303. [DOI] [PubMed] [Google Scholar]
- 3.Coffey, T. J., S. Berron, M. Daniels, M. E. GarciaLeoni, E. Cercenado, E. Bouza, A. Fenoll, and B. G. Spratt. 1996. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988-1994): novel major clones of serotypes 14, 19F and 15F. Microbiology 142:2747-2757. [DOI] [PubMed] [Google Scholar]
- 4.Coffey, T. J., M. Daniels, M. C. Enright, and B. G. Spratt. 1999. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology 145:2023-2031. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 6.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Patton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 7.Dagan, R., N. Givon-Lavi, O. Zamir, and D. Fraser. 2003. Effect of a nonavalent conjugate vaccine on carriage of antibiotic-resistant Streptococcus pneumoniae in day-care centers. Pediatr. Infect. Dis. J. 22:532-540. [DOI] [PubMed] [Google Scholar]
- 8.Dowson, C. G., A. Hutchinson, J. A. Brannigan, R. C. George, D. Hansman, J. Liñares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.duPlessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echaniz-Aviles, G., M. E. Velazquez-Meza, M. N. Carnalla-Barajas, A. Soto-Nogueron, J. L. DiFabio, F. Solorzano-Santos, Y. Jimenez-Tapia, and A. Tomasz. 1998. Predominance of the multiresistant 23F international clone of Streptococcus pneumoniae among isolates from Mexico. Microb. Drug Resist. 4:241-246. [DOI] [PubMed] [Google Scholar]
- 11.Filipe, S. R., and A. Tomasz. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc. Natl. Acad. Sci. USA 97:4891-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finland, M., and M. W. Barnes. 1977. Changes in occurrence of capsular serotypes of Streptococcus pneumoniae at Boston City Hospital during selected years between 1935 and 1974. J. Clin. Microbiol. 5:154-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming, A. 1929. On the antibacterial action of cultures of penicillium with special reference to their use in the isolation of B. haemophilus. Br. J. Exp. Pathol. 10:226-236. [Google Scholar]
- 14.Garcia, E., P. Garcia, and R. Lopez. 1993. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniae type 3. Mol. Gen. Genet. 239:188-195. [DOI] [PubMed] [Google Scholar]
- 15.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Bell. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, F. 1928. The significance of pneumococcal types. J. Hyg. 27:113-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerra, B., S. Soto, R. Helmuth, and M. C. Mendoza. 2002. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob. Agents Chemother. 46:2977-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakenbeck, R. 2000. Transformation in Streptococcus pneumoniae: mosaic genes and the regulation of competence. Res. Microbiol. 151:453-456. [DOI] [PubMed] [Google Scholar]
- 19.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. deSaizieu. 2001. Mosaic genes and mosaic chromosome: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakenbeck, R., H. Ellerbrok, T. Briese, S. Handwerger, and A. Tomasz. 1986. Penicillin-binding proteins of penicillin-susceptible and -resistant pneumococci: immunological relatedness of altered proteins and changes in peptides carrying the β-lactam binding site. Antimicrob. Agents Chemother. 30:553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakenbeck, R., and M. Kohiyama. 1982. Purification of penicillin-binding protein 3 from Streptococcus pneumoniae. Eur. J. Biochem. 127:231-236. [DOI] [PubMed] [Google Scholar]
- 22.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, S.-M., L., Wang, and P. R. Reeves. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 69:1244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kell, C. M., U. K. Sharma, C. G. Dowson, C. Town, T. S. Balganesh, and B. G. Spratt. 1993. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B, and 2X of Streptococcus pneumoniae. FEMS Microbiol. Lett. 106:171-175. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence, E. R., C. A. Arias, B. Duke, D. Beste, K. Broughton, A. Efstratiou, R. C. George, and L. M. Hall. 2000. Evaluation of serotype prediction of cpsA-cpsB gene polymorphism in Streptococcus pneumoniae. J. Clin. Microbiol. 38:1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lund, E., and J. Henrichsen. 1978. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae. Methods Microbiol. 12:241-262. [Google Scholar]
- 28.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muñoz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz, R., J. M. Musser, M. Crain, D. E. Briles, A. Marton, A. J. Parkinson, U. Sorensen, and A. Tomasz. 1992. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin. Infect. Dis. 15:112-118. [DOI] [PubMed] [Google Scholar]
- 31.Nesin, M., M. Ramirez, and A. Tomasz. 1998. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J. Infect. Dis. 177:707-713. [DOI] [PubMed] [Google Scholar]
- 32.Porter, R. D., and W. R. Guild. 1969. Number of transformable units per cell in Diplococcus pneumoniae. J. Bacteriol. 97:1033-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pozzi, G., L. Masala, F. Iannelli, R. Mangenelli, L. S. Håvarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shoemaker, N. B., M. D. Smith, and W. R. Guild. 1979. Organization and transfer of heterologous chloramphenicol and tetracycline resistance genes in pneumococcus. J. Bacteriol. 139:432-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sibold, C., J. Wang, J. Henrichsen, and R. Hakenbeck. 1992. Genetic relationship of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect. Immun. 60:4119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, A. M., and K. P. Klugman. 1998. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:1329-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, A. M., and K. P. Klugman. 2001. Alterations in MurM, a cell wall muropeptide branching enzyme, increase high-level penicillin and cephalosporin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:2393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 41.Trzciński, K., C. M. Thompson, and M. Lipsitch. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl. Environ. Microbiol. 69:7364-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayakumar, M. N., S. D. Priebe, and W. R. Guild. 1986. Structure of conjugative element in Streptococcus pneumoniae. J. Bacteriol. 166:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, A. Schuchat, and the Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]