Abstract
The effect of the transformational competence-specific Streptococcus pneumoniae single-stranded DNA binding protein, SpSsbB, on the ATP-dependent three-strand exchange activity of the SpRecA protein was investigated. Although SpRecA exhibited only a trace level of strand exchange activity in the absence of SpSsbB, an extensive strand exchange reaction was observed when SpSsbB was added to the reaction solution after SpRecA. A more limited strand exchange reaction was observed, however, when SpSsbB was added to the reaction solution before SpRecA. This dependence on the order of addition, together with additional DNA-dependent ATP hydrolysis experiments, indicated that the mechanism of stimulation involved the postsynaptic binding of SpSsbB to the displaced linear single-stranded DNA reaction product. When dATP was provided in place of ATP as the nucleotide cofactor (to suppress a potentially inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA reaction substrate), the stimulatory effect of SpSsbB on the strand exchange reaction was apparent regardless of the order in which it was added to the reaction solution. These findings suggest that SpSsbB may be able to facilitate SpRecA-promoted DNA recombination reactions during natural transformation in S. pneumoniae.
Keywords: RecA protein, SSB protein, DNA strand exchange, natural transformation, Streptococcus pneumoniae
1. Introduction
The naturally transformable bacterium Streptococcus pneumoniae is able to take up DNA from its environment and incorporate it into its own chromosome [1]. This process serves as a general mutational mechanism that allows S. pneumoniae to change its genomic composition in response to environmental changes and stresses [2]. In the first step, an exogenous DNA molecule binds to the surface of the S. pneumoniae cell and one of the strands is degraded while the remaining complementary strand is transported into the cell interior. The internalized single-stranded DNA is then assimilated into a homologous region of the double-stranded S. pneumoniae chromosome [1]. Genetic studies indicate that the transformational recombination reaction is carried out by the S. pneumoniae RecA protein (SpRecA), a DNA recombinase analogous to the well-characterized RecA protein from E. coli (EcRecA) [3–4].
The EcRecA protein has single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA)-dependent ATP hydrolysis activity, and is able to promote a variety of ATP-dependent DNA pairing reactions that reflect its cellular recombination functions [5–6]. The most extensively studied reaction is the three-strand exchange reaction between a circular ssDNA and a homologous linear dsDNA [5–7]. In this reaction, EcRecA first polymerizes onto the circular ssDNA to form a filament-like structure known as the presynaptic complex. The presynaptic complex then interacts with the linear dsDNA and new base pairing interactions are established between the circular ssDNA and the complementary strand of the linear dsDNA. The complementary linear strand is then transferred to the circular ssDNA to form a nicked circular dsDNA and a displaced linear ssDNA as the final reaction products. This reaction is stimulated by the E. coli single-stranded DNA binding protein (EcSSB), which is generally included in assays as an accessory factor. During the presynaptic phase of the reaction, EcSSB binds to the circular ssDNA substrate and removes regions of secondary structure which otherwise impede the binding of EcRecA. EcRecA then displaces EcSSB from the circular ssDNA to form the presynaptic complex [8]. During the postsynaptic phase of the reaction, EcSSB facilitates the formation of the fully-exchanged products by binding to the displaced strand that is generated when the circular ssDNA is paired with the linear dsDNA [9].
We previously used the ATP-dependent three-strand exchange reaction to characterize the strand exchange activity of the SpRecA protein [10]. In our earlier study, the S. pneumoniae SsbA protein (SpSsbA), a single-stranded DNA binding protein (SSB) analogous to the EcSSB protein, was provided as an accessory factor [10–11]. We found that SpRecA differed from EcRecA in that presynaptic complex formation appeared to be inhibited (rather than enhanced) by the SpSsbA protein. An extensive strand exchange reaction was observed when SpSsbA was added to the reaction solution after SpRecA had been allowed to interact with the circular ssDNA and linear dsDNA substrates, however, suggesting that SpSsbA was able to facilitate the postsynaptic phase of the reaction [10].
In addition to SpSsbA, which is a constitutively-expressed protein that may function in routine cellular functions in a manner analogous to that of the EcSSB protein in E. coli, there is a second SSB protein in S. pneumoniae, SpSsbB, whose expression is strongly induced when the cells become competent for natural transformation (there is no counterpart to the SpSsbB protein in E. coli) [1, 12–13]. It has recently been shown that SpSsbB binds to the internalized exogenous single-stranded DNA and protects it from degradation by cellular nucleases [13–14]. In view of its central role in natural transformation, we have now investigated the effect of the SpSsbB protein on the three-strand exchange activity of the SpRecA protein.
2. Materials and Methods
2.1. Materials
S. pneumoniae RecA protein [4] and S. pneumoniae SsbB protein [12] were prepared as described. All SpRecA concentrations are expressed as total monomers, and all SpSsbB concentrations are expressed as total tetramers. ATP, dATP, [γ-32P]ATP, and [α-32P]dATP were from Amersham Biosciences. Circular φX ssDNA (+ strand) and circular φX dsDNA were from New England Biolabs. Linear φX dsDNA was prepared from circular φX dsDNA by PstI digestion as described [7]. Single- and double-stranded φX DNA concentrations were determined by absorbance at 260 nm using the conversion factors 36 and 50 µg ml−1 A260−1, respectively. All DNA concentrations are expressed as total nucleotides.
2.2. Three-strand exchange assay
The reaction solutions contained 25 mM Tris acetate (pH 7.5), 10 mM magnesium acetate, 5% glycerol, 1 mM dithiothreitol, and the concentrations of circular φX ssDNA, linear φX dsDNA, SpRecA, SpSsbB, and ATP or dATP given in the relevant figure legends. At the indicated times, aliquots (20 µL) were removed from the reaction solutions and quenched with SDS (1% final concentration)/EDTA (15 mM final concentration). The quenched aliquots were analyzed by electrophoresis on a 0.8% agarose gel using a Tris acetate-EDTA buffer system. The substrates and products of the reactions were visualized by ethidium bromide staining [7].
2.3. ATP and dATP hydrolysis assay
The reaction solutions contained 25 mM Tris acetate (pH 7.5), 10 mM magnesium acetate, 5% glycerol, 1 mM dithiothreitol, and the concentrations of circular φX ssDNA or linear φX dsDNA, SpRecA, SpSsbB, and [γ-32P]ATP/ATP or [α-32P]dATP/dATP given in the relevant figure legends. The ATP and dATP hydrolysis reactions were monitored using a thin layer chromatography method as previously described [15].
3. Results
3.1. ATP-dependent three-strand exchange
The ATP-dependent three-strand exchange activity of the SpRecA protein was examined in the absence and presence of SpSsbB protein. In the three-strand exchange assay, a circular φX ssDNA (5386 nucleotides) and a homologous linear φX dsDNA (5386 base pairs) are recombined to form a nicked circular φX dsDNA and a linear φX ssDNA. The substrates and products of this reaction are readily monitored by agarose gel electrophoresis [7].
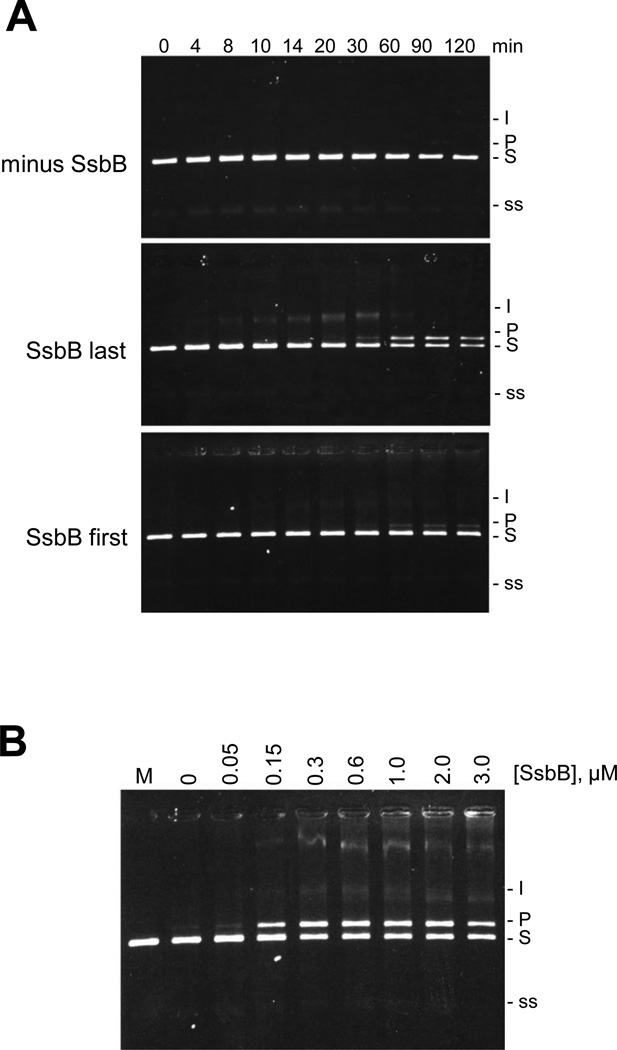
The ATP-dependent three-strand exchange reactions are shown in Fig. 1. SpRecA exhibited only a trace level of strand exchange activity in the absence of SpSsbB (Fig. 1A, minus SsbB). When SpSsbB was added to the reaction solution after the SpRecA, however, an extensive strand exchange reaction was observed in which most of the circular ssDNA substrate was converted into the nicked circular dsDNA product within 60 min (Fig. 1A, SsbB last). When SpSsbB was added to the reaction solution before the SpRecA, in contrast, only a small amount of nicked circular dsDNA product was formed, even after 120 min (Fig. 1A, SsbB first). These results showed that SpSsbB was able to stimulate the ATP-dependent strand exchange reaction, but that the degree of stimulation was strongly dependent on the order in which the SpSsbB was added to the reaction solution.
Figure 1. Effect of SpSsbB on SpRecA-promoted ATP-dependent three-strand exchange.
(A) Dependence on order of SpSsbB addition. The reaction solutions contained 5 µM circular φX ssDNA, 15 µM linear φX dsDNA, 5 mM ATP, 6 µM SpRecA, and no SpSsbB (minus SsbB), or 0.3 µM SpSsbB added 10 min after SpRecA (SsbB last), or 0.3 µM SpSsbB added 10 min before SpRecA (SsbB first). The reactions were carried out at 37 °C for the indicated times and then analyzed by agarose gel electrophoresis. In these reactions, the circular φX ssDNA (5 µM total nucleotide) was limiting relative to the linear φX dsDNA (15 µM total nucleotide = 7.5 µM base pairs) and therefore the maximum amount of linear dsDNA that could be converted to nicked circular dsDNA product was 67%. (B) Dependence on SpSsbB concentration. The reactions were carried out as described above (using the SsbB last order of addition) except that the concentration of SpSsbB was varied from 0 to 3.0 µM. The 60 min time points are shown. Labels: Slinear φX dsDNA substrate; I, partially exchanged intermediates; Pnicked circular φX dsDNA product; ssφX ssDNA substrate and product.
An additional set of reactions was carried out under the same conditions as those in Fig. 1A (using the SsbB last order of addition), except that the SpSsbB concentration was varied from 0 to 3.0 µM. As shown in Fig. 1B, the efficiency of the strand exchange reaction increased with increasing SpSsbB concentration until reaching a maximal level at 0.3 µM SpSsbB (this concentration was used for the reactions shown in Fig. 1A). Previous studies have shown that this concentration of SpSsbB would be sufficient to saturate the total amount of φX ssDNA (5 µM, circular substrate or linear product) that would be present in the strand exchange reaction solutions [16].
3.2. ATP hydrolysis
To explore the mechanistic basis for the pronounced dependence of the ATP-dependent strand exchange reaction on the order of SpSsbB addition, the effect of SpSsbB on the circular φX ssDNA and linear φX dsDNA-dependent ATP hydrolysis activities of the SpRecA protein was examined. The reactions were carried out under the same conditions as the strand exchange reactions shown in Fig. 1A.
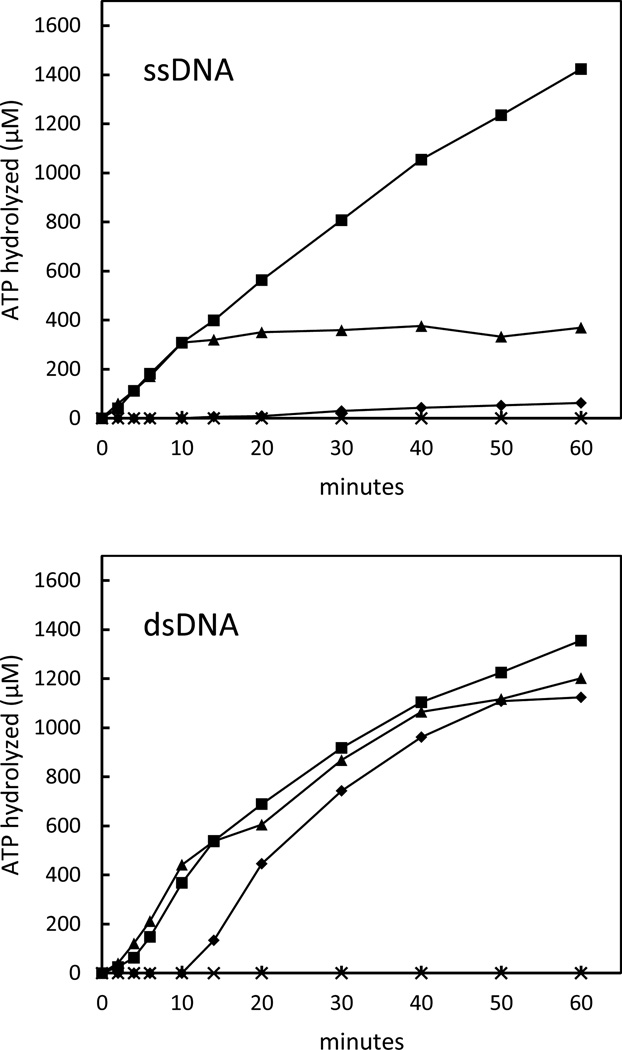
The circular ssDNA-dependent ATP hydrolysis reactions are shown in Fig. 2 (ssDNA). An initial rate of ATP hydrolysis of 31 µM min−1 was observed when SpRecA was added to the circular ssDNA in the absence of SpSsbB. This result demonstrated that SpRecA was able to form an active complex on the circular ssDNA substrate under these reaction conditions (see reference 10 for further analysis). When SpSsbB was added to the reaction solution before the SpRecA, however, virtually no ATP hydrolysis was observed. Moreover, when SpSsbB was added to an ongoing ATP hydrolysis reaction, the reaction was immediately terminated. These results showed that SpSsbB has a strong inhibitory effect on the circular ssDNA-dependent ATP hydrolysis activity of the SpRecA protein.
Figure 2. Effect of SpSsbB on SpRecA-catalyzed ATP hydrolysis.
The reaction solutions contained 5 µM circular φX ssDNA (ssDNA) or 15 µM linear φX dsDNA (dsDNA), 5 mM ATP, and 6 µM SpRecA, with or without 0.3 µM SpSsbB. The reactions were initiated by adding SpRecA at 0 min with no SpSsbB (■), by adding SpSsbB at 0 min and SpRecA at 10 min (♦), or by adding SpRecA at 0 min and SpSsbB at 10 min (▲). The reactions were carried out at 37 °C for the indicated times. SpRecA exhibited no ATP hydrolysis activity under these reaction conditions when DNA was omitted from the reaction solution (×).
The linear dsDNA-dependent ATP hydrolysis reactions are shown in Fig. 2 (dsDNA). An initial rate of ATP hydrolysis of 40 µM min−1 was observed when SpRecA was added to the linear dsDNA in the absence of SpSsbB. This result was similar to that obtained with the circular ssDNA, and demonstrated that SpRecA was also able to interact with the linear dsDNA under these conditions. In contrast to the results that were obtained with the circular ssDNA, however, the linear dsDNA-dependent ATP hydrolysis reaction was unchanged when SpSsbB was added to the reaction solution, either before or after SpRecA. These results were consistent with the expected inability of SpSsbB to compete with SpRecA for binding to the linear dsDNA (SpSsbB has no detectable linear φX dsDNA binding activity under these reaction conditions; data not shown). Furthermore, these results showed that SpSsbB does not have a general inhibitory effect on the activity of the SpRecA protein and therefore indicated that the inhibition that was observed in the circular ssDNA-dependent ATP hydrolysis reaction was due to the binding of SpSsbB to the circular ssDNA.
The results in Fig. 2 suggested that SpSsbB will have no effect on the interaction of SpRecA with the linear dsDNA substrate, but may bind to the circular ssDNA substrate and interfere with the formation of a presynaptic SpRecA-circular ssDNA complex, when it is added to a strand exchange reaction solution before SpRecA. These findings could potentially account for the results in Fig. 1A, which showed that SpSsbB stimulated the ATP-dependent strand exchange reaction most effectively when it was added to the reaction solution after SpRecA had been allowed to interact with the reaction substrates.
3.3. dATP hydrolysis
To determine if the dependence of the ATP-dependent strand exchange reaction on the order of SpSsbB addition was due to an inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA substrate, an additional series of experiments was carried out in which dATP used in place of ATP as the nucleotide cofactor. It has been shown that RecA proteins bind more tightly to ssDNA in the presence of dATP than with ATP [17–19]. It was therefore anticipated that an inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA substrate would be suppressed if dATP was provided as the nucleotide cofactor, and that this, in turn, would reduce or eliminate the dependence of the strand exchange reaction on the order of SpSsbB addition.
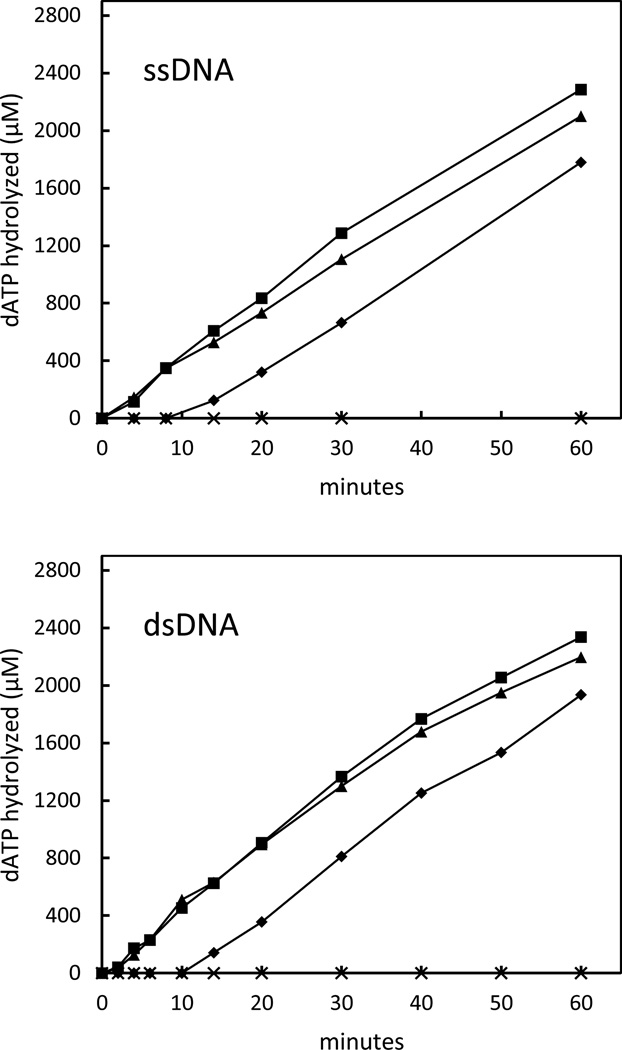
The DNA-dependent dATP hydrolysis reactions that were carried out in the absence and presence of SpSsbB are shown in Fig. 3. The initial rates of circular ssDNA-dependent dATP hydrolysis (44 µM min−1) and linear dsDNA-dependent dATP hydrolysis (45 µM min−1) that were observed in the absence of SpSsbB were similar to those for the ATP hydrolysis reactions. Furthermore, the linear dsDNA-dependent dATP hydrolysis reaction was unaffected by the addition of SpSsbB, again consistent with the expected inability of SpSsbB to compete with SpRecA for binding to the linear dsDNA. In contrast to the results that were obtained with ATP, however, the circular ssDNA-dependent dATP hydrolysis reaction was also unchanged when SpSsbB was added to the reaction solution. These results showed that the potentially inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA could indeed be suppressed by providing dATP as the nucleotide cofactor.
Figure 3. Effect of SpSsbB on SpRecA-catalyzed dATP hydrolysis.
The reaction solutions contained 5 µM circular φX ssDNA (ssDNA) or 15 µM linear φX dsDNA (dsDNA), 5 mM dATP, and 6 µM SpRecA, with or without 0.3 µM SpSsbB. The reactions were initiated by adding SpRecA at 0 min with no SpSsbB (■), by adding SpSsbB at 0 min and SpRecA at 10 min (♦), or by adding SpRecA at 0 min and SpSsbB at 10 min (▲). The reactions were carried out at 37 °C for the indicated times. SpRecA exhibited no dATP hydrolysis activity under these reaction conditions when DNA was omitted from the reaction solution (×).
3.4. dATP-dependent three-strand exchange
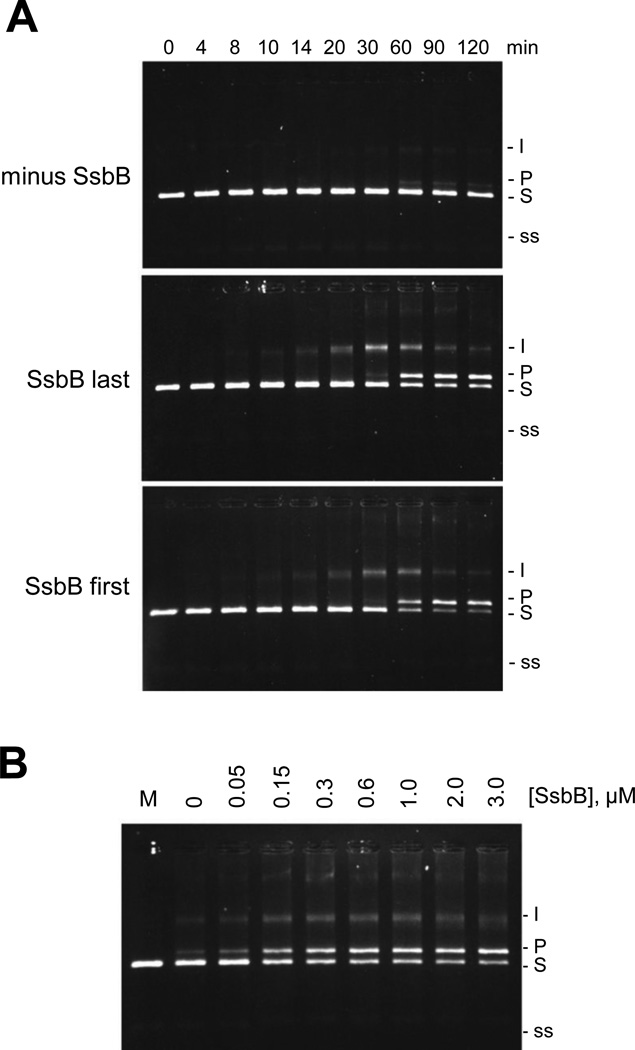
The three-strand exchange reactions that were carried out using dATP as the nucleotide cofactor are shown in Fig. 4. SpRecA exhibited a low level of dATP-dependent strand exchange activity even in the absence of SpSsbB (Fig. 4A, minus SsbB). A more extensive strand exchange reaction was observed, however, when SpSsbB was added to the reaction solution after SpRecA, with most of circular ssDNA substrate being converted into the nicked circular dsDNA product within 60 min (Fig. 4A, SsbB last). Moreover, in contrast to the results that were obtained with ATP, an equally extensive strand exchange reaction was observed when SpSsbB was added to the reaction solution before SpRecA (Fig. 4A, SsbB first). These results showed that when the potentially inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA is suppressed by using dATP as the nucleotide cofactor, the stimulatory effect of SpSsbB can be observed regardless of the order in which it is added to the strand exchange reaction solution.
Figure 4. Effect of SpSsbB on SpRecA-promoted dATP-dependent three-strand exchange.
(A) Dependence on order of SpSsbB addition. The reaction solutions contained 5 µM circular φX ssDNA, 15 µM linear φX dsDNA, 5 mM dATP, 6 µM SpRecA, and no SpSsbB (minus SsbB), or 0.3 µM SpSsbB added 10 min after SpRecA (SsbB last), or 0.3 µM SpSsbB added 10 min before SpRecA (SsbB first). The reactions were carried out at 37 °C for the indicated times and then analyzed by agarose gel electrophoresis. In these reactions, the circular φX ssDNA (5 µM total nucleotide) was limiting relative to the linear φX dsDNA (15 µM total nucleotide = 7.5 µM base pairs) and therefore the maximum amount of linear dsDNA that could be converted to nicked circular dsDNA product was 67%. (B) Dependence on SpSsbB concentration. The reactions were carried out as described above (using the SsbB last order of addition) except that the concentration of SpSsbB was varied from 0 to 3.0 µM. The 60 min time points are shown. Labels: Slinear φX dsDNA substrate; I, partially exchanged intermediates; Pnicked circular φX dsDNA product; ssφX ssDNA substrate and product.
A final set of reactions was carried out under the same conditions as those in Figure 4A (using the SsbB last order of addition), except that the concentration of SpSsbB was varied from 0 to 3.0 µM. As shown in Fig. 4B, the results were similar to those that were obtained for the ATP-dependent reaction (Fig. 1B), with optimal stimulation occurring at approximately 0.3 µM SpSsbB (this concentration was used for the reactions shown in Fig. 4A). These results suggested that the dATP and ATP-dependent strand exchange reactions may be stimulated in a similar manner, when SpSsbB is added to the reaction solution after SpRecA.
4. Discussion
The ATP-dependent three-strand exchange reaction is strongly stimulated when SpSsbB is added to the reaction solution after SpRecA, whereas only a modest stimulation is observed when it is added before SpRecA. This pronounced dependence on the order of addition, together with the demonstrated inhibitory effect of SpSsbB on the circular ssDNA-dependent ATP hydrolysis activity of SpRecA, indicates that the mechanism of stimulation involves a postsynaptic binding of SpSsbB to the displaced linear ssDNA reaction product rather than a presynaptic binding to the circular ssDNA substrate. When the potentially inhibitory effect of SpSsbB on the interaction of SpRecA with the circular ssDNA substrate is suppressed by using dATP as the nucleotide cofactor, the stimulatory effect of SpSsbB on the strand exchange reaction is apparent when it is added to the reaction solution either before or after SpRecA. The dATP-dependent strand exchange reaction, like the ATP-dependent (SpSsbB added last) reaction, proceeds optimally at SpSsbB concentrations that would be sufficient to saturate the displaced linear ssDNA product in a postsynaptic stimulation mechanism [9].
The effects of the SpSsbB protein on the strand exchange activity of the SpRecA protein are similar to those that were described previously for the SpSsbA protein [10]. This finding is consistent with previous results which showed that SpSsbA and SpSsbB have similar DNA binding properties under the solution conditions that were used for the strand exchange reactions [20], and suggests that SpSsbA and SpSsbB may have comparable effects on SpRecA during natural transformation. However, although they have similar amino-terminal DNA binding domains, the carboxy-terminal domains of SpSsbA and SpSsbB differ significantly in length and composition [16], and studies of other bacterial SSB proteins have shown that the carboxy-terminal domains can serve as binding sites for other proteins involved in DNA metabolism [21]. It is therefore conceivable that SpSsbA and SpSsbB will interact with different subsets of proteins in the S. pneumoniae cell, and this may determine the extent to which SpSsbA and SpSsbB would be functionally interchangeable during natural transformation.
The dependence of the ATP-dependent three-strand exchange reaction on the order of SpSsbB addition suggests that additional accessory proteins may be required in order for SpRecA to initiate a transformational recombination reaction between the SpSsbB-covered exogenous single-stranded DNA and the double-stranded S. pneumoniae chromosome. In this regard, it has been reported that the S. pneumoniae DprA protein is able to promote the binding of RecA protein to SSB-covered single-stranded DNA [22]. That study was carried out with the EcRecA and EcSSB proteins, however, and the relevance of the findings to the mechanism of transformational recombination in S. pneumoniae remains to be confirmed. In any case, our results suggest that once an SpRecA-ssDNA complex has been formed and strand pairing between the single strand and a homologous region of the S. pneumoniae chromosome has been initiated, SpSsbB would be able to facilitate the ensuing strand exchange reaction.
Highlights.
The effect of SpSsbB on the SpRecA-promoted strand exchange reaction was investigated.
SpRecA had only a minimal level of strand exchange activity in the absence of SpSsbB.
An extensive strand exchange reaction was observed if SpSsbB was added after SpRecA.
SpSsbB may serve as a stimulatory factor for SpRecA-mediated recombination reactions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Claverys JP, Martin B, Polard P. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 2009;33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 2.Claverys JP, Prudhomme M, Mortier-Barriere I, Martin B. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 2000;35:251–259. doi: 10.1046/j.1365-2958.2000.01718.x. [DOI] [PubMed] [Google Scholar]
- 3.Mortier-Barriere I, deSaizieu A, Claverys JP, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 4.Steffen SE, Bryant FR. Purification and characterization of the RecA protein from Streptococcus pneumoniae. Arch. Biochem. Biophys. 2000;382:303–309. doi: 10.1006/abbi.2000.2029. [DOI] [PubMed] [Google Scholar]
- 5.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;94:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 7.Cox MM, Lehman IR. RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl. Acad. Sci. USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 9.Lavery PE, Kowalczykowski SC. A postsynaptic role for single-stranded DNA-binding protein in RecA protein-promoted DNA strand exchange. J. Biol. Chem. 1992;267:9315–9320. [PubMed] [Google Scholar]
- 10.Steffen SE, Katz FS, Bryant FR. Complete inhibition of Streptococcus pneumoniae RecA protein-catalyzed ATP hydrolysis by single-stranded DNA-binding protein (SSB protein): Implications for the mechanism of SSB protein-stimulated DNA strand exchange. J. Biol. Chem. 2002;277:14493–14500. doi: 10.1074/jbc.M112444200. [DOI] [PubMed] [Google Scholar]
- 11.Steffen SE, Bryant FR. Purification and characterization of the single-stranded DNA binding protein from Streptococcus pneumoniae. Arch. Biochem. Biophys. 2001;388:165–170. doi: 10.1006/abbi.2001.2286. [DOI] [PubMed] [Google Scholar]
- 12.Hedayati MA, Grove DE, Steffen SE, Bryant FR. Expression and purification of the SsbB protein from Streptococcus pneumoniae. Protein Expr. Purif. 2005;43:133–139. doi: 10.1016/j.pep.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 13.Attaiech L, Olivier A, Mortier-Barriere I, Soulet AL, Granadel C, Martin B, Polard P, Claverys JP. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: Maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7:e1003156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison DA, Mortier-Barriere I, Attaiech L, Claverys JP. Identification of the major protein component of the pneumococcal eclipse complex. J. Bacteriol. 2007;189:6497–6500. doi: 10.1128/JB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstock GM, McEntee K, Lehman IR. ATP-dependent renaturation of DNA catalyzed by the recA protein of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1979;76:126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grove DE, Willcox S, Griffith JD, Bryant FR. Differential single-stranded DNA binding properties of the paralogous SsbA and SsbB proteins from Streptococcus pneumoniae. J. Biol. Chem. 2005;280:11067–11073. doi: 10.1074/jbc.M414057200. [DOI] [PubMed] [Google Scholar]
- 17.Menetski JP, Kowalczykowski SC. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry. 1989;28:5871–5881. doi: 10.1021/bi00440a025. [DOI] [PubMed] [Google Scholar]
- 18.Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 19.Katz FS, Bryant FR. Interdependence of the kinetics of NTP hydrolysis and the stability of the RecA-ssDNA complex. Biochemistry. 2001;40:11082–11089. doi: 10.1021/bi011030x. [DOI] [PubMed] [Google Scholar]
- 20.Grove DE, Bryant FR. Effect of Mg2+ on the DNA binding modes of the Streptococcus pneumoniae SsbA and SsbB proteins. J. Biol. Chem. 2006;281:2087–2094. doi: 10.1074/jbc.M510884200. [DOI] [PubMed] [Google Scholar]
- 21.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. >Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, Polard P, Claverys JP. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]