Abstract
Objective
The role for interferon (IFN)-α in systemic lupus erythematosus (SLE) pathogenesis is strongly supported by gene expression studies. The aim of this study was to improve characterization of the blood-IFN signature in adult SLE patients.
Methods
Consecutive patients were enrolled and followed-up prospectively. Microarray data were generated using Illumina beadchips. A modular transcriptional repertoire was employed as a framework for the analysis.
Results
Our repertoire of 260 modules, which consist of co-clustered gene sets, included 3 IFN-annotated modules (M1.2, M3.4 and M5.12) that were strongly up-regulated in SLE patients. A modular IFN signature (mIS) was observed in 54/62 (87%) patients or 131/157 (83%) longitudinal samples. The IFN signature was more complex than expected with each module displaying a distinct activation threshold (M1.2<M3.4<M5.12), thus providing a modular score to stratify SLE patients based on the presence of 0, 1, 2 or 3 active IFN modules. A similar gradient in mIS was observed within clinically quiescent patients, for whom moderate/strong modular scores (2 or 3 active IFN modules) were associated with higher anti-dsDNA titers and lower lymphocyte count than patients with absent/mild modular scores (0 or 1 active IFN modules). Longitudinal analyses revealed both stable (M1.2) and variable (M3.4 and M5.12) components of mIS over time in single patients. Interestingly, mining of other datasets suggested that M3.4 and M5.12 could be also driven by INF-β and γ.
Conclusion
Modular repertoire analysis reveals complex IFN signatures in SLE, not restricted to the previous IFN-α signature, but involving also β and γ IFNs.
Keywords: Systemic Lupus Erythematosus, Interferon, Modular analyses, Transcriptional signature
Interferons (IFNs) regulate the function of most immune cells. Type-I (including IFN-α and IFN-β) and type-II (IFN-γ) IFNs are the most studied members of the family. Many cell types, especially macrophages and dendritic cells, secrete type-I IFN, whereas type-II IFN is mostly produced by T lymphocytes and natural-killer cells. IFN-regulated genes (IRGs) (1) play a major role in controlling viral infections, and non-viral inflammatory and autoimmune disorders, such as systemic lupus erythematosus (SLE) (2).
SLE is a chronic autoimmune disease characterized by the breakdown of tolerance to nuclear antigens, especially nucleic acids, resulting in widespread organ damage. Its broad-spectrum manifestations and waning and waxing course make evaluation of disease activity challenging for clinicians. Some laboratory tests, including anti-double-stranded DNA (anti-dsDNA) antibody titers and complement factor levels, are often monitored, but several longitudinal studies have demonstrated their shortcomings (3) and no surrogate biomarker for disease activity has been validated to date. Hence, there is an important need to develop objective, simple, and robust SLE biomarkers (4).
Defining the molecular pathways responsible for the pathogenesis of SLE could aid diagnosis, biomarker development, and therapy. A pivotal role for type-I IFN (IFN-α) in the pathogenesis of SLE is strongly supported by many data, including gene-expression studies (5). Over the past decade, several groups have reported increased expression of type-I IRGs in SLE: i.e., the so-called "type-I IFN signature" (5,6). This discovery prompted the initiation of therapeutic trials aimed at evaluating the benefits of anti-IFN-α therapy in SLE patients (7), and of IFN-related biomarkers or “scores” to assess SLE disease activity or response to therapy (5,6).
Although type-I IFN activation has been previously correlated with SLE activity (5,6), this association has not been validated in recent longitudinal studies (8,9). This is probably because few longitudinal gene-expression studies have been conducted (10), and the contribution of type-II IFN to the “IFN signature” in blood and tissues may have been overlooked, as recently shown in dermatomyositis (11), Sjögren's syndrome (12), or even in SLE (13–15). In addition, because the transition from genome-wide RNA-expression analysis (thousands of genes) to only a few target genes is challenging, scores may have been biased by a knowledge-driven data-reduction process.
Our group has developed an original approach based on modules that correspond to co-clustered gene sets built via an unbiased data-driven approach (16). This approach has shown promising results in pediatric SLE (17). The aim of this study was to use modular transcriptional repertoire analysis to improve characterization of the blood-IFN signature in adult patients with SLE.
Methods
Ethics statements
The LUPUCE study (NCT00920114) was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved in France by the Comité de Protection des Personnes Sud Méditerranée 1 (IDRCB 2009-A00257-50) and in the USA by the Institutional Review Boards of the Baylor Institute of Immunology Research (IRB 011-173) and the Benaroya Research Institute (IRB 12085). Informed written consent was obtained from all patients enrolled prior to any study-related procedure.
Study design and patients' classification
Sixty-two consecutive patients with SLE fulfilling the 1997 ACR criteria were enrolled between 2009 and 2011 in the Departments of Internal Medicine and Nephrology at a French reference center for autoimmune diseases (Hôpital de la Conception, Marseille, France) and followed-up prospectively. Blood was collected by peripheral venipuncture using Tempus tubes at inclusion and at each follow-up visit. At each visit, disease activity was assessed with the SELENA-SLEDAI score. Flares were defined as ≥3 points in SELENA-SLEDAI score (improvement: a decrease of ≤2 points) (18).
Immunological analyses (auto-antibodies and complement fractions) were performed in the same laboratory (see Supplemental data: Methods). Healthy volunteers comprised 20 adult donors matched for age, gender, and ethnicity (Table S1) with no personal or family history of lupus or other autoimmune conditions, who were sampled once.
SLE patients were split into three groups (Table S1). The “at inclusion” group included all SLE patients at their first visit, irrespective of SLE disease activity at that time. The “quiescent” group included SLE patients at their first available visit with low disease activity, defined by no flare or treatment modifications for at least 60 days prior to the visit, and a SLEDAI of ≤4. The “longitudinal” group included SLE patients who had at least three consecutive visits during the study.
RNA preparation and microarray hybridization
RNA was processed as described elsewhere (19) (see Supplemental data: Methods). Data are deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo, GEO Series accession number GSE49454). PCR analyses were performed on the same samples using a Fluidigm Real-Time PCR platform (see Supplemental data: Methods).
Microarray analyses
Analyses were performed using R software environment. Background subtracted data were preprocessed with quantile normalization, flooring to 10, log2 calculation, and PAL10% (selection of transcripts that were “present” in at least 10% of samples). Probes that passed the filters (Benjamini–Hochberg multiple testing correction with False Discovery Rate (FDR) ≤0.05 and fold change (FC) ≥2 or ≤½) were considered for further gene ontology and pathway analyses.
Modular transcriptional repertoire analyses
Analyses were performed using the second generation of a modular framework as previously described (16,17). Three of 260 modules (M1.2, M3.4, M5.12) are annotated “Interferon”. Module transcript content and annotations are available online (http://www.biir.net/public_wikis/module_annotation/V2_Trial_8_Modules).
Group-level analyses showing disease fingerprints (vs. healthy baseline), as linear models, were run at the probe-level using the R package Limma (20) to determine which probes were statistically significant (Benjamini–Hochberg FDR <0.05). Positive percentages for up-regulation and negative percentages for down-regulation were calculated based on the number of statistically significant probes assigned to each module. Because probes assigned to a module typically show a consistent pattern, a percentage difference (%positive – %negative) was calculated to represent the module with one metric. The percentage difference of each of the modules was mapped on a grid where each position corresponded to one of the 62 main modules. Modules containing transcripts with increased expression were represented on a red scale while those with decreased expression were represented on a blue scale.
For individual analyses, each sample was compared to the average of the controls for each probe. Filtering comprised a fold change (≥2) and a difference in gene expression level (≥100). The level of regulation of each module was calculated as the percent difference: % up-regulated probes – % down-regulated probes. A module was considered “active” when the percent difference was ≥20%. A modular IFN signature (mIS) was considered present if at least one of the three IFN-annotated modules was active. A modular IFN score was defined according to the number of IFN-annotated modules as absent, mild, moderate, or strong if, respectively, 0, 1, 2, or 3 of these IFN-related modules were active.
Other gene-expression datasets and scores
Publicly available blood gene-expression profiles from pediatric and adult independent SLE cohorts (19,21) were used to validate modular IFN signatures observed in our cohort. Blood gene-expression profiles from patients receiving IFN-α or IFN-β (22,23) were used to evaluate the influence of various types of IFN on the modular IFN signature. Interferome v2.0 (24), an online resource containing data on interferon-inducible genes from more than 20 in vitro studies conducted on various human cells (http://interferome.its.monash.edu.au/interferome/home.jspx), was used to identify IFN-related genes and to evaluate the weight of type-I versus type-II IFN, as well as α versus β IFN, on the expression of these genes (see Supplemental data, Methods). An “IFN molecular distance to health” was defined as the number of genes with >2 fold-change for each sample compared to the healthy controls, where only genes were included that had evidence of IFN regulation from the Interferome database. Previously published IFN scores, using different sets of IFN-regulated genes, were calculated from gene-expression microarray data of our samples according to published algorithms (8, 25–27).
Statistical analyses
Numerical data were processed and analyzed using R statistical software. For continuous data, comparisons between groups were conducted using ANOVA (assuming normality was appropriate) or the non-parametric Kruskal–Wallis test. The Student's t-test or Wilcoxon's test was conducted if further testing was needed to determine which group was different. Linear models were used to test for trend. For categorical variables, Fisher’s exact test was used to determine differences in contingency tables, and the chi-squared test for trends in proportions if the categorical variable was ordinal. Correlations were assessed by Pearson's (assuming normality was appropriate) or Spearman's correlation test. Random forest analysis (randomForest package from CRAN) was used to build a classifier based on a 9-gene IFN panel. Other packages from CRAN were used for the bee-swarm plots (beeswarm package) and the heatmaps (lattice, latticeExtra, gridpackages). A Circos plot was used to represent the longitudinal IFN modular signature (28).
Results
Characteristics of SLE patients
Characteristics of the 62 SLE patients are detailed in Table S1. Median age was 38 (range: 18–70) years, 85% of patients were women, and 89% were White. Mean duration of SLE was 7.8 (range: 0–40) years. At inclusion, anti-nuclear antibodies and anti-dsDNA antibodies were positive in, respectively, 97% and 63% of patients. Fifty-two (84%) patients were receiving oral corticosteroids, 35 (56%) were receiving antimalarials, and 31 (50%) received immunosuppressive drugs. At inclusion, 27 (44%) patients were clinically quiescent whereas 35 (56%) presented with a flare with a mean SLEDAI of 12 (range: 4–26). Median follow-up time per patient was 5.9 (range: 0–28) months; data on 157 visits were collected (mean number of visits/patient 2.5 [1–6]).
Modular repertoire analysis recapitulates whole transcriptome gene-expression analyses and confirms the prevalent IFN signature in adult SLE patients
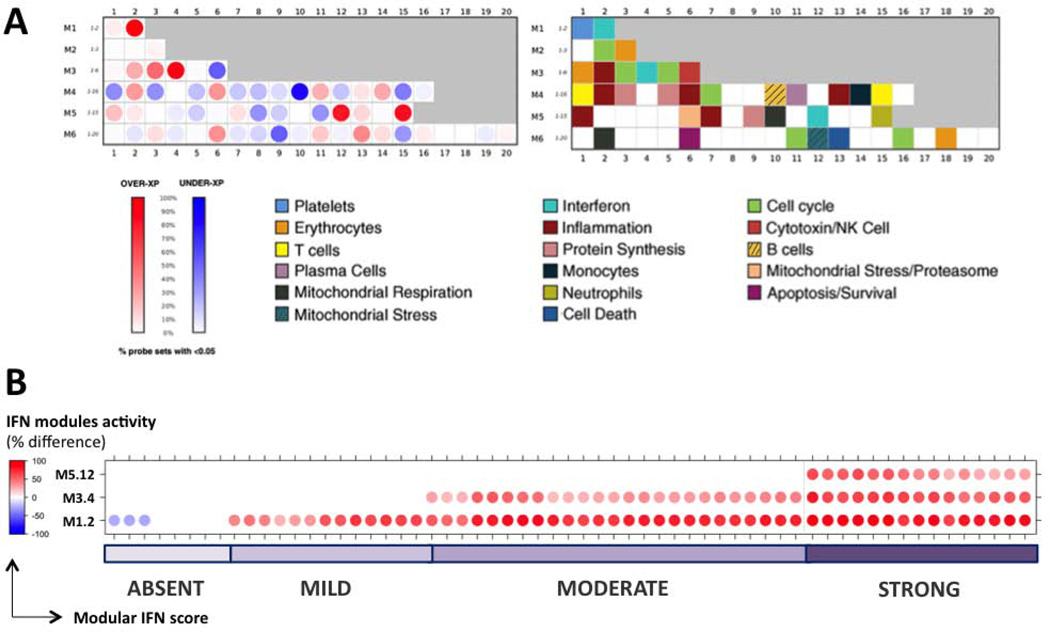
Modular repertoire analysis was performed at the group level (all SLE samples vs. all healthy controls) and at individual level (each individual SLE sample vs. all healthy controls). At the group level, this approach identified the three previously described IFN modules among the most up-regulated in SLE patient samples (n=157) compared to matched healthy controls. There were no down-regulated probes, and 36/36 (100%), 56/62 (90%), and 49/63 (78%) of significantly up-regulated probes, respectively, for M1.2, M3.4, and M5.12 (Figure 1A). Other modules corresponding to signatures previously established in SLE patients (e.g., neutrophil signature and its corresponding module M5.15) were also identified (Figure 1A). At the individual level, a modular IFN signature (at least one active IFN module) was observed in 54/62 (87%) SLE patients at inclusion (Figure 1B) and 131/157 (83%) SLE longitudinal samples (data not shown).
Figure 1. Modular repertoire analysis of SLE patients compared to healthy controls.
A: Modular analysis at the group level. Samples from SLE patients (n=157) are compared to matched healthy controls (n=20). Each module is assigned a position on the grid. The percent difference between probes significantly upregulated and down-regulated within each module determines the color and intensity of the spot (red for upregulation, blue for down-regulation in SLE). Modules annotations are provided in the second grid. A strong upregulation in SLE was observed for 4 modules, including the 3 IFN-annotated modules M1.2 (100% probes upregulated in SLE patients), M3.4 (90% upregulated, 0% downregulated) and M5.12 (78% upregulated, 0% downregulated).B Modular IFN signature at the individual level. Each sample from SLE patients at inclusion (n=62) is compared to the average of healthy controls (n=20). The percent difference between probes up- and down-regulated (FC ≥ 2 or ≤1/2 and difference ≥ 100) determines the color and intensity of each IFN module for each sample. Modular IFN signature was present (at least one active IFN module, i.e. percent difference ≥ 20) in 54/62 SLE patients. A modular IFN score was assigned to samples (“absent”, “mild”, “moderate”, or “strong”) according to the number (0, 1, 2 or 3) of active IFN modules.
Results obtained using this modular approach matched those observed with a gene-level approach (Figure S1). Applying published IFN scores to our cohort revealed the presence of an IFN signature (e.g., a “positive” IFN score) in 71–88.5% of the 157 SLE samples (Table S2).
Individual modular repertoire analysis reveals a dynamic modular IFN signature within and across patients
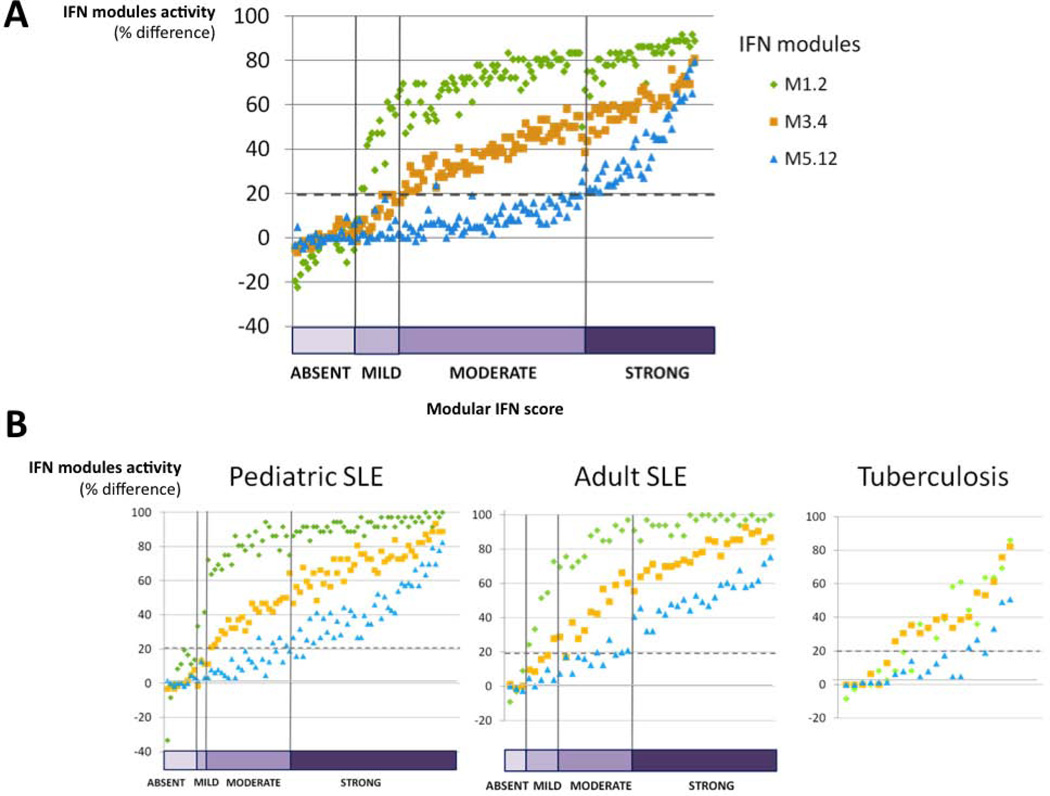
Strikingly, we observed a coordinated gradient of IFN modules across samples, each module displaying a distinct activation threshold. Indeed, when only one of the three IFN modules was up-regulated, it always corresponded to M1.2. Module M3.4 appeared next, and there was no M5.12 module up-regulation in the absence of the two others (Figure 1B). Samples could then be classified according to a “qualitative” modular IFN score based on the number of active IFN modules: “absent” for 0, “mild” for 1, “moderate” for 2, and “strong” for 3 active IFN modules. Among the 62 SLE patients, only 8 (13%) had an “absent” modular IFN signature at inclusion, whereas 8 (13%), 24 (39%), and 22 (35%) exhibited “mild”, “moderate”, or “strong” modular IFN scores, respectively (Figure 1B). Similar patterns were observed across longitudinal (n=157) SLE samples (Figure 2A. In addition, a similar coordinated gradient of the 3 IFN modules was observed in two independent cohorts of SLE patients involving both children and adults, while a different gradient was observed in a cohort of patients with tuberculosis, although they did exhibit an interferon signature (Figure 2B).
Figure 2. Repartition of SLE samples according to modular IFN score.
A: The 157 SLE samples were ordered and classified according to the modular IFN score (number of active IFN modules, from 0 to 3). IFN signature was “absent” in 26 samples, “mild” in 17 samples, “moderate” in 68 samples and “strong” in 46 samples. A dynamic IFN signature, from M1.2 to M3.4 and M5.12, was observed: when only one of the three IFN modules was up-regulated, it always corresponded to M1.2. Module M3.4 appeared next, and there were no M5.12 modules up-regulated in the absence of the two others. B: The same dynamic IFN signature was observed in 2 independent cohorts of SLE patients: a cohort of 82 pediatric patients (19), with Hispanic (57%), Black (23%), White (15%) and Asian (5%) ethnicities, and a cohort of 43 adult patients (21), with Black (54%), White (44%) and Asian (2%) ethnicities. On the contrary, although a modular IFN signature was observed in active tuberculosis (19), no such gradient was observed in IFN modules.
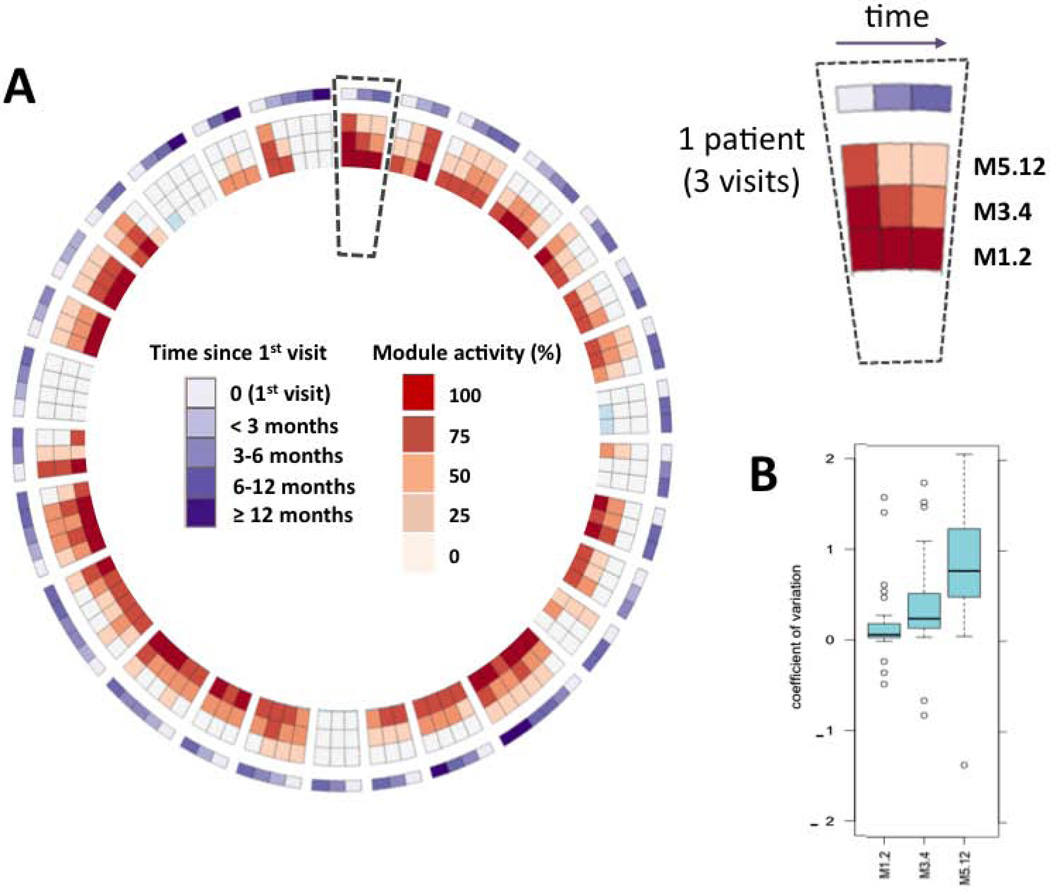
Longitudinal analyses were performed on 29 SLE patients who had at least three visits (Table S1). Median follow-up time was 8.3 (range: 2–28) months, with a median interval between visits of 3.2 (0.5–19) months. Although module M1.2 was very stable over time within individual patients (coefficient of variation [CV] 0.05), significantly much greater variation was seen for modules M3.4 (CV 0.39), and especially M5.12 (CV 0.91) (Figure 3), supporting that a complex regulation of biological pathways underlies the IFN signature of SLE and that the three modules represent distinct interferon signatures.
Figure 3. Longitudinal intra-individual variation of IFN modules in SLE patients.
Longitudinal analyses were obtained for 29 SLE patients with at least 3 consecutive visits. A: The level of upregulation of each IFN module at each visit is plotted on the circos figure, representing from center to periphery M1.2, M3.4, M5.12 and time elapsed since 1st visit. Spaces separate different patients (e.g., 3 visits of the same patient are framed by the dotted line). B: Coefficient of variation (mean CV ± SD), corresponding to intra-individual variability of IFN modules, indicates that while M1.2 is stable over time for a given patient (CV = 0.05 ± 0.88), M3.4 (CV = 0.39 ± 0.56) and even more M5.12 (CV = 0.91 ± 0.82) show fluctuations across time, reflecting the complexity of IFN signature. These differences of variability between modules are significant (M1.2 CV vs. M3.4 CV, p= 0.0033; M1.2 CV vs. M5.12 CV, p = <0.0001; M3.4 CV vs. M5.12 CV, p = 0.00065).
IFN modules and modular IFN score are associated with disease activity in SLE patients
At the individual level, the levels of IFN modules upregulation (percent difference between up-regulated and down-regulated probes in SLE patients versus controls) were correlated with anti-dsDNA titers (Table 1). M1.2 levels did not correlate with the SLEDAI score or with the presence of a flare (Table 1). Conversely, a weak but significant correlation was observed between expression of M3.4 and M5.12 modules and SLEDAI and/or the presence of a flare. Both M3.4 and M5.12 modules correlated with cutaneous flares whereas only M5.12 correlated with renal flares (Table 1).
Table 1. Correlations of IFN modules with clinical and biological markers of SLE activity.
The levels of IFN modules upregulation (% difference) were correlated with clinical and biological markers of activity in the 157 SLE samples.
| IFN modules | M1.2 | M3.4 | M5.12 | |
|---|---|---|---|---|
| SLEDAI score | ||||
| spearman correlation | 0.05 | 0.12 | 0.21 | |
| p-value | 0.54 | 0.13 | 0.008 | |
| Flare (no flare, mild/moderate flare, or severe flare) | ||||
| spearman correlation | 0.12 | 0.2 | 0.28 | |
| p-value | 0.13 | 0.012 | 0.0003 | |
| Cutaneous flare | ||||
| spearman correlation | 0.11 | 0.15 | 0.21 | |
| p-value | 0.17 | 0.05 | 0.01 | |
| Articular flare | ||||
| spearman correlation | −0.13 | −0.024 | 0.023 | |
| p-value | 0.11 | 0.77 | 0.77 | |
| Hematological flare | ||||
| spearman correlation | −0.012 | −0.043 | 0.055 | |
| p-value | 0.13 | 0.6 | 0.5 | |
| Renal flare | ||||
| spearman correlation | 0.066 | 0.13 | 0.22 | |
| p-value | 0.41 | 0.1 | 0.007 | |
| Anti-dsDNA titer | ||||
| spearman correlation | 0.28 | 0.26 | 0.19 | |
| p-value | 0.0007 | 0.0015 | 0.002 | |
| Low C3 or C4 | ||||
| spearman correlation | 0.072 | 0.086 | 0.12 | |
| p-value | 0.37 | 0.29 | 0.14 | |
Individual clinical and biological parameters were also compared at the group level according to modular IFN scores. No differences were observed concerning patient age, gender, ethnicity, or disease duration in the four groups (Table S3). Anti-dsDNA titers were significantly higher (p=0.03) and lymphocyte counts lower (p<0.0001) in patients with moderate or strong modular IFN scores compared to patients with absent or mild scores (Figure S2, Table S4). The SLEDAI score did not significantly vary between these groups, though a linear trend was observed (p=0.06). Only cutaneous flares were significantly more frequent in patients with moderate or strong modular IFN scores compared to patients with absent or mild scores (Table S4). Patients with moderate or strong modular IFN scores were less likely to receive antimalarial (p=0.002) or combined immunosuppressant plus antimalarial (p=0.0006) therapies.
Modular IFN scores allow stratification of clinically quiescent patients
Because IFN signatures can be rapidly influenced by recent treatment modifications (e.g., a severe flare and/or receiving high-dose steroid pulses before sampling (33)), the modular IFN signature was investigated in the clinically quiescent group, which included 64 visits corresponding to 34 SLE patients (Table S1. At the group level, the modular IFN signature was comparable to that of non-quiescent patients. At the individual level, 51/64 (80%) of patients exhibited a modular IFN signature (Figure S3). No differences were observed concerning age, gender, ethnicity or disease duration among these 34 quiescent patients according to IFN score groups (Table S5). Anti-dsDNA titers were significantly higher (p=0.007) and lymphocyte counts lower (p<0.0001) in patients with moderate or strong modular IFN scores compared to patients with absent or mild scores (Table S6). No significant differences in therapy were observed between patients with moderate or strong modular IFN scores compared to patients with absent or mild signatures.
Modular IFN signature and molecular insight into SLE pathogenesis
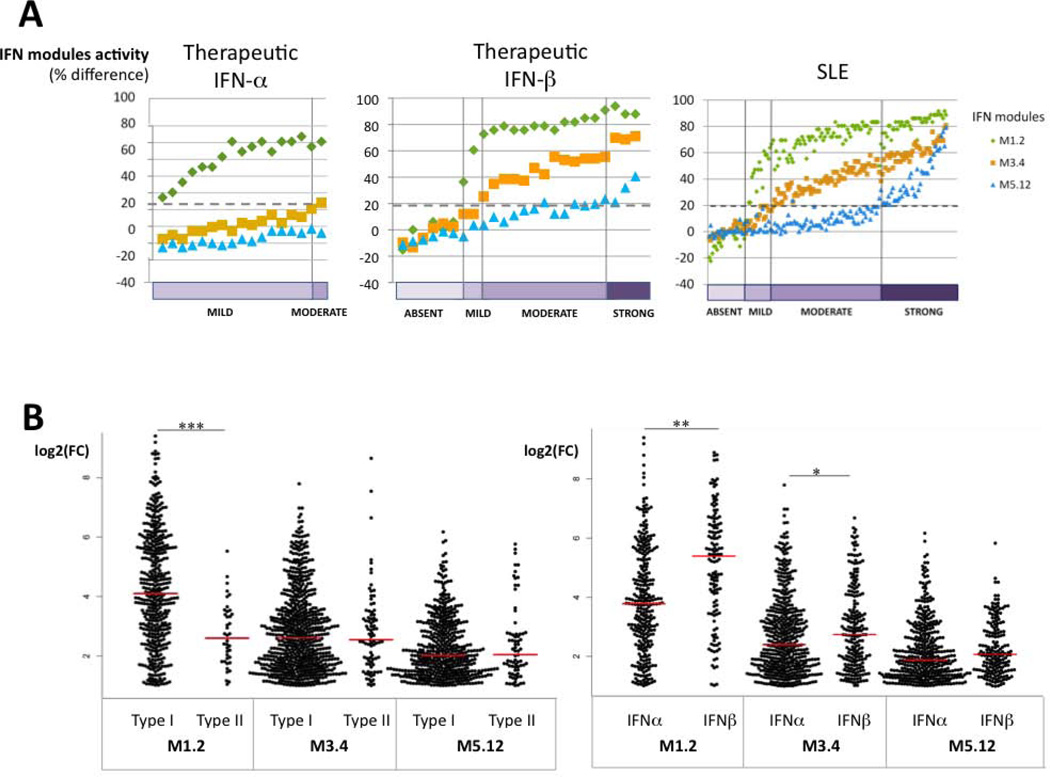
Variations in modular IFN signature across patients and within individual patients followed-up longitudinally, especially for those who were clinically quiescent with no therapeutic modifications, could reflect disease-specific pathogenic events. Specifically, the different IFN modular patterns could partially reflect the involvement of different types of interferons. To test this hypothesis, we first analyzed the effects of exogenous IFN administration using publicly available datasets. The effects of therapeutic IFNs on modular IFN signatures in patients treated with IFN-α (for hepatitis C virus infection) (23) or with IFN-β (for multiple sclerosis) (22) were analyzed. Strikingly, administration of IFN-α or -β alone in these patients could not completely reproduce the modular IFN signature observed in SLE. Indeed, treatment with IFN-α resulted in the upregulation of M1.2 only, while treatment with IFN-β was associated in most patients with both a strong upregulation of M1.2 and an upregulation of M3.4. Conversely, transcripts belonging to M5.12 were poorly induced by IFN-α or IFNβ alone (Figure 4A).
Figure 4. Effect of different types of IFN on IFN modular patterns.
Modular analysis was performed on gene expression data from 2 public domain datasets (22,23). A: At the individual level, treatment with IFN-α results in the upregulation of M1.2 only, while treatment with IFN-β is associated in most patients with upregulation of both M1.2 and M3.4, as well as M5.12 in some patients. B: The responsiveness to different types of IFN of the genes from the 3 IFN modules was evaluated using the Interferome database: the log2(FC) observed in each experiment for each gene after in vitro stimulation was compared between type I and type II IFN, as well as between IFNα and β and represented on the Beeswarm plots. Transcripts belonging to M1.2 were induced significantly more by type I than type II IFN (median log2(FC) of 4.10 vs 2.60, ***p<0.0001), while transcripts belonging to M3.4 and M5.12 were similarly induced by type I and type II IFN (respectively, 2.60 vs 2.54, p=0.87 and 2.01 vs 2.04, p=0.68). In addition, M1.2 and M3.4 transcripts were induced significantly more by IFN-β than by IFN-α (5.39 versus 3.78, **p=0.0001 and 2.74 versus 2.40, *p=0.034 respectively), while transcripts belonging to M5.12 only exhibited a non significant trend (2.07 versus 1.89, p=0.051).
We also used the Interferome database to determine to what extent genes belonging to IFN modules were inducible in vitro by the different IFNs. Genes significantly upregulated in SLE patients compared to healthy controls were identified in each IFN-annotated module (27 genes in M1.2, 48 in M3.4, 48 in M5.12), and filtered to select genes registered as “interferon-related” (Figure 4B). Comparison of log-2(fold-change) after stimulation with different types of IFNs in this database showed that M1.2 transcripts were markedly induced more by type-I than type-II IFN (p<0.0001). In addition, and in agreement with what was observed with in vivo data, M1.2 and M3.4 transcripts were induced significantly more by IFN-β than by IFN-α (p=0.0001 and p=0.034, respectively). Finally, transcripts belonging to M3.4 and M5.12 were similarly induced in vitro by type-I and type-II IFNs. Overall, these results suggest that the dynamic modular IFN signature observed in SLE patients is not exclusively driven by IFN-α; IFN-β could play a role in the upregulation of these modules, and type-II IFN, in combination with type-I IFNs, could contribute to the full-blown response of M3.4 and M5.12.
We next used a gene-level whole-transcriptome approach to characterize differences between modular IFN score groups in clinically quiescent patients. There were 209 differentially expressed transcripts in these patients compared to healthy controls; these corresponded to 171 unique gene symbols, of which 75 were registered as “IFN-related” in Interferome. Corresponding transcripts were used to calculate an “IFN molecular distance to health” for each sample, this was strongly correlated to modular IFN score (r= 0.946, p <0.0001) (Figure S4A). Quiescent SLE samples were listed according to their modular IFN score, and a hierarchical clustering of the 209 differentially expressed transcripts was performed. Five clusters were identified (genes and detailed pathway annotations are provided in Table S9 and Figure S4B). Three of them (C3, C4, C5) were annotated “IFN-related”, but exhibited various patterns of expression in the different modular IFN score groups. C4, “IFN early signaling-related”, was upregulated similarly in patients with mild, moderate, or strong modular IFN scores (but not in patients from the “absent” group); upregulation of transcripts belonging to C3 and C5 only occurred in the “moderate” and “strong” groups. Interestingly, C3 was annotated “IFN-regulated chemotaxis”, and C5 as “IFN-downstream signaling-related”, which suggests that the modular IFN score could reflect sequential pathogenic events that occur from a state of immunological quiescence (absent modular signature), to dynamic activation of transcripts from the early to downstream parts of the IFN pathway.
Modular analysis: a new approach for longitudinal analyses and biomarker development in SLE?
For clinical applicability, an approximation of modular IFN score should be obtainable using focused assays such as PCR and targeting only a few genes. First, to validate the microarray data, we confirmed the overexpression of 16 genes belonging to the three IFN modules using whole-blood QRT-PCR in the same SLE samples (Figure S5A). There were excellent correlations between microarray and TaqMan assays across samples (Figure S5B). We also used a random-forest approach to reduce this to the best three classifiers of each of the modular IFN signatures. We could accurately classify samples according to the predefined categories of modular IFN score (absent/mild vs. moderate/strong) with an average overall error of 4.7% (Table S7).
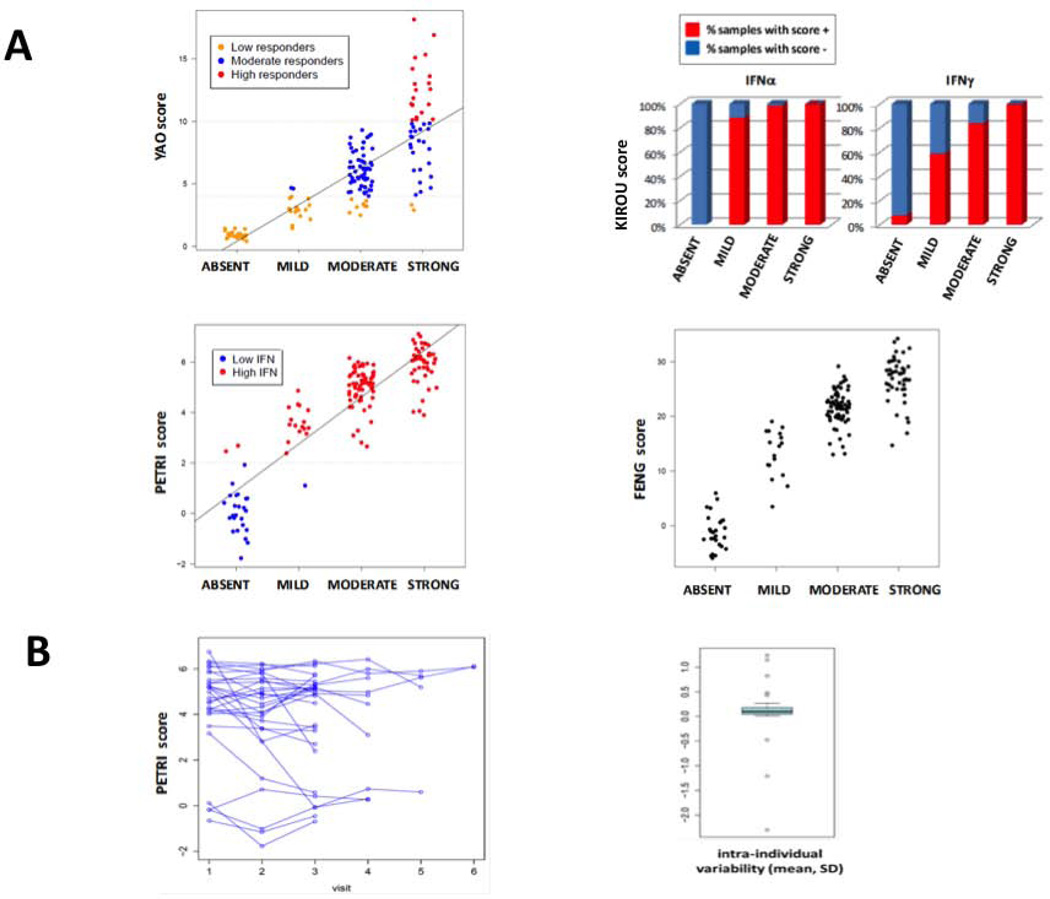
We compared our modular IFN signatures to other previously established IFN scores from the literature. Although all scores were strongly correlated with modular IFN signature (Figure 5A), they were not redundant. For example, although most samples were considered “high IFN” using Petri IFN score, these samples could belong to mild, moderate, or strong groups according to our modular IFN scores. Similarly, patients considered as “low responders” using the Yao IFN score could belong to different groups according to our modular IFN score (mostly absent or mild, but also moderate or strong). Interestingly, all patients with an “absent” modular IFN score had a negative IFN-α “Kirou score”, whereas all patients with a strong modular IFN score had both IFN-α- and IFN-γ-positive Kirou scores (Figure 5A), which is in agreement with the possible implication of Type II IFN in the activity of modules M3.4 and M5.12. Concerning longitudinal variations in IFN signatures, there was no significant variation in Petri IFN score across time in our samples (mean per/patient CV 0.07) (Figure 5B), whereas a variation was identified using the modular IFN score (Figure 3). Notably, genes selected for IFN scores according to the literature are mostly from M1.2 (the least variable module) and to a lesser extent M3.4, but very few belong to M5.12 (the more variable IFN module)Table S8.
Figure 5. Inter-scores correlations in SLE patients.
A: Modular IFN score (absent/mild/moderate/strong) is compared to 4 scores from the literature using the 157 SLE samples of the present cohort. Linear model shows an increase in the Yao score of 2.9 points per category of modular IFN score (p<0.0001), an increase in the Petri score of 1.85 points per category (p<0.0001) and an increase in the Feng score of 9.14 points per category (p<0.0001). Both IFN-α and IFN-γ Kirou’s scores were linked (chi-squared test for trend in proportions p<0.0001) with modular IFN score. B: Petri IFN score in 29 patients with ≥ 3 visits (longitudinal cohort) across time. A limited inter-individual variability of the Petri IFN score across time is observed (coefficient of variability = 0.07). This score is based on the expression of 3 IFN-inducible genes (IFI27, IFI44 and OAS3).
Discussion
Microarray technology is a valuable tool to gain insight into the molecular complexity of autoimmune diseases and to identify new therapeutic targets and biomarkers (15). Just a decade ago, the first SLE transcriptomic studies revealed a blood IFN-α signature (29,30). These data were supported by many additional studies (8,9,17,26,27,31,32) and led to the development of IFN-α blockers that are currently being tested in clinical trials (7). Herein, we report original results from data-driven constructed framework models of gene-sets (16) to assess, cross-sectionally and longitudinally, the blood-transcriptional profiles of adult SLE patients. This modular approach has confirmed the high prevalence of IFN signatures in these patients, as reported using various IFN scoring systems (5,6). But, strikingly, the IFN signature was more complex than expected: a dynamic modular IFN signature was observed in SLE patients with each component module displaying a distinct activation threshold. Corresponding transcripts, reflecting distinct aspects of the IFN signaling pathway, were not exclusively IFN-α-inducible, but might have also been induced by IFN-β and IFN-γ. In addition, the modular IFN signature correlated with serological disease activity (anti-dsDNA titer and lymphocyte count) and, to some extent, clinical activity. Interestingly, this modular signature was variable across time in single individuals: M1.2 was stable, whereas M3.4 and M5.12 did vary. Importantly, the modular signature allowed stratification of quiescent patients and we observed correlations of the modular score with SLE biological activity parameters as well as intensity of maintenance treatments. So, additional studies are needed to test the usefulness of the IFN modular score to predict future flares in these clinically quiescent SLE patients.
These results differ from recent publications, which show no significant variation in IFN scores across time and no correlations with disease activity (8,9). Interestingly, most published transcripts selected to build IFN scores belong to M1.2, the least variable of the IFN modules (Table S8). Because therapies for flares, especially high-dose corticosteroids, can rapidly “extinguish” the IFN signature (33), we repeated our analyses on quiescent patients with no recent treatment modifications and were able to show the same variability. Although our modular IFN score was strongly correlated with previously reported IFN scores, it differed inherently from them, perhaps allowing a more global and qualitative overview of IFN-pathway activation. Thus, a modular approach that does not just rely on knowledge (choice of genes previously identified as IFN-inducible) and/or expression intensity (genes with the highest fold changes), but on hypothesis-free and data-driven descriptions, could result in a “qualitative” appreciation of the IFN signature in individual SLE patients and aid the construction of new SLE biomarkers.
Having observed this dynamic IFN signature in SLE, we used the modular IFN score to stratify patients and compare gene expression across these groups. Indeed, combining our modular approach with the Interferome database, we conclude that not only IFN-α but also IFN-β and IFN-γ may contribute to the IFN signature in SLE. Indeed, as recently stated by others in the context of infectious (34–36) and autoimmune systemic diseases (11,12), there is a large overlap between type-I and type-II IFN-inducible signatures. Interestingly, the transcripts selected by Hall et al. (12) to discriminate between type-I (IFIT3 and MDA5) and type-II (GBP1 and GBP2) IFNs in tissue biopsies from patients with Sjögren's syndrome reflect the type-I to type-II transition observed between our three IFN modules: IFIT3 belongs to M1.2, MDA5 and GBP1 to M3.4, and GBP2 to M5.12. Also, IFN-inducible chemokines (CCL2, CXCL10, CCL19) and Blyss/BAFF, whose circulating levels have been associated with disease activity (15,37), are both type-I and type-II IFN-inducible (13).
Finally, recent reports of overlapping downstream signaling events (1,14) demonstrate cross-talk between type-I and type-II IFN-signaling pathways. Some experiments support the role of low-dose type-I IFNs in priming cells to secrete type-II IFNs (38). Similarly, the ability of IFN-γ to enhance signaling through Toll-like receptors may enhance type-I IFN secretion (39). Studying gene expression in the context of viral infection, Su et al. have demonstrated that type-I IFN-induced genes dominate the early phase of viral infection, whereas IFN-γ, triggered by IFN-α, and its targets were responsible for the effector phase of the anti-viral responses (40). Our results suggest a dynamic IFN response in SLE, driven by the interaction of genes induced by various IFNs. The theory of a sequential role of type-II IFN following type-I IFNs is supported by murine data on the development of lupus nephritis (41,42), and previously suggested by Crow et al. (43,44).
We show that only the 3rd IFN module, M5.12, correlated with renal flares in our cohort. Disappointing results for the IFN-α blockade have been reported recently in clinical trials in SLE and psoriasis (45–47). In a phase-II clinical trial in SLE patients, a significant response was observed only in patients with a “low-level” IFN signature (46). These surprising results may suggest a need for stronger inhibition of IFN-α in patients with a “high-level” signature (7), or may indicate that IFNs other than IFN-α are involved in SLE disease pathogenesis and that targeting IFN-α alone in SLE may not be sufficient to control disease activity.
This study has several limitations. First, whole-blood gene-expression analysis does not permit identification of cell-specific components of the IFN signature (48). However, it allows easy sample collection and preparation which are fundamental elements for clinical applicability. Second, modular IFN signatures may differ according to ethnic backgrounds, while most patients in our cohort were Caucasian. However, we observed similar patterns in the pediatric cohort of Pascual (19), which contained mostly Hispanic and African American patients (only 15% non-Hispanic White patients) and in the adult cohort of Arasappan (21), which included only 44% White patients. Finally, the dynamic modular signature we observed could have been linked to the use of SLE samples, as SLE was included in the building of the original modular framework (17). However, the different IFN signature pattern observed in tuberculosis samples, also used in the original framework construction, rules out this possibility and instead supports the diversity of IFN pathways in different human diseases.
Together, the results obtained using our modular framework support the hypothesis that the IFN signature observed in SLE patients corresponds to more IFN types than just IFN-α. This approach may enable a better molecular stratification of SLE patients. Ultimately, these modular signatures may aid in the design of biomarkers to assess disease activity and in the selection and monitoring of therapies for SLE, as well as other possible IFN-related autoimmune (31) or infectious (49) conditions.
Supplementary Material
Acknowledgements
We thank Nathalie Bardin, Nicolas Schleinitz, Jean-Marc Durand, Karin Mazodier, Bertrand Dussol, Philippe Brunet, Valérie Moal, Radj Purgus, Julie Frances, Catherine Farnarier, Quynh-Anh Nguyen and Peter Lindsey for their help on this project. We thank Nicole Cesarale and nurses from the outpatient clinics of Nephrology and Internal Medicine for their help in the screening of patients.
Funding sources: The LUPUCE study that was funded by Assistance Publique Hôpitaux de Marseille (AORC-APHM), ADEREM (Association pour le Développement des Recherches Biologiques et Médicales) and CRN (Centre de Recherche en Nephrologie, La Conception, Marseille) and received support from NIAID Autoimmunity Centers of Excellence (U19 AIO82715).
Footnotes
Disclosures: VP declares Consultant/Speaker fees from Novo-Nordisk and Roche. There are no relevant disclosures to report for other co-authors.
References
- 1.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rönnblom L, Eloranta ML. The interferon signature in autoimmune diseases. Curr Opin Rheumatol. 2013 Mar;25(2):248–253. doi: 10.1097/BOR.0b013e32835c7e32. [DOI] [PubMed] [Google Scholar]
- 3.Isenberg DA, Manson JJ, Ehrenstein MR, Rahman A. Fifty years of anti-ds DNA antibodies: are we approaching journey's end? Rheumatology (Oxford) 2007 Jul;46(7):1052–1056. doi: 10.1093/rheumatology/kem112. Epub 2007 May 11. [DOI] [PubMed] [Google Scholar]
- 4.Ahearn JM, Liu CC, Kao AH, Manzi S. Biomarkers for systemic lupus erythematosus. Transl Res. 2012 Apr;159(4):326–342. doi: 10.1016/j.trsl.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiche L, Jourde-Chiche N, Pascual V, Chaussabel D. Current perspectives on systems immunology approaches to rheumatic diseases. Arthritis Rheum. 2013;65:1407–1417. doi: 10.1002/art.37909. [DOI] [PubMed] [Google Scholar]
- 7.Kirou KA, Gkrouzman E. Anti-interferon alpha treatment in SLE. Clin Immunol. 2013;148:303–312. doi: 10.1016/j.clim.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, Lal P, et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009 Oct;18(11):980–989. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landolt-Marticorena C, Bonventi G, Lubovich A, Ferguson C, Unnithan T, Su J, et al. Lack of association between the interferon-α signature and longitudinal changes in disease activity in systemic lupus erythematosus. Ann Rheum Dis. 2009;68:1440–1446. doi: 10.1136/ard.2008.093146. [DOI] [PubMed] [Google Scholar]
- 10.Tektonidou MG, Ward MM. Validity of clinical associations of biomarkers in translational research studies: the case of systemic autoimmune diseases. Arthritis Res Ther. 2010;12:R179. doi: 10.1186/ar3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong D, Kea B, Pesich R, Higgs BW, Zhu W, Brown P, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PLoS One. 2012;7(1):e29161. doi: 10.1371/journal.pone.0029161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall JC, Casciola-Rosen L, Berger AE, Kapsogeorgou EK, Cheadle C, Tzioufas AG, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci USA. 2012 Oct 23;109(43):17609–17614. doi: 10.1073/pnas.1209724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harigai M, Kawamoto M, Hara M, Kubota T, Kamatani N, Miyasaka N. Excessive production of IFN-gamma in patients with systemic lupus erythematosus and its contribution to induction of B lymphocyte stimulator/B cell-activating factor/TNF ligand superfamily-13B. J Immunol. 2008 Aug 1;181(3):2211–2219. doi: 10.4049/jimmunol.181.3.2211. [DOI] [PubMed] [Google Scholar]
- 14.Karonitsch T, Feierl E, Steiner CW, Dalwigk K, Korb A, Binder N, et al. Activation of the interferon-gamma signaling pathway in systemic lupus erythematosus peripheral blood mononuclear cells. Arthritis Rheum. 2009 May;60(5):1463–1471. doi: 10.1002/art.24449. [DOI] [PubMed] [Google Scholar]
- 15.Bauer JW, Petri M, Batliwalla FM, Koeuth T, Wilson J, Slattery C, et al. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis Rheum. 2009 Oct;60(10):3098–3107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013 Apr 18;38(4):831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petri M, Buyon J, Kim M. Classification and definition of major flares in SLE clinical trials. Lupus. 1999;8(8):685–691. doi: 10.1191/096120399680411281. [DOI] [PubMed] [Google Scholar]
- 19.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010 Aug 19;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 21.Arasappan D, Tong W, Mummaneni P, Fang H, Amur S. Meta-analysis of microarray data using a pathway-based approach identifies a 37-gene expression signature for systemic lupus erythematosus in human peripheral blood mononuclear cells. BMC Med. 2011;9:65. doi: 10.1186/1741-7015-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra S, Bustamante MF, Pérez-Miralles F, Rio J, Ruiz de Villa MC, et al. Search for specific biomarkers of IFNβ bioactivity in patients with multiple sclerosis. PLoS One. 2011;6(8):e23634. doi: 10.1371/journal.pone.0023634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MW, Tsukahara T, McClintick JN, Edenberg HJ, Kwo P. Cyclic changes ingene expression induced by Peg-interferon alfa-2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patients during the first 10 weeks of treatment. J Transl Med. 2008 Nov 5;6:66. doi: 10.1186/1479-5876-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013 Jan;41(Database issue):D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, Brohawn P, et al. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009 Jun;60(6):1785–1796. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 26.Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 27.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 28.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs BW, Liu Z, White B, Zhu W, White WI, Morehouse C, et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis. 2011;70:2029–2036. doi: 10.1136/ard.2011.150326. [DOI] [PubMed] [Google Scholar]
- 32.Nikpour M, Dempsey AA, Urowitz MB, Gladman DD, Barnes DA. Association of a gene expression profile from whole blood with disease activity in systemic lupus erythaematosus. Ann Rheum Dis. 2008;67:1069–1075. doi: 10.1136/ard.2007.074765. [DOI] [PubMed] [Google Scholar]
- 33.Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, Meeker T, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010 Jun 17;465(7300):937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh GC, Schreiber MF, Bautista R, Maude RR, Dunachie S, Limmathurotsakul D, et al. Host responses to melioidosis and tuberculosis are both dominated by interferon-mediated signaling. PLoS One. 2013;8(1):e54961. doi: 10.1371/journal.pone.0054961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kropp KA, Robertson KA, Sing G, Rodriguez-Martin S, Blanc M, Lacaze P, et al. Reversible inhibition of murine cytomegalovirus replication by gamma interferon (IFN-γ) in primary macrophages involves a primed type I IFN-signaling subnetwork for full establishment of an immediate-early antiviral state. J Virol. 2011 Oct;85(19):10286–10299. doi: 10.1128/JVI.00373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification oftype I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA. 2012 Mar 13;109(11):4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petri MA, van Vollenhoven RF, Buyon J, Levy RA, Navarra SV, Cervera R, et al. BLISS-52 and BLISS-76 Study Groups. Baseline predictors of systemic lupus erythematosus flares: data from the combined placebo groups in the phase III belimumab trials. Arthrit Rheum. 2013 Aug;65(8):2143–2153. doi: 10.1002/art.37995. [DOI] [PubMed] [Google Scholar]
- 38.Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, et al. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. 2010 Apr 27;8(4):e1000361. doi: 10.1371/journal.pbio.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, Koshiba R, et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci USA. 2008 Dec 23;105(51):20446–20451. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng SL, Moslehi J, Craft J. Roles of interferon-gamma and interleukin-4 in murine lupus. J Clin Invest. 1997 Apr 15;99(8):1936–1946. doi: 10.1172/JCI119361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas C, Ryffel B, Le Hir M. IFN-gamma receptor deletion prevents autoantibody production and glomerulonephritis in lupus-prone (NZB×NZW)F1 mice. J Immunol. 1998 Apr 15;160(8):3713–3718. [PubMed] [Google Scholar]
- 43.Crow MK. Type I interferon and autoimmune disease. Autoimmunity. 2003 Dec;36(8):445–446. doi: 10.1080/08916930310001625961. [DOI] [PubMed] [Google Scholar]
- 44.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 45.Bissonnette R, Papp K, Maari C, Yao Y, Robbie G, White WI, et al. A randomized, double-blind, placebo-controlled, phase I study of MEDI-545, an anti-interferon-alfa monoclonal antibody, in subjects with chronic psoriasis. J Am Acad Dermatol. 2010 Mar;62(3):427–436. doi: 10.1016/j.jaad.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 46.Kalunian K, Merrill JT, Maciuca R, Ouyang WJ, McBride JM, Townsend MJ, et al. Efficacy and safety of rontalizumab (anti-interferon alpha) in SLE subjects with restricted immunosuppressant use: results of a randomized, double-blind, placebo-controlled phase 2 study. Arthritis Rheum. 2012;64:S1111–S1111. [Google Scholar]
- 47.Petri M, Wallace DJ, Spindler A, Chindalore V, Kalunian K, Mysler E, et al. Sifalimumab, a human anti-interferon-α monoclonal antibody, in systemic lupus erythematosus: a phase I randomized, controlled, dose-escalation study. Arthritis Rheum. 2013 Apr;65(4):1011–1021. doi: 10.1002/art.37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyons PA, McKinney EF, Rayner TF, Hatton A, Woffendin HB, Koukoulaki M, et al. Novel expression signatures identified by transcriptional analysis of separated leucocyte subsets in systemic lupus erythematosus and vasculitis. Ann Rheum Dis. 2010;69:1208–1213. doi: 10.1136/ard.2009.108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013 Aug 5;8(8):e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.