Abstract
Stem cells are normally maintained in a quiescent state and proliferate only under certain conditions; however, little is known about the biological stimuli that initiate the proliferation and differentiation of stem cells. In this study, we found that functional Toll-like receptors (TLRs) are expressed on mouse embryonic stem (ES) cells and that TLR ligands stimulate ES cell proliferation and promote their hematopoietic differentiation. TLR ligands activate TLR-mediated signaling pathways, leading to the altered expression of numerous genes in ES cells. Moreover, TLR ligands efficiently stimulate the proliferation and expansion of adult stem cells and progenitors of nonhematopoietic tissues, such as mammary glands and intestine as well. We further found that mammary luminal progenitor cells (Lin−CD29+CD61+) express TLR4-MD2 complex and actively proliferate, resulting in the enhanced growth of mammospheres in response to TLR ligands. Thus, mouse ES cells and adult tissue-specific stem cells/ progenitors directly sense and respond to microbial products, which function as a class of foreign, but biological stimuli for stem cell/progenitor proliferation. This finding expands the biological role of TLRs and has implications in understanding stem cell biology, tissue repair/homeostasis, and the role of infection and inflammation in malignant transformation.
Keywords: Adult stem cells, Embryoid bodies, Embryonic stem cells, siRNA
Introduction
Mammalian Toll-like receptors (TLRs) are a family of at least 12 germline-encoded, evolutionarily conserved membrane proteins that are critical in the activation of innate immunity [1]. The immune function of Toll was first identified on the basis of its crucial role in producing antimicrobial proteins and determining the resistance of Drosophila melanogaster to infection with fungi and Gram-positive bacteria [2]. Mammalian TLRs were subsequently discovered on the basis of their homology with Drosophila Toll [3, 4]. The TLR-family members are expressed either on the cell surface or intracellular organelles and endocytic vesicles, and recognize unique microbial molecular structures, such as lipopolysaccharide (LPS), lipopeptides, and virus-associated nucleic acids [1]. TLRs are composed of an ectodomain of leucine-rich repeats, which are involved directly or through accessory molecules in ligand binding, and a cytoplasmic Toll/interleukin-1 (IL-1) receptor (TIR) domain that interacts with TIR-domain-containing adaptor molecules. Upon the interaction with their ligands, TLRs trigger a cascade of nuclear factor κB (NF-κB)-dependent and interferon (IFN)-regulatory factor-dependent signaling pathways, leading to the activation of a large number of proinflammatory genes [1]. In addition to their biological roles in immunity, Drosophila Toll was originally recognized as a transmembrane receptor required for the establishment of dorso-ventral polarity in the developing embryo [5] and was further found to play broad roles in morphogenetic movement, muscle attachment, and hematopoiesis during embryo development [2].
Stem cells comprise embryonic stem (ES) cells that are responsible for embryonic development and adult stem cells that reside in different organs and tissues. ES cells possess an unlimited self-renewal capacity as well as the ability to differentiate into virtually any cell type of an organism. Adult tissue-specific stem cells are usually multipotent and have the capacity for limited lineage differentiation by replicating themselves and producing daughter cells committed to terminal differentiation for the tissue homeostasis and regeneration [6, 7]. Tissue injury is also accompanied by the expansion of tissue-specific stem cells through renewal divisions in order to repair the injury. After the wound is repaired, cell numbers resulting from the amplification of stem cells and the differentiation of their progeny are restored, and subsequently stem cell compartment returns to quiescence [8]. However, little is known about the biological stimuli that initiate the proliferation and differentiation of stem cells. In this study, we reveal the important role of TLRs in regulating the proliferation and differentiation of mouse ES cells, as well as adult tissue-specific stem cells/progenitors.
Materials and Methods
Mice and In Vivo Experiments
C57BL/6J, BALB/c, C3HeB/FeJ, and C3H/HeJ [9], nonobese diabetic/severe combined immunodeficient (NOD/SCID), and MMTV-neu mice were purchased from the Jackson Laboratory (West Grove, PA, http://www.jacksonimmuno.com) and maintained in a pathogen-free mouse facility at Baylor College of Medicine and the University of Southern California Keck School of Medicine according to institutional guidelines. Approval for performing these mice experiments was obtained from the institutional review board. For teratoma formation, approximately 5 × 105 mouse ES cells were subcutaneously injected into the dorsal flank of anesthetized NOD/SCID mouse. Five weeks after inoculation, teratomas were surgically dissected from the mice. Samples were weighed and fixed in zinc formalin solution for tissue sample preparation [10]. For examining tissue-specific progenitor populations in vivo, groups of mice were intraperitoneally injected with LPS from E. coli 055:B5 (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) for indicated days. Various tissues were harvested for the preparation of single-cell suspension with mechanic or enzymatic dissociation for multicolor fluorescence-activated cell sorting (FACS) analysis, or for histology and immunohistochemistric staining according to previous reports [11, 12].
Antibodies and Flow Cytometric Analysis
A panel of antibodies and matched isotype controls were used for staining mouse ES cells and single-cell suspensions of mouse tissues. Anti-CD16/ CD32 Fcγ III/II receptor antibodies (BD Pharmingen, San Diego, CA, http://www.bdbiosciences.com/index_us.shtml) were routinely used to pretreat cells at 4°C for 30 minutes to block nonspecific Fcγ receptor binding in some experiments. The antibodies used in this study include phycoerythrin (PE)-conjugated anti-stage-specific embryonic antigen (SSEA)-1, goat anti-Oct 3/4 antibodies (clone N-19; Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com), allophycocyanin (APC)-conjugated anti-TLR4-MD2 (MTS510; eBioscience, San Diego, CA, http://www.ebioscience.com), anti-TLR2 (6C2; eBioscience), fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (RM4-5), CD8α (53-6.7), B220 (RA3-6B2), Ter-119 (TER-119), Gr-1 (RB6-8C5), and Mac-1 mAb (WT.5; BD Pharmingen) for lineage (Lin) subpopulation. For FACS analyses of tissues samples, PE-Cy7-conjugated anti-Sca-1 (D7; BD Pharmingen), biotinlyated anti-EphB3 Abs (R&D Systems, Minneapolis, http://www.rndsystems.com)-intestinal sample, PE-conjugated anti-CD24, FITC-conjugated anti-CD31 and CD45 mAbs, biotinlyated anti-CD61 (BD Pharmingen), anti-CD29 (MB1.2; Chemicon, Temecula, CA, http://www.chemicon.com), anti-cytokeratin 14 (AF64; Covance, Princeton, NJ, http://www.covance.com), anti-cytokeratin six (Abcam, Cambridge, UK, http://www.abcam.com) mAbs, FITC-conjugated anti-PCNA (FL-261), and anti-Bcl-2 Abs (C-2; Santa Cruz Biotechnology)-mammary gland sample. For intracellular staining, cells were first fixed and permeablized [11, 12]. For indirect staining, APCcy7-conjugated streptavidin (BD Pharmingen), fluorchrome-congugated secondary antibodies included APC-conjugated anti-rat IgG (Caltech Labs, San Francisco, CA, http://www.caltechlabs.com), anti-rabbit IgG (Molecular Probes Inc., Eugene, OR, http://probes.invitrogen.com), or anti-goat IgG-Alexa488 (Molecular Probes) were used to detect the primary antibodies. Flow cytometry analyses were conducted on a FACSAria (Becton, Dickinson and Company, Franklin Lakes, NJ, http://www.bd.com), and data were analyzed using the FlowJo software.
Mouse ES Cell Culture and In Vitro Differentiation
Mouse ES, D3 cells (CRL-1934, ATCC), were grown on gelatin-coated tissue culture dishes (100 mm; BD Biosciences Falcon, San Jose, CA, http://www.bdbiosciences.com) with knockout Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) supplemented with 15% fetal bovine serum (StemCell Technologies, Vancouver, Canada, www.stem-cell.com), 2 mM L-glutamine, 100 μM monothioglycerol, 100 U/ ml penicillin, 10 μg/ml streptomycin, and 1,000 U/ml leukemia inhibitory factor (LIF; ESGRO, Chemicon). Alkaline phosphatase staining was performed using a detection kit from Chemicon. For in vitro hematopoietic differentiation, ES cells were differentiated into hematopoietic cells in vitro by using the two-step differentiation procedure in semisolid methylcellulose-based medium according to the manufacturer’s protocol (StemCell Technologies). As the first step, undifferentiated ES cells were dissociated, and single ES cells (500 cells/plate) were plated into methylcellulose-based medium containing a stem cell factor (SCF; StemCell Technologies) with or without LPS (10 μg/ml; InVivogene) for 10 days. In the second step, embryonic bodies (EBs) were disrupted by collagenase to prepare a single-cell suspension, and single cells were replated in methycellulose-based medium containing growth factors (erythropoietin, IL-3, IL-6, and SCF) with or without LPS as well. After 14 days of incubation, the numbers of differentiated colonies were counted and cell suspensions of these differentiated cells were subjected to FACS analysis.
Mammary Cell Preparation and Mammosphere Formation Assays
Primary mammary epithelial cells were isolated from freshly dissected mammary glands by enzymatic dissociation, as previously described [11, 12]. Briefly, mammary glands were dissected from 5 to 10 weeks old virgin female mice. Mammary glands were chopped manually by razor blaze. The tissue was washed with phosphate buffered-saline (PBS) and then placed in culture medium DMEM/F12 (Invitrogen) supplemented with 1 mM glutamine, 5% bovine calf serum, 10 ng/ml epidermal growth factor (EGF), 5 μg/ml insulin, 500 ng/ml hydrocortisone and containing collagenase (2 mg/l; Roche Diagnostics, Basel, Switzerland, http://www.roche-applied-science.com) and hyaluronidase (100U/ml; Sigma-Aldrich) at 37 for 3 hours. The resultant organoids were resuspended in 0.25% trypsin-ethylenediaminete-traacetic acid (Invitrogen) for 10 minutes before being filtered through a 40-μm strainer to yield a single-cell suspension. Isolated mammary epithelial cell suspensions were stained with FITC-conjugated with CD31, CD45, anti-CD29 mAb, and biotin-lyated anti-CD61 followed by APC-conjugated streptavidin (BD Pharmingen), PE-conjugated anti-rat IgG (Caltech) as secondary antibodies. For mammoshpere formation assays, these single cells with or without FACS sorting were plated onto ultra-low attachment six-well plates (Corning Life Sciences, Acton, MA, http://www.corning.com/lifesciences) at a cell density of 10,000 cells/ well in triplicate. Cells were grown in serum-free mammary epithelial growth medium supplemented with B27 (Invitrogen), EGF (10 ng/ml), and fibroblast growth factor (10 ng/ml) growth factors. Cells were fed every 3–4 days for 10–14 days. Primary mammospheres were counted and collected by gentle centrifugation (800 rpm). Primary mammospheres were then dissociated enzymatically and mechanically, and cultured them for the secondary mammosphere formation assays, as described [11–13].
Reverse Transcription-Polymerase Chain Reaction and Microarray Assays
Total cellular RNA was isolated using RNeasy kit (Qiagen, Hilden, Germany, http://www1.qiagen.com) and reverse-transcribed using the SuperScript kits (Invitrogen) as described [14, 15]. All primers and probes were purchased from the Operon (Alameda, CA, http://www.operon.com) for testing TLRs expression and Applied Biosystems (Foster City, CA, http://www.appliedbiosystems.com) for quantitative reverse transcription-polymerase chain reaction (RT-PCR), respectively. Quantitative RT-PCR was performed using gene-specific double fluorescence-labeled probes and an ABI PRISM 7900HT Sequence Detector (Applied Biosystems). For each sample, Taq-Man PCR reaction was done in quadruplicate for each probe of interesting genes and for a reference gene, 18S rRNA, to normalize for input sample levels. The PCR parameters were these recommended for the TaqMan Universal PCR Master Mix kit (Applied Biosystems). The ratio between the values obtained provided relative gene expression levels [14, 15]. A list of primers used in this study was provided in the supporting information. For microarray assays, the biotin-labeled RNAs were used directly as target for the Affymetrix (Santa Clara, CA, http://www.affymetrix.com) oligonucleotide GeneChip Mouse Genome (MG) 430 2.0 Array according to the Affymetrix standard protocol. Fluorescence intensities were scanned with an Affymetrix GeneArray 3000 Scanner, and signal intensities for each probe were collected with Microarray Suite version 5.0 software from Affymetrix. Expression levels of each of the genes represented in the array was computed in the R statistical programming environment and analyzed using BRB ArrayTool as well.
Proliferation Assays
mES cells were seeded in gelatin-coated 96-well plate, 500 cells/per well, with 2% fetal bovine serum-ES culture medium in the presence or absence of different concentration of TLR ligands [16]. Proliferative rates of these cells were evaluated 72 hours after LPS or Poly I:C treatment using an BrdU incorporation assay kit (Roche).
Immunohistochemical Staining
For bromodeoxyurdine (BrdU) immunodetection to test the proliferation of mammary cells, mice were injected with 0.5 mg per 10g body weight 5-BrdU cell labeling reagent (Amersham Biosciences, Piscataway, NJ, http://www.gelifesciences.com) 4 hours before tissue collection. Detection of BrdU in this report was performed using BrdU Staining kit (Zymed Lab., San Francisco, CA, http:www.zymed.com) as described previously [17]. For quantitative comparison of BrdU stained cells between mammary glands, at least eight individual 40× fields per group were captured for counting. The number of BrdU stained cells was expressed as a percentage of the total cell number of each terminal end bud (TEB).
Statistical Analysis
We used two-tailed Student’s t test for statistical significance analysis (InStat 2.01 program, Graph Pad Software, San Diego, CA), and a 95% confidence limit was taken to be significant, defined as p < .05. Results are typically presented as mean ± SE as indicated.
Results
Mouse ES Cells Express Functional TLRs and Directly Respond to TLR Ligands
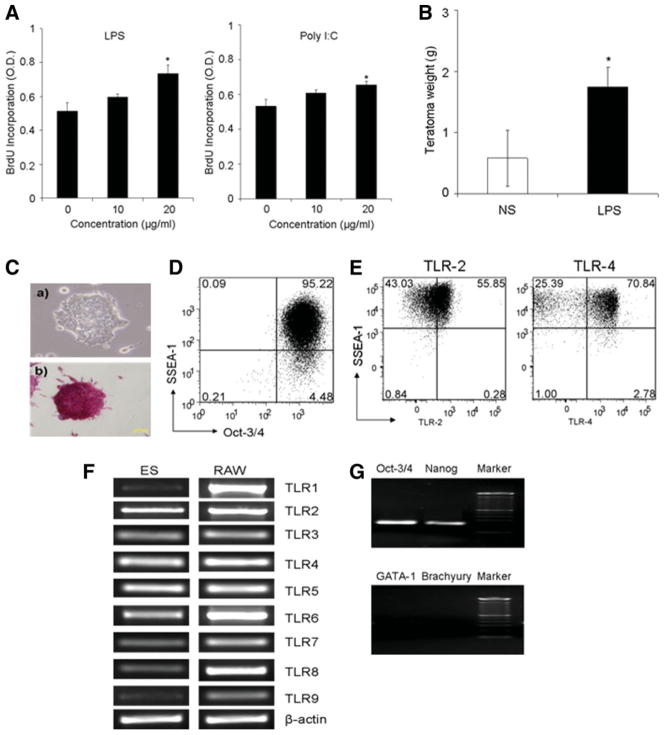
In investigating the regulation of signal transduction of cytokines in ES cells, we unexpectedly found that TLR ligands such as LPS (ultrapure; InvivoGen, San Diego, CA, www.invivogen.com), a ligand of TLR4, and Poly(I:C), a ligand of TLR3, stimulated the proliferation of murine ES cells (ES-D3) grown on gelatin-coated dishes in the standard ES cell culture medium containing LIF (Fig. 1A). TLR ligands, LPS and Poly (I:C), were also found to promote ES cells to form primary embryoid bodies (EB) (supporting information Fig. 1A). To further evaluate the stimulatory effect of TLR ligands on ES cells, we performed secondary EB formation assays. It was shown that the cells from the LPS-stimulated primary EBs were still able to more efficiently form the secondary EBs in the presence of LPS (supporting information Fig. 1B), suggesting that the ES cells after stimulation with LPS retained the capability for self-renewal [18]. Moreover, we in vivo examined the effect of TLR ligands on teratoma development from undifferentiated ES cells in immunodeficient NOD/SCID mice. ES-D3 cells were subcutaneously transplanted into groups (n = 4) of NOD/SCID mice (5 × 105 cells/mouse). On day 4 after inoculation, the mice were treated with LPS (5 μg/mouse) once every 2 days for 5 weeks. Figure 1B shows that the teratomas growth was significantly more progressive developed in LPS-treated mice than those in untreated mice (p < .05; Student’s t test). These teratomas from both groups of mice comprised various differentiated tissues, including skeletal muscle, squamous epithelium, and vascular tissues (data not shown). The cultured mouse ES cells were demonstrated to be positive for alkaline phosphatase as well as the ES cells markers, the POU transcription factors Oct-3/4 and the surface glycolipid SSEA-1 antigen (Fig. 1C, 1D). Flow cytometric assays further showed the surface protein expression of representative TLRs such as TLR2 and TLR4 on SSEA-1+ ES cells (Fig. 1E). In addition, the expression of various TLRs in the mouse ES cells at various levels was confirmed by RT-PCR (Fig. 1F). The possibility of genomic DNA contamination in these RNA samples was largely eliminated, because genomic PCR assays without reverse transcription failed to yield detectable PCR products (data not shown). The “undifferentiated” status of these TLR+ mouse ES cells was further confirmed by demonstrating the transcription of the ES markers Nanog and Oct-3/4, as well as the lack of the transcription of differentiation markers such as GATA-1 and Brachyury transcription factors [19] (Fig. 1G).
Figure 1.
TLR ligands stimulated mouse ES cell proliferation in vitro and in vivo. (A): TLR ligands stimulated the proliferation of the ES cells in vitro. ES proliferation rates were measured in the ES cell (ES-D3) culture after the treatment of LPS or Poly(I:C) for 72 hours by BrdU incorporation. Data are presented as the mean ± SEM of three independent experiments (*, p < .05). (B): LPS stimulated the growth of teratoma in vivo. Teratoma growth in NOD/SCID mice 5 weeks after ES cells subcutaneously inoculation (5 × 105 cells/mouse) with or without intraperitoneal administration of LPS (5 μg/mouse, n = 4 mice per group) once every 2 days was compared. Data are shown as mean ± SEM of two independent experiments (*, p < .05, two-tail p value). (C): Mouse ES cells (not treated with a TLR agonist) cultured in gelatin-coated plates with the medium supplemented with leukemia inhibitory factor (1,000 U/ml; ESGRO, Chemicon) without (a) or with (b) staining for the ES cell marker alkaline phosphatase using Chemicon’s detection kits. (D): Flow cytometric analysis of the expression of ES cell markers SSEA-1 and Oct-3/4 in the cultured ES cells. (E): The surface expression of TLR-2 and TLR-4 on SSEA-1-positive ES cells by flow cytometric analysis. (F): Expression of TLRs mRNA in ES cells by semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) with the TLR1-TLR9-spe-cific primers. (G): The expression of ES cell marker genes, Oct-3/4 and Nanog, and the lack of the expression of differentiating markers GATA-1 and Brachyury transcription factors in the cultured ES cells by RT-PCR analysis. Marker: 100 bp DNA Ladder (New England BioLabs, Ipswich, MA, http://www.neb.com). Abbreviations: BrdU, bromodeoxyurdine; ES, embryonic stem; LPS, lipopolysaccharide; NS, no stimulation; RAW, RAW mouse monocyte macrophage cell line; SSEA, stage-specific embryonic antigen; TLR, Toll-like receptor.
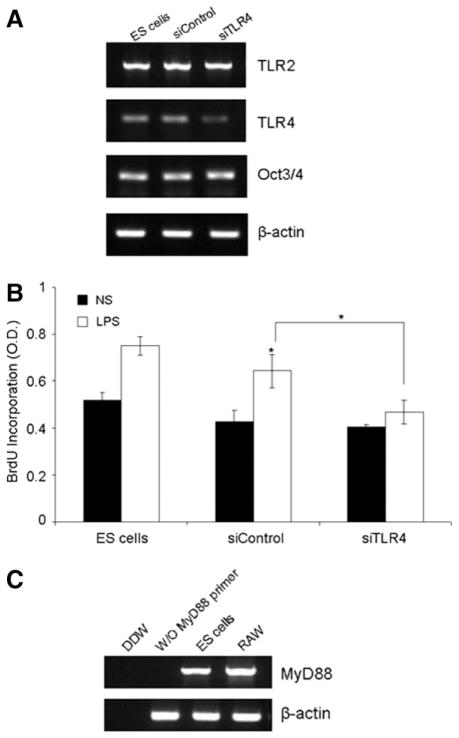
To test whether the stimulatory effects by LPS is directly mediated by TLR4 on ES cells, we examined the proliferation of wild-type and TLR4-silenced ES cells in response to LPS stimulation. It was demonstrated that mouse ES cells were efficiently transfected with Cy3-labeled siRNA oligonucleotide duplexes by nucleofection (>80%) (supporting information Fig. 2A) and the TLR4 mRNA level was significantly downregulated by mouse TLR4-siRNA oligo (sc-40261; Santa Cruz Biotechnology) (Fig. 2A). Figure 2B shows that TLR4-siRNA nucleofection, but not control-siRNA (Santa Cruz Biotechnology) nucleofection, reduced the stimulatory effect of LPS on ES cells (*, p < .05; two-tail p value) (Fig. 2B). In addition, we found that the gene of MyD88, a key adaptor molecule in the TLR4-mediated signal transduction [20, 21], was expressed in ES cells, as demonstrated by RT-PCR assay (Fig. 2C). Collectively, these data suggest the direct effect of LPS on ES cells via TLR.
Figure 2.
Reduced responsiveness of TLR4-silenced ES cells to LPS stimulation. (A): Semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) analyses showing the selective downregulation of TLR4 expression in ES cells 24 hours after nucleofection of 20 nM mouse TLR4-siRNA (sc-40261) and control siRNA oligo (Santa Cruz Biotechnology) by Nucleofector II (Lonza Amaxa, Walkersville, MD, http://www.amaxa.com). (B): BrdU incorporation assay of wild-type and siRNA-transfected ES cells 72 hours after LPS (20 μg/ml) stimulation or NS. LPS was added at 24 hours after siRNA transfection. Data are shown as mean ± SEM of three independent experiments. *, p < .05, versus NS, *, p < .05, siControl versus siTLR4 (two-tail p value). (C): MyD88 mRNA expression in ES cells by RT-PCR. RAW cells mRNA was used as positive control. Abbreviations: BrdU, bromodeoxyurdine; ES, embryonic stem; LPS, lipopolysaccharide; NS, no stimulation; RAW, RAW mouse monocyte macrophage cell line; TLR, Toll-like receptor.
TLR Ligands Activate TLR-Mediated Signaling Pathways in ES Cells and Promote Hematopoietic Differentiation
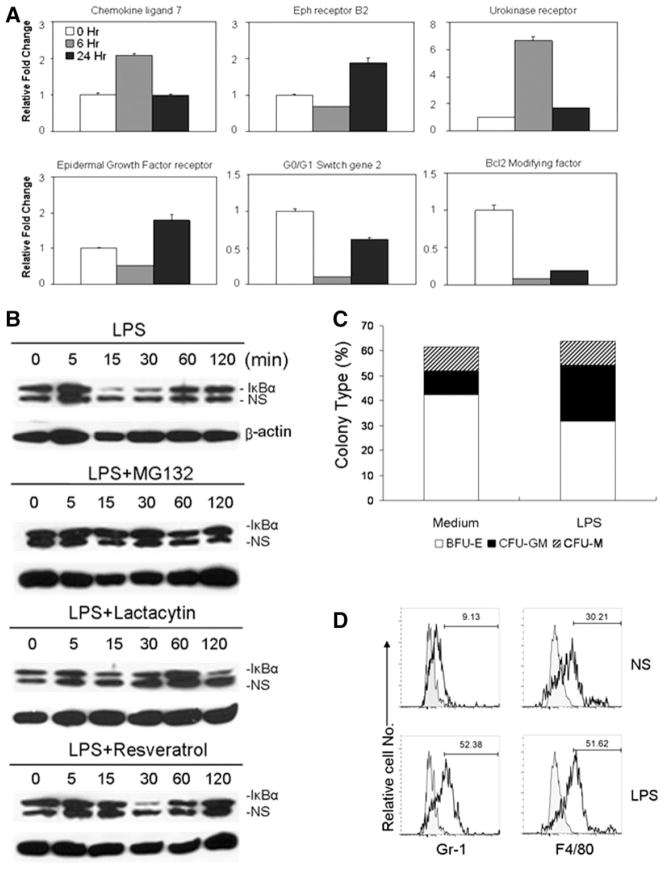
To examine whether the effects on ES cells by TLR ligands are mediated by TLR-mediated signaling, we examined the activation of signal transduction pathways in ES cells after exposure to TLR ligands. We used Affymetrix GeneChip Mouse Genome (MG) 430 2.0 Arrays to examine the overall gene expression changes of ES cells at different time points after LPS stimulation. In general, approximately 115 genes were upregulated at least threefold, whereas 175 genes were downregulated at least threefold in ES cells 6 hours after LPS stimulation, suggesting a direct activation of an array of genes in ES cells by LPS. Notably, the expression of genes belonged to transcription factors/regulators, cell cycle and apoptosis regulators, such as G0G1 switch gene2, and Bcl2 modifying factor, was significantly upregulated or downregulated, more than threefold, in response to LPS stimulation (supporting information Table 1). Twenty four hours after stimulation with LPS, the expression of 86 genes in ES cells was significantly upregulated, whereas relatively fewer genes (approximately 30) were downregulated in the ES cells (data not shown). To confirm these microarray data, we used quantitative RT-PCR to examine the mRNA levels of representative genes. Figure 3A shows the increased or reduced expression of the mRNA of representative genes in ES cells after LPS stimulation, supporting the microarray data of the enhanced and reduced expression of an array of genes in ES after LPS stimulation. Furthermore, we used Western blotting to examine the activation of TLR-mediated signal pathways. Figure 3B shows that the amount of the inhibitory protein IkBα transiently decreased in LPS-stimulated ES cells between 15 and 30 minutes after LPS stimulation. Proteasome inhibitors, both MG132 and Lactocystin, blocked LPS-induced IkBα degradation. Interestingly, Resveratrol, which blocks MyD88-independent, Toll/IL-1R domain-containing adaptor inducing IFN-beta signaling in the TLR4 pathway [22], inhibited IkBα degradation at the early time point, but not at a later time point (30 minutes) after LPS treatment. Thus, these data demonstrate that LPS activates TLR-mediated signaling pathways, resulting in the upregulation and downregulation of the expression of numerous genes, including cell cycle and apoptosis regulators, and growth factors and their receptors, in ES cells.
Figure 3.
Toll-like receptor (TLR) ligands activated TLR-mediated signaling pathways in embryonic stem (ES) cells and promote hematopoietic differentiation. (A): Quantitative RT-PCR analysis of mRNA levels of representative genes in mouse ES cells at different time points after stimulation with LPS (10 μg/ml). (B): Proteasome-dependent transient IkBα degradation in ES cells induced by LPS. Total cell extracts of ES cells that were treated with MG132 (10 μM; Sigma-Aldrich), lactacystin (10 μM; Sigma-Aldrich), and Resveratrol (50 μM; Sigma-Aldrich) at different time points after LPS stimulation (1 μg/ml) were subjected to Western blotting with an anti-IkBα antibody. The same blots were reprobed with an anti-β-actin antibody. (C, D): LPS stimulated the hematopoietic differentiation of EBs in vitro. In vitro hematopoietic differentiation assays of ES-D3 cells in the presence or absence of LPS (10 μg/ml) were performed using the two-step differentiation procedure. The percentages of each colony populations (C) in total colonies numbers of each group (n = 5 plates/per group) and histograms of myeloid cell markers (F4/80 and Gr-1) from the pooled differentiated cells (D) after 14 days of in vitro culture are presented from one representative of three independent experiments. Abbreviations: BFU-E, burst forming unit-erythroid; CFU-GM, colony-forming unit-granulocyte-macrophage; CFU-M, colony-forming unit macrophage; LPS, lipopolysaccharide; NS, nonspecific protein band as an additional sample loading control.
We also tested whether TLR signaling promotes ES differentiation into hematopoietic progenitors using a two-step differentiation protocol in vitro. Figure 3C shows that adding LPS to the ES hematopoietic differentiation culture preferentially increased the differentiation of myeloid-lineage progenitors, predominantly colony-forming unit-granulocyte-macrophage colonies. FACS analyses showed that the increased percentages of Gr-1+ and F4/80+ cells, markers of monocytes and macrophages, in LPS-treated cultures (Fig. 3D). Taken together, these data demonstrate that TLR ligands stimulate ES cell proliferation as well as promote the hematopoietic differentiation from ES cells.
TLR Ligands Stimulate the Expansion of Tissue-Specific Stem Cells/Progenitors of Nonhematopoietic Tissues
The stimulatory effect of TLR ligands on mouse ES differentiation prompted us to investigate whether TLRs also function in adult tissue-specific stem cells/progenitors. Groups of mice were intraperitoneally injected with LPS (50 μg/mouse) once a day for 2 days and various tissues/organs were then harvested to prepare cell suspension for multiple color fluorescence - activated cell sorting (FACS) analysis. To examine a subpopulation of intestinal stem/progenitor cells, we used EphB-3 receptor, which was reported as a key receptor for the migration and proliferation of intestinal progenitors in the stem cell niche, as a marker on Lin− and Sca-1+ population for intestinal stem cells/progenitors [23]. It was shown that that Lin− Sca-1+EphB-3+ subpopulation was significantly increased in LPS-treated wild-type (wt) mice (C3HeB/FeJ). In contrast, there was no apparent increase in the percentages of Lin− Sca-1+EphB-3+ cell subpopulations between TLR4 mutant C3H/HeJ mice with or without stimulation with LPS (supporting information Fig. 3). Thus, these results hint a biological role of TLRs in the adult stem cells/progenitors of nonhematological tissues.
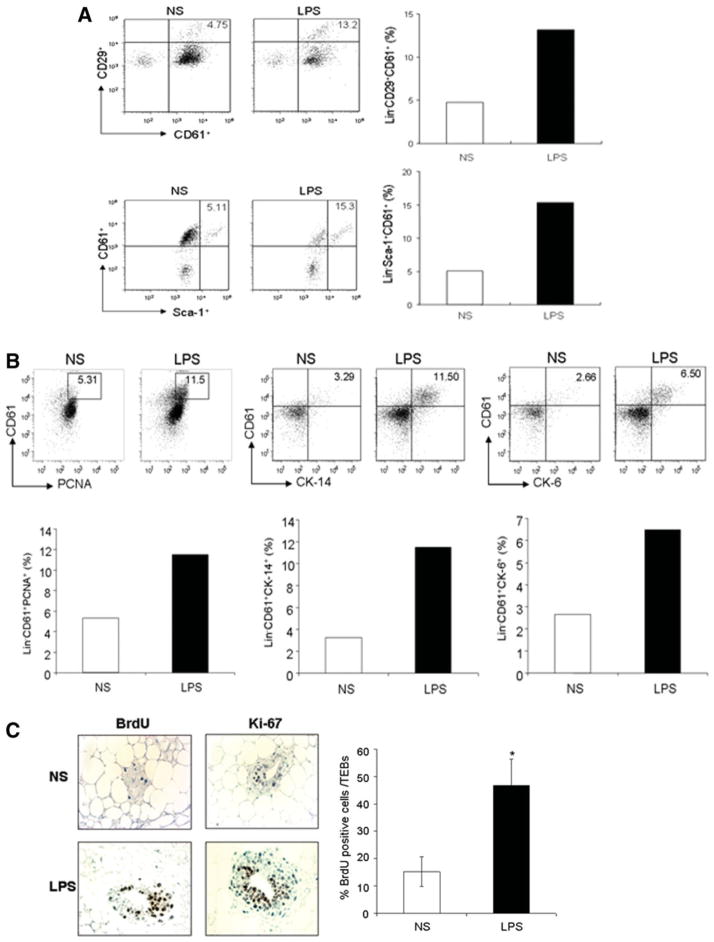
To test whether TLRs also function in stem cells/progenitors of other nonhematopoietic tissues, we examined the effect of TLR ligands on mammary gland stem cells/progenitors, using recently identified surface markers such as CD61 (β3-integrin), CD24, and CD29 that are associated with mammary gland stem cell or progenitor-enriched populations [11, 24, 25]. In the mammary glands tissue, mammary ducts are composed of two primary epithelial cell lineages—myoepithelial and luminal epithelial cells—that are presumably differentiated from common stem cells. We found that the Lin− CD61+CD29+ subpopulation was significantly elevated in LPS-treated wt mice, compared with untreated wt mice (13.2 vs. 4.75%) (Fig. 4A). Moreover, Sca-1, which is used as a marker for mammary gland progenitor cells, positive cells in Lin− CD61+ population were enriched in the mice as well (Fig. 4A). In contrast, there was no apparent increase of this population in LPS-stimulated TLR4 mutant mice (C3H/HeJ). Increases in the Lin− CD24+ or CD24+CD29+ subpopulations were less apparent in LPS-treated wt mice (data not shown). To further investigate the effect of TLR ligands on mammary gland stem cells/progenitor cells, we analyzed the cell population positive for cytokeratin 14 (CK-14), which is expressed on mammary stem cell-enriched population [11]. The CK-14+CD61+Lin− cell population was elevated approximately fourfold after LPS-stimulation. Furthermore, there was also an increase in the cell population positive for CK-6, which is a marker for mammary progenitors [26, 27], in LPS-treated wt mice (Fig. 4B). To test whether the increase/enrichment of the mammary progenitor population is due to the stimulatory effect of LPS, we examined the percentage of proliferating cells in mammary cell populations by costaining CD61 with the proliferating cell nuclear antigen (PCNA), a proliferation marker [23]. Figure 4B shows an approximately twofold increase in the percentage of PCNA+ proliferating cells in the Lin− CD61+ progenitor population of LPS-stimulated wt mice. In addition, we treated groups of wt mice at 6 weeks of age with LPS or PBS every 2 days for 4 weeks and then in vivo administered the mice with BrdU for 4 hours. Figure 4C shows that the average percentages of BrdU-incorporated, proliferating cells in the section of TEBs-like structure, which are considered to be temporary stem cell niches of mammary glands [28], were significantly higher in LPS-treated mice than in PBS-treated mice (*, p < .05). In agreement, the average percentages of the cells positive for the proliferation marker Ki-67 in the section of TEBs-like structure of mammary glands were also increased in LPS-treated mice as well (Fig. 4C).
Figure 4.
In vivo expansion of mouse adult mammary gland stem/progenitor cells by Toll-like receptor (TLR) ligand stimulation. (A): Fluorescence-activated cell sorting (FACS) dot plots and histograms showing the increased percentage of CD61+ cells in Lin−CD29+ population (upper panel), as well as Sca-1+ cell in Lin−CD61+ population (low panel) of mammary gland suspension prepared from mice 2 days after in vivo LPS stimulation (50 μg/mouse; Sigma-Aldrich). Data shown are one representative of three independent experiments. (B): FACS dot plots and histograms showing the increased percentages of PCNA+, CK-14+, or CK-6+ cells in Lin−CD61+ mammary gland stem cell/progenitor-enriched population in mice 2 days after in vivo LPS stimulation (50 μg/mouse) from one representative experiments of 3. (C): Immune histochemical staining of BrdU and Ki-67 in mammary glands of mice (n = 4 mice, 6-week-old) in vivo stimulated with LPS (50 μg/mouse) once every 2 days for 4 weeks. Data are presented as means percentage of BrdU-positive cell that are in the section of each terminal end bud-like structure (±SEM, n = 8) from two independent experiments (*, p < .05; two-tailed p value). Abbreviations: BrdU, bromodeoxyurdine; LPS, lipopolysaccharide; NS, no stimulation.
TLR Ligands Preferentially Stimulate Mammary Luminal Progenitor Cells
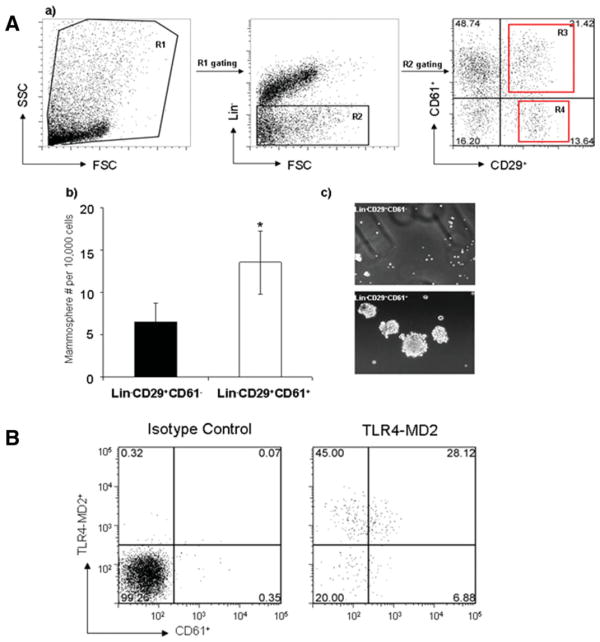
Mammospheres are composed of stem cells and progenitors capable of self-renewal and multilineage differentiation [29]. We compared the efficiency of mammosphere formation by CD61 negative or positive mammary cells, since CD61 was recently reported to be a marker associated with mammary luminal progenitor cells [24]. We observed that efficiency of mammosphere formation in sorted Lin−CD29+CD61+ cell population was significantly higher than sorted CD61− subpopulation in mammary gland cell suspension (Fig. 5A; b and c), confirming that mammary gland stem/progenitors cells are enriched in CD61+ cell fraction. Moreover, we found that CD61+ cell population in Lin− mammary gland cell subpopulation expressed TLR4-MD2 complex on the cell surface (Fig. 5B), suggesting the ability of CD61+ luminal progenitor cells to respond to TLR ligands.
Figure 5.
TLR4-MD2 expression on mammary CD61+ progenitor cells. (A): (a) Gating strategy used to select Lin−(R1 gating) and CD61+ cells. (b) The numbers of mammospheres per 10,000 CD61+ or CD61− mammary cells sorted from Lin−CD29+ cell subpopulation of mammary gland cell suspensions. Data are presented as mean ± SEM of two independent experiments (p < .05). (c) Photomicrographs of representative primary mammospheres derived from Lin–CD29+CD61+ and Lin−CD29+CD61− subpopulation. (B): Flow cytometry showing the surface expression of TLR4-MD2 on mammary Lin−CD61+ cell population. Abbreviations: FSC, forward scatter; SSC, side scatter.
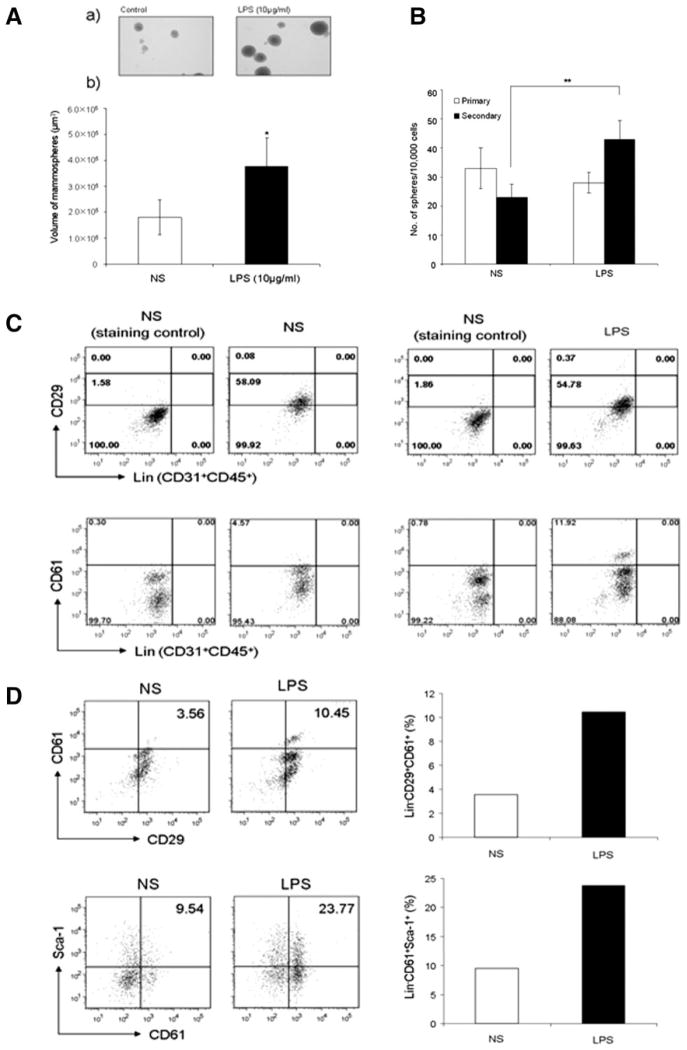
We then performed in vitro primary and secondary mammosphere formation assays in the presence or absence of TLR ligands in order to directly examine the stimulatory effect of TLR ligands on mammary stem cells/progenitors. Single cells of mammary epithelial cells prepared from mammary tissues of wt female mice (5–7 weeks old) were grown in the conditions described in “Materials and Methods” with or without LPS (10 μg/ml). Figure 6A shows that a significant increase in the average size/volume of mammospheres in LPS-treated mammary cell cultures, in comparison to the cultures without LPS (3.7 × 105 ± 1.0 × 105 vs. 1.8 × 105 ± 6.6 × 104 μm3; *, p < .05), suggesting an increase in proliferation of mammary gland stem cells/progenitors in LPS-treated cultures [30]. We also noticed that the numbers of mammospheres in the LPS-treated and untreated cell cultures were comparable (Fig. 6B), suggesting that LPS stimulation did not affect primary mammosphere-forming efficiency.
Figure 6.
Toll-like receptor (TLR) ligands stimulated the proliferation of mammary gland stem cells/progenitors in vitro. (A): TLR ligands stimulated the growth of primary mammospheres in vitro. Mammo-sphere formation assays from mammary cell suspensions were performed in the presence or absence of LPS (10 μg/ml). (a) Photographs of primary mammospheres after 14 days of in vitro cultivation in the presence or absence of LPS. (b) The average volumes of mammospheres in the cultures using the Scion Image software program (NIH, Image). Data are shown as means size of mammospheres (±SEM) of each group from three independent experiments. *, p < .05, versus NS. (B): TLR ligands increased the formation of secondary mammospheres in vitro. Cell suspensions prepared from the primary mammosphere formation assays in the presence or absence of LPS were subjected to the secondary mammosphere formation assays in the presence or absence of LPS (10 μg/ml). Data are presented as mean (±SEM) of the numbers of mammospheres per 10,000 single mammary cells in each groups from three independent experiments **, p < .01, versus NS. (C, D): Fluorescence-activated cell sorting dot plots showing the expansion of Lin−CD29+ (top) and Lin−CD61+ (bottom) subpopulations (C), and Lin−CD61+CD29+ (upper panel) and Lin−CD61+Sca-I+ (low panel) subpopulations (D) in the secondary mammosphere formation assays in the presence of LPS from one representative experiment of two. Histograms (right panels of D) showing the percentage of Lin−CD61+CD29+ population and Lin−CD61+Sca-I+ population in the single-cell suspensions from the secondary mammosphere formation assays. Abbreviations: LPS, lipopoly-saccharide; NS, no stimulation.
To test whether the increase in the average size/volume of mammospheres was due to the increased proliferation and numbers of mammary stem cells/progenitors in response to TLR ligands, we performed secondary mammosphere assays in vitro. In contrast to the primary mammosphere formation, the numbers of secondary mammospheres were significantly increased in the LPS-treated cultures, compared with untreated cultures (43 ± 6.51 vs. 23 ± 4.52; *, p < .05) (Fig. 6B). We confirmed that cell numbers in the single-cell suspensions derived from the secondary mammospheres increased in the cultures treated with LPS (data not shown). To further confirm the in vitro expansion of mammary stem cells/progenitors by TLR ligands, we stained the cell suspensions of the secondary mammospheres with antibodies against various surface markers. Figure 6C shows a significant increase in the Lin−CD61+ subpopulation in LPS-stimulated cell cultures, compared with untreated cell cultures (11.92 vs. 4.57%). We also observed an increase in the Lin−CD29high subpopulation after LPS stimulation (Fig. 6C). Moreover, we found that the CD61+CD29+ double positive stem cell/progenitor subpopulation in the Lin− population was enriched in LPS-stimulated cultures, more than two times than that in the untreated cell cultures (Fig. 6D). Lin−CD61+Sca-1+ population in the mammosphere cultures was enriched after LPS stimulation (Fig. 6D). Taken together, these in vivo and in vitro data demonstrate a direct effect of TLR ligands on the proliferation and expansion of mammary luminal progenitor-enriched populations.
TLR Ligands Stimulate Mammary Luminal Progenitor Cells in MMTV-Neu Transgenic Mice
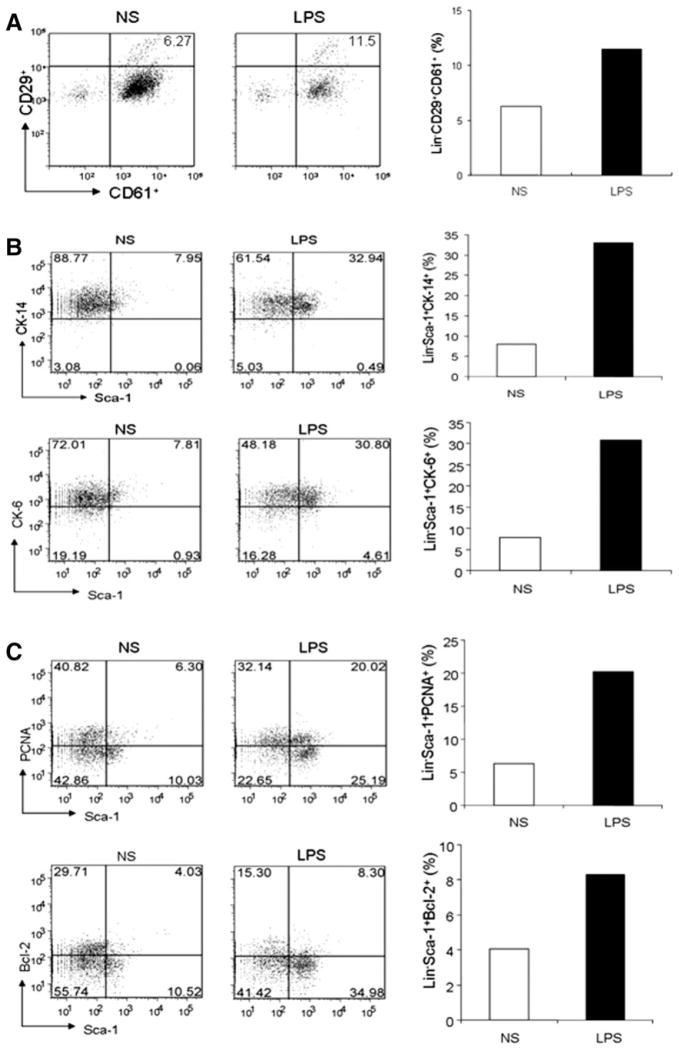
We further examined the effect of TLR ligands on mammary stem cell/progenitor-enriched populations in MMTV-neu transgenic mice that overexpresses ErbB2/Neu proto-oncogene under control of the mouse mammary tumor virus (MMTV) promoter and develop spontaneous mammary tumors. As observed in wt mice, LPS administration in MMTV-neu transgenic mice resulted in an increase of Lin−CD29+CD61+ subpopulation, approximately twofold in comparison with untreated mice (Fig. 7A). Furthermore, we found that the sub-population positive for CK-14, a marker for mammary stem cells/progenitors, in Lin−Sca-1+ population was elevated approximately threefold in LPS-treated mice, compared with untreated mice (Fig. 7B). The Lin− cell population that was positive in both Sca-1 and CK-6, which are early progenitor cell markers in mammary hyperplasias and tumors [26, 27], was also enriched in LPS-treated mice (Fig. 7B). Interestingly, the percentage of both Sca-1+ and PCNA+ cells in Lin− population was significantly increased (fourfold) in the LPS-stimulated mice, compared with untreated mice (Fig. 7C), suggesting the enhanced proliferation of mammary stem cell/progenitor-enriched populations. Furthermore, the Bcl-2+Sca-1+ cells in the Lin− population were enriched approximately twofold in the LPS-stimulated mice (Fig. 7C). These results demonstrated the stimulatory effect of TLR ligands on the proliferation and expansion of mammary stem cell/progenitor-enriched populations in both wt mice and MMTV-neu transgenic mice.
Figure 7.
Increased mammary gland progenitor subpopulation in tumorigenic MMTV-neu mice stimulated with Toll-like receptor (TLR) ligands in vivo. (A): Fluorescence-activated cell sorting (FACS) dot and histogram showing the elevated percentage of Lin−CD29+ CD61+ subpopulation in the mammary gland cell suspension prepared from 5-week-old MMTV-neu mice in vivo stimulated with or without LPS (50 μg/ml) for 2 days. (B): FACS analyses showing the increase percentages of Lin−Sca-I+CK-14+ (upper panel) and Lin−Sca-I+CK-6+ subpopulation in the cell suspension of mammary glands derived from 8-week-old MMTV-neu mice in vivo stimulated with or without LPS for 2 days. (C): FACS analyses showing the increase percentages of proliferating Lin−Sca-I+PCNA+ subpopulation (upper panel) and Lin−Sca-I+Bcl-2+ subpopulation in the cell suspension of mammary glands derived from MMTV-neu mice in vivo stimulated with or without LPS for 2 days. All data are shown as one representative of two independent experiments. Abbreviations: LPS, lipopolysaccharide; NS, no stimulation.
Discussion
The results of this study demonstrate that mouse ES cells and the adult tissue-specific stem cells/progenitors of nonhematopoietic tissues directly sense and respond to microbial products via TLRs, expanding the role of TLRs in the activation of immunity to that in regulating the proliferation and differentiation of mammalian stem cells. This finding should have profound implications in understanding stem cell biology, tissue repair/homeostasis, and the role of inflammation and infection in malignant transformation. In supporting our finding, it has been recently reported that TLR signaling stimulated the proliferation of fibroblasts [31], hematopoietic progenitors [32], and bone marrow mesenchymal stem cells [16], as well as controlled intestinal epithelial homeostasis [33, 34]. Furthermore, it was found that TLR signaling directly modulates self-renewal of neural stem/progenitor cells as well [35].
It is well known that many malignancies are initiated by infections, although the molecular mechanism behind the tumorigenesis remains elusive. For example, chronic bacterium Helicobacter pylori infection is the leading cause of stomach cancer [36], whereas chronic hepatitis C and B infection in the liver predisposes to liver carcinoma. NF-κB, which is activated by TLR ligands, has been demonstrated to play an important role in tumorigenesis, promoting cell proliferation and inhibiting apoptosis [37]. In addition to microbial products, a number of endogenous proteins, such as myeloid-related protein (Mrp) -8 and -14 [38], high-mobility-group box (HMGB)1 alarmin protein [39], and β-defensin [8], were found to bind and activate TLRs as endogenous ligands. Moreover, Rakoff-Nahoum and Medzhitov [33] recently found that MyD88-dependent signaling plays a critical role in both spontaneous and carcinogen-induced tumor development. Interestingly, Kelly et al. [40] recently reported that TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. This study provides evidence that links TLR-4 signaling, inflammation, and chemoresistance in ovarian cancer cells and suggests that the TLR-4-MyD88 signaling pathway may be a risk factor for developing cancer [41].
Stem cells are appealing candidates as the original cells of cancer [42, 43], since they are relatively long-lived and have the capacity for unlimited self-renewal [7, 8, 44]. The data of this study, together with the findings of others, hint a possible novel mechanism by which chronic infection and inflammation causes tumorigenesis by directly stimulating the proliferation of tissue-specific stem cells, thus providing the opportunity to accumulate the multiple mutations that are required to convert an activated tissue-specific stem cell into an uncontrolled cancer-initiating stem cell [45]. At present, the biological significance of functional TLRs in ES cells is not yet apparent, imploring further investigation. In summary, the finding of this study provides new insights into the biology of stem cells and TLRs, as well as the understanding of the roles of infection and inflammation in tumorigenesis, tissue homeostasis, and other chronic diseases.
Conclusion
Collectively, this study demonstrates that mouse ES cells and adult tissue-specific stem cells/progenitors express functional TLRs and directly sense and respond to microbial products. This study indicates that microbial products function as a class of foreign, but biological stimuli for stem cell/progenitor proliferation. This finding expands the biological role of TLRs and has implications in understanding stem cell biology, tissue repair/homeostasis, and the role of infection and inflammation in malignant transformation.
Supplementary Material
Acknowledgments
We thank Drs. Bingya Liu, Melissa Aldrich, and Xiao-Tong Song in the lab for technical assistance and valuable suggestions. We also thank Drs. Daniel Medina, Michael T. Lewis, and Thomas Zwaka for helpful suggestions and assistance. In addition, we thank Frances Kittrell and Ricardo Moraes for technical help; Jeannie L. Zhong and Kalli Antalis for technical support with microscopy and data analysis; Xuejun Zhang for flow cytometry; and Laure liles Garza for the Microarray assays. This work was supported by grants from the National Institute of Health (R01 CA90427 and R01 CA116677), and the Leukemia and Lymphoma Society SCOR.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author contributions: S.-H.L., B.H., A.S., X.F.H.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; S.Y.C.: conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
see www.StemCells.com for supporting information available online
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B. The road to Toll. Nat Rev Immunol. 2004;4:521–527. doi: 10.1038/nri1390. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 6.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 7.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 8.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 9.Jude BA, Pobezinskaya Y, Bishop J, et al. Subversion of the innate immune system by a retrovirus. Nat Immunol. 2003;4:573–578. doi: 10.1038/ni926. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 12.Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129:1567–1580. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen L, Evel-Kabler K, Strube R, Chen SY. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat Biotechnol. 2004;22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- 15.Song X-T, Kabler KE, Shen L, Rollins L, Huang XF, Chen S-Y. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–265. doi: 10.1038/nm1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pevsner-Fischer M, Morad V, Cohen-Sfady M, et al. Toll-like receptors and their ligands control mesenchymal stem cell functions. Blood. 2007;109:1422–1432. doi: 10.1182/blood-2006-06-028704. [DOI] [PubMed] [Google Scholar]
- 17.He XC, Zhang J, Tong WG, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 18.Chan RJ, Johnson SA, Li Y, Yoder MC, Feng GS. A definitive role of Shp-2 tyrosine phosphatase in mediating embryonic stem cell differentiation and hematopoiesis. Blood. 2003;102:2074–2080. doi: 10.1182/blood-2003-04-1171. [DOI] [PubMed] [Google Scholar]
- 19.Kubo A, Shinozaki K, Shannon JM, et al. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 20.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 21.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Youn HS, Lee JY, Fitzgerald KA, Young HA, Akira S, Hwang DH. Specific inhibition of MyD88-independent signaling pathways of TLR3 and TLR4 by resveratrol: molecular targets are TBK1 and RIP1 in TRIF complex. J Immunol. 2005;175:3339–3346. doi: 10.4049/jimmunol.175.5.3339. [DOI] [PubMed] [Google Scholar]
- 23.Holmberg J, Genander M, Halford MM, et al. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 24.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 25.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Welm B, Podsypanina K, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welm AL, Kim S, Welm BE, Bishop JM. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci USA. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moraes RC, Zhang X, Harrington N, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 29.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasan UA, Trinchieri G, Vlach J. Toll-like receptor signaling stimulates cell cycle entry and progression in fibroblasts. J Biol Chem. 2005;280:20620–20627. doi: 10.1074/jbc.M500877200. [DOI] [PubMed] [Google Scholar]
- 32.Nagai Y, Garrett KP, Ohta S, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 34.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Rolls A, Shechter R, London A, et al. Toll-like receptors modulate adult hippocampal neurogenesis. Nat Cell Biol. 2007;9:1081–1088. doi: 10.1038/ncb1629. [DOI] [PubMed] [Google Scholar]
- 36.Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–640. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 37.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 38.Vogl T, Tenbrock K, Ludwig S, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 39.Apetoh L, Ghiringhelli F, Tesniere A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 40.Kelly MG, Alvero AB, Chen R, et al. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006;66:3859–3868. doi: 10.1158/0008-5472.CAN-05-3948. [DOI] [PubMed] [Google Scholar]
- 41.Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their Toll—the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27:225–233. doi: 10.1038/sj.onc.1210907. [DOI] [PubMed] [Google Scholar]
- 42.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 43.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 44.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 45.Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118 (Part 16):3585–3594. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.