Abstract
Bacterial spores have long been recognized as the sturdiest known life forms on earth, revealing extraordinary resistance to a broad range of environmental assaults. A family of highly conserved spore-specific DNA-binding proteins, termed α/β-type small, acid-soluble spore proteins (SASP), plays a major role in mediating spore resistance. The mechanism by which these proteins exert their protective activity remains poorly understood, in part due to the lack of structural data on the DNA-SASP complex. By using cryoelectron microscopy, we have determined the structure of the helical complex formed between DNA and SspC, a characteristic member of the α/β-type SASP family. The protein is found to fully coat the DNA, forming distinct protruding domains, and to modify DNA structure such that it adopts a 3.2-nm pitch. The protruding SspC motifs allow for interdigitation of adjacent DNA-SspC filaments into a tightly packed assembly of nucleoprotein helices. By effectively sequestering DNA molecules, this dense assembly of filaments is proposed to enhance and complement DNA protection obtained by DNA saturation with the α/β-type SASP.
Bacterial spores have recently been brought to public attention, following the use of these organisms for warfare purposes and the exciting report of viable 250-million-year-old spores (35). Both developments highlight the unmatched ability of dormant spores to endure highly adverse environments and to remain viable for extremely long periods. Because spores may well be able to survive the loss of practically the entire complement of any specific enzyme (32), their unique survival properties primarily reflect their ability to preserve their DNA in an undamaged form. Such effective long-term DNA preservation is intriguing, because spores usually carry only one copy of their genome (2) and hence are unable to promote DNA repair based on homologous recombination. Efficient repair processes are also precluded in dormant spores by the absence of high-energy compounds such as nucleoside triphosphates and the virtually complete inactivation of enzymes within the spore core (32). Evidently, spore DNA durability relies mainly upon a passive, physical, and continuous protection, rather than on ongoing enzymatic repair processes, although DNA repair during spore germination can be important in spore survival in some conditions (28).
Spore DNA protection from damaging effects of desiccation, heat, oxidizing agents, and UV irradiation has been shown to derive primarily from a group of α/β-type small acid-soluble proteins (SASPs) (60 to 75 residues), termed α/β-type SASP, which bind DNA in a nonspecific manner (5, 20, 29, 32). These α/β-type SASPs accumulate during sporulation of Bacillus and Clostridium species to levels that are high enough to saturate spore DNA, and in all Bacillus species yet examined two proteins make up ≥80% of the α/β-type SASP pool (32). The critical role of α/β-type SASP in mediating spore DNA protection has been indicated by in vitro studies, as well as by the finding that spores lacking these proteins (termed α−β− spores) are much more sensitive to DNA damage than are wild-type spores (16, 32). The attenuated resistance of α−β− spores can be restored if these spores are provided with sufficient levels of any major or minor α/β-type SASP, thus indicating that these proteins can complement each other. Indeed, amino acid sequences of all α/β-type SASPs are highly conserved both within and between species (32, 33).
Although α/β-type SASPs and their DNA-binding properties have been extensively studied, the actual mechanism by which these proteins confer DNA resistance remains poorly understood, mainly due to the lack of detailed structural data on DNA-SASP complexes. Circular dichroism spectroscopy of α/β-type SASPs indicated that although these proteins adopt a random coil configuration in the absence of DNA, their secondary structure is dominated by α-helices upon binding to double-stranded DNA (10). High-resolution structural studies of the resulting complexes have heretofore been hampered by the large cooperative nature of the binding, as well as by the tendency of the nucleoprotein complexes to form massive aggregates (8).
Previous electron microscopy studies have shown a helical complex formed between DNA and SspC, a well-characterized representative member of the highly conserved α/β-type SASP family (32), in which SspC binds cooperatively along the DNA (8). Such helical filaments are amenable to structural analysis by cryoelectron microscopy (cryoEM) techniques. Here we present the three-dimensional (3D) structure of the DNA-SspC complex determined by a modified version of the iterative helical real-space reconstruction (IHRSR) method that combines single-particle and helical-reconstruction techniques (3). A novel mode of nucleoprotein organization is identified, in which tight packaging of DNA-SspC filaments is mediated by interdigitation of protein domains from adjacent helices. By promoting DNA dehydration and enabling efficient DNA sequestration, this tight toroid-like assembly is proposed to extend protection of spore DNA beyond the level achieved by DNA saturation with SspC, as well as to facilitate DNA repair.
MATERIALS AND METHODS
DNA-SspC complex formation and cryoEM.
SspC, an α/β-type SASP from Bacillus subtilis (32), was purified as described previously (9) from Escherichia coli BL21(DE3) containing plasmid pET11d that carries the sspC gene under control of the T7 promoter. DNA-SspC complexes were formed at a 10:1 (wt/wt) ratio (protein/DNA) in 10 mM Tris-maleate (pH 7.0), using pBluescript plasmid DNA (0.5 mg/ml) linearized with EcoRI. The protein/DNA ratio used in these experiments was approximately twofold higher than that needed for theoretical saturation of DNA with SASP, in order to ensure that DNA was saturated with the protein. Samples were incubated for 5 to 10 min at room temperature and deposited on glow-discharged Quantifoil grids. Specimens were embedded in 16% ammonium molybdate for cryonegative staining (1) and vitrified by plunging into liquid ethane. Images were recorded using low-dose cryo-conditions on Kodak SO163 plates either on a CM12 microscope operating at 100 kV and a magnification of ×60,000 or on an FEI Tecnai 12 (T12) microscope operating at 120 kV and a magnification of ×67,000.
Image reconstruction.
Electron micrographs were digitized with an Imacon FlexTightII scanner, at 16-bit dynamic range and 1,800 dpi (corresponding to 0.235 or 0.221 nm/pixel for images recorded on the CM12 or T12 microscope, respectively). Images from the T12 microscope were interpolated to 0.235 nm/pixel to allow data merging with images taken on the CM12 microscope. Electron micrographs were acquired at a defocus range of −500 nm to −3 μm, and prior to reconstruction, their phases were CTF (contrast transfer function) corrected. Image processing was conducted using the Spider software package (version 7.01) (6), except for the real-space helical search and reconstruction (IHRSR) method (3) that was performed with Fortran routines provided by E. Egelman and revised for application to DNA-SspC helical filaments. Helical segments (i.e., “particles”) 16 nm in length were picked from 22 electron micrographs with 70% overlap, while preserving information on filament polarity and membership within filament groups. Particles were subsequently aligned to an initial smooth bipolar model with uniform densities that was constructed by assuming a 3.4-nm helical pitch and 5.5-nm filament width. No additional structural features were introduced.
Multireference alignment to this model was performed without restriction of rotational search. Subsequently, a search for helical symmetry was performed on the reconstruction, and an improved symmetrized 3D model served as reference for the next iteration. A stable solution converged after ∼60 iterations. At this point, a polarity was observed to have evolved in the model, so this solution was used as a reference for 10 further iterations, which established the explicit polarity of the filaments. This was performed by a “voting” procedure: after multireference alignment to the reference model projections, particles from each filament were grouped according to their polarity, compared to the model. The majority vote was checked for divergence from a binomial distribution, and only those groups with 90% confidence of divergence were added to the reconstruction. The final reconstruction included 2,224 particles, corresponding to ∼13,500 protein subunits and the length (25 μm) of the helical fiber. The resolution, as determined by Fourier shell correlation with a cutoff of 0.5 (27, 34), reached 1.1 nm. The computed transform of a projection from the final reconstruction exhibited layerline reflections up to 0.94 nm. The errors in the pitch and subunit/turn (= 211.75° ± 0.04° and z = 1.872 ± 0.03 nm, respectively) were calculated from the baseline noise in the refinement plots of helical parameters versus refinement cycle. The isosurface level of the reconstruction in all figures was chosen to be 0.5 standard deviation (SD) above the average background intensity.
The significance of the polarity observed in the final model was assessed in the following way. Two independent reconstructions (each using half of the data) were made after the last cycle of refinement. The reconstructions were helically symmetrized, and projections were calculated. The program HLXFIT from the Brandeis Helical Package (22) was used to compare the best phase matching obtained when the two reconstructions were oriented one to the other either in parallel or antiparallel orientations. Comparison of the four possible permutations (front and back sides of one reconstruction compared to those of the other) showed a highly significant preference for polar orientations (phase residual = 20.4° ± 2.6°) versus nonpolar orientations (62.9° ± 3.4°).
Docking of adjacent filaments.
The program MolFit (4, 11) was used for docking analysis. An atomic representation of the electron microscopy-reconstructed electron density map was obtained by packing spheres into a 21.85-nm-long fragment of the DNA-SspC filament at the isosurface level described above. The electron density map for this fragment was calculated on a cubic grid with a 0.235-nm interval and converted to a list of “virtual atoms” whose centers were at the positions of the grid points.
Two search procedures were performed, starting with identical filaments either parallel or antiparallel to each other. During the search, one fiber was kept fixed while the other was allowed to rotate and translate about three perpendicular axes (the z axis coincided with the long axis of the filament) in steps of 10°. The translation interval was 0.1 nm in all search procedures. A full range of rotations was allowed for the z axis, but for the x and y axes only rotations of ±30° were allowed, producing 1,764 relative orientations. For each relative orientation, five docking solutions with the highest geometric complementarity score were saved. A high complementarity score indicates that surfaces align with minimal gaps and clashes. The mean score and SD were derived by fitting the observed distribution of scores to an extreme value distribution function (14). Local searches, at 1° intervals, were performed around the best parallel and antiparallel solutions. A third filament was then docked to the best two-filament antiparallel solution, employing identical translation and rotation intervals. This procedure resulted in the formation of 3D assemblies of filaments with both parallel and antiparallel contacts.
Thin-section transmission electron microscopy of bacterial spores.
B. subtilis cultures were grown in Luria-Bertani medium at 30°C for 72 h. The resulting spores were harvested and transferred to cellulose capillaries (inner diameter, 200 μm). The samples were frozen in liquid nitrogen in a Balzers HPM 010 apparatus under a pressure of 2.1 × 108 Pa for 500 ms (estimated freezing time, 20 ms). Following cryofixation samples were cryo-substituted with anhydrous acetone containing1% OsO4. Samples were then embedded in Epon; thin sections were stained with 1% uranyl acetate and examined on a Philips CM12 electron microscope operating at 100 kV. Intracellular localization of DNA was performed with the DNA-specific stain osmium ammine-SO2 (36). Grid-mounted thin sections of Epon-embedded bacteria were floated on 5 N HCl for 30 min at room temperature, washed with distilled water, and treated with osmium ammine-B (Polysciences) in 8 N acetic acid and 40 mM sodium metabisulfite for 1 h at 37°C. Sections were then thoroughly rinsed with distilled water, dried, and studied without additional staining. Three independent experiments were conducted for each set of staining conditions, and in each experiment more than 1,000 cell slices were screened.
RESULTS
3D reconstruction of DNA-SspC helical complex.
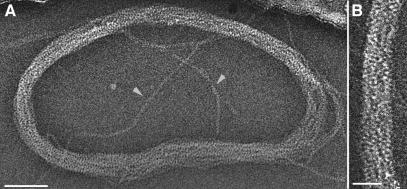
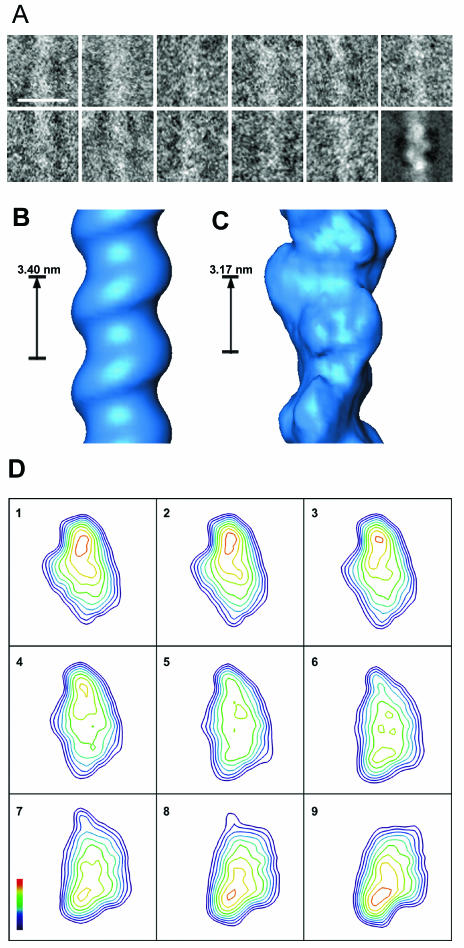
The 3D structure of the DNA-SspC helical filament was determined using cryoEM. The relatively small diameter (∼5.5 nm) and curved morphology of the ice-embedded DNA-SspC filaments imaged under low-dose conditions lead to particularly noisy images. In order to improve the signal-to-noise ratio, DNA-SspC filaments were imaged using the cryonegative staining technique (1). Filaments tended to aggregate into massive bundles and swirls (Fig. 1), as previously reported (8). Although the DNA-SspC complex is substantially stiffer than naked DNA, the filaments exhibited a highly curved morphology, thus preventing helical processing of segments longer than ∼95 nm. Collectively, these characteristics hampered the use of a classical helical-reconstruction approach. Therefore, we employed the IHRSR method (3). This method involves iterative cycles of a two-step process: multireference-based 3D reconstruction of helical segments (such as in Fig. 2A) using single-particle image-processing methods (with no symmetry imposed) followed by a search for helical symmetry in the resulting 3D model (see Materials and Methods).
FIG. 1.
Transmission electron microscopy of DNA-SspC complex. (A) Image of a typical toroidal aggregate formed by DNA-SspC filaments. Arrowheads point toward single filaments spreading out of the circle. (B) High magnification of a region from panel A, highlighting the tight packing of adjacent filaments. Scale bars are 50 nm (A) and 10 nm (B).
FIG. 2.
DNA-SspC 3D reconstruction. (A) Gallery of raw “particles” (helix sections) that have been aligned by rotation and translation. The last frame shows the average of all aligned particles belonging to one representative class from the reconstruction. (B and C) Surface representations of the initial model (B) and final reconstruction (C). (D) Slices through the reconstruction (perpendicular to the helical axis). Consecutive slices are separated by 0.235 nm and range over one helical subunit. Density values are contoured every 0.5 SD, where the outermost contour is 0.5 SD above the average background intensity.
The initial model was a smooth helix where all density values were identical (Fig. 2B). The initial structural features were the value for the pitch (rise per turn), which was taken as the pitch of B-form DNA (3.4 nm), and the filament width (5.5 nm), as measured in the raw images. The final reconstructed volume (Fig. 2C and D) contains 1.7 helical subunits (the unique volume that is repeated along the helical filament) per turn with a pitch of 3.18 nm, leading to a 1.87-nm rise and 211.8° rotation per helical subunit. These values for pitch and rise per subunit could be observed in the Fourier transform of the averages of aligned filaments. The reconstructed 3D map showed two protrusions for each helical subunit. The protrusions vary in size and radial distance from the filament axis, each apparently corresponding to an SspC molecule. The resolution of the map was 1.1 nm, as measured by Fourier shell correlation (see Materials and Methods). The handedness of the helical DNA-SspC complex could not be determined independently, and was assumed to adopt the DNA right-handed conformation. It was not possible to locate the placement of DNA within the reconstructed volume, due to lack of sufficient detail in the internal density variation.
Geometric docking of DNA-SASP filaments.
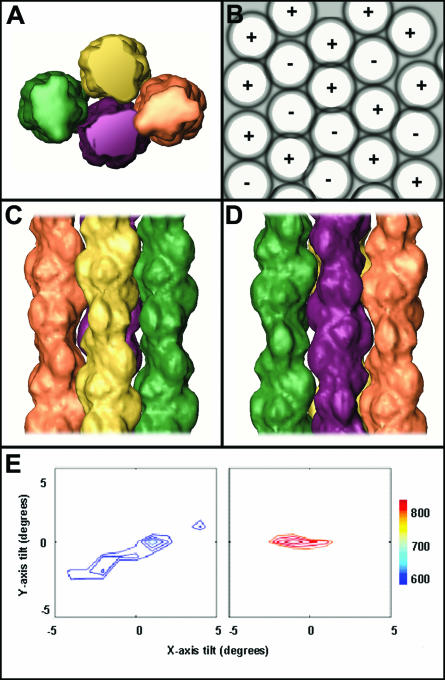
DNA-SspC helical filaments were previously shown to form bundles and swirls in solution (8) (Fig. 1). In vivo, bundles must consist of both parallel and antiparallel orientations, because of the circular nature of the bacterial chromatin. We therefore investigated packaging of adjacent DNA-SspC filaments by geometrical docking (11) in both parallel and antiparallel orientations. The maximal score for parallel fitting was at 56° rotation about the filament (z) axis, 1° rotation about the perpendicular x axis, and no rotation around the y axis (Fig. 3C). The score of the best antiparallel solution was at 56° rotation about the filament (z) axis, and no rotation about the perpendicular (x and y) axes (Fig. 3D).
FIG. 3.
Geometrical docking of DNA-SspC filaments: 3D packaging of four adjacent DNA-SspC filaments. (A) Top view of the filaments. The purple filament forms antiparallel contacts with the other three. (B) Schematic top view showing how multiple filaments form a hexagon-like packing arrangement, with both antiparallel and parallel contacts between neighbors. (C) Shown are the same four filaments from panel A, where the orange, yellow, and green filaments make parallel contacts. (D) The purple filament makes antiparallel contacts with the green and orange filaments. (E) Contour plots of parallel (left) and antiparallel (right) geometrical docking score results. The color bar represents the complementarity score (arbitrary scale) in the range of the parallel and antiparallel solutions, where contour levels of 1 SD from the best solution are plotted every 0.25 SD.
The fitting of antiparallel pairs of filaments resulted in tight docking (Fig. 3D), leaving essentially no gaps between the filaments. The best complementarity score was 9.1 SDs above the mean, in the distribution of solutions. It is a highly unique solution as well, since small rotations about the z axis (±3° from optimum geometry), or ±1° about one of the x or y axes, reduced the complementarity score by 1 SD (Fig. 3E, right). In contrast, fitting of parallel pairs of filaments resulted in significant gaps and a relatively low complementarity (Fig. 3C). The best score, at only 6.2 SDs above the mean, is also less unique, as small rotations of the parallel-packed filaments do not significantly affect the complementarity score (Fig. 3E, left). The addition of additional filaments in the fitting reveals a plausible model for 3D packaging of DNA-SASP filaments (Fig. 3A). The additional filament is found to form good antiparallel contacts with one fiber and additional, considerably less extensive parallel contacts with the other fiber. The overall packing resembles close hexagonal, with each filament surrounded by six other fibers, three parallel and three antiparallel, on average (Fig. 3B).
DNA packaging in bacterial spores.
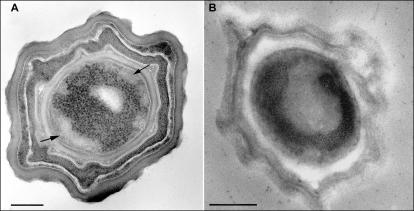
Previous studies indicated that chromatin in developing forespores, as well as in germinating spores of B. subtilis, adopts a tightly packed ring-shaped morphology (24, 25). Although the nucleoid structure in dormant spores was not determined due to the impermeability of the spore core to fixative and stains, it has been suggested that this tight toroidal organization is also present in dormant spores. We used cryofixation techniques and specific DNA staining methods in order to assess DNA structure in mature spores. Cryofixed B. subtilis spores exhibit layers consisting of thick lamellar coat, cortex, and inner membrane that engulf the inner core. Within this core, ribosomes that appear as darkly stained particles, as well as chromatin demarcated as ribosome-free regions, are detected (Fig. 4A and B). As indicated by a specific DNA staining method that was previously shown to be particularly effective for labeling condensed DNA regions (36), spore chromatin is localized in the periphery of the core, forming a tightly packed toroidal structure. Notably, a complete DNA toroid was observed in ∼15% of the spores. The other specimens revealed incomplete toroidal structures, as a portion of the toroid was sliced out in the thin sections probed in transmission electron microscopy.
FIG. 4.
Electron microscopy of dormant B. subtilis spores. (A) B. subtilis spore stained with uranyl acetate. The densely stained particles are ribosomes; ribosome-free spaces in the periphery of the spore core are indicated by arrows and contain chromatin. (B) Specific DNA staining of a dormant spore, which results in darkly stained DNA, highlights the toroid morphology of the chromatin. Since the sections probed are ∼70 nm thick, only a segment of the toroid can be detected. Scale bars, 200 nm. Dormant spores were prepared for transmission electron microscopy as described in Materials and Methods.
DISCUSSION
A major determinant of spore DNA protection against heat, desiccation, oxidizing agents and UV irradiation is the saturation of spore DNA with α/β-type SASP that are expressed in large amounts during spore formation (30). Due to the lack of structural data on the resulting DNA-SASP complex, many aspects of the mechanisms by which these conserved DNA-binding proteins promote DNA protection remain enigmatic.
Previous circular dichroism and Fourier transform infrared studies implied that binding of DNA to SspC, a characteristic member of the α/β-type SASP family, promotes a conformational transition of DNA molecules from a B form to an A-like structure (19, 31). It was proposed that this major conformational modification underlies the ability of α/β-type SASP to protect spore DNA, by substantially modulating DNA photochemistry (19, 31). It was also suggested that the change in DNA supercoiling detected following the interaction with SspC in vitro (20), as well as by DNA in spores (21), derives from the large alteration in the number of base pairs per helical turn that accompanies B-to-A DNA transition (31). Subsequent electron microscopy studies of DNA-SspC filaments indicated, however, that the length of DNA molecules, and hence the rise per base pair, are not significantly altered upon SspC binding, thus implying that DNA conformational state within the DNA-SspC complex remains similar to the canonical B form (8). Moreover, it has been shown that the altered photochemical properties of DNA molecules within DNA-SspC filaments cannot be straightforwardly ascribed to an A-like DNA conformation, but, at least partially, to DNA dehydration (23).
The structural analysis presented here indicates that DNA helical parameters sustain only minor conformational changes upon binding to SspC. The final structure reveals that the pitch (3.18 nm) remains close to that of canonical B-form DNA. These results raise the following question: what are the mechanisms responsible for DNA protection and the alteration of DNA supercoiling (20, 21)?
On the basis of our structural analysis, we propose that the quaternary organization of the DNA-SspC filaments plays an important role in mediating both DNA protection and the increased DNA supercoiling. Within this tight packaging, which derives from a near-to-perfect interdigitation of adjacent filaments, water content is substantially reduced. The attenuated water activity is likely to result in a decreased probability of formation of reactive oxidizing radicals as well as in altered DNA photochemical properties (23), whereby UV-induced production of thymine dimers is restricted. DNA molecules saturated with SspC appear to preferentially adopt a tightly packed toroidal conformation both in vitro (8) (Fig. 1) and in dormant spores (Fig. 4). This particular conformation is significant in light of the recent proposal according to which the exceptional resistance of the bacterium Deinococcus radiodurans towards irradiation and desiccation is promoted by the tight toroidal packaging of its genome (12). Within this rigid matrix, free DNA ends generated by double-stranded breaks are kept firmly together as a result of restricted diffusion, thus allowing for error-free repair through template-independent joining of DNA fragments through nonhomologous end joining (NHEJ). Notably, DNA repair through homologous recombination cannot occur in germinating spores because they regularly carry only one copy of their genome (2). As the ring-shaped organization of chromatin has been shown to persist upon spore germination for several hours (25), it is tempting to suggest that this tight morphology contributes to spore resistance by facilitating repair of double-stranded DNA breaks. Indeed, it has recently been proposed that NHEJ represents an important repair pathway of double-stranded DNA breaks in B. subtilis (36).
Within the 3D toroidal structure, both antiparallel and parallel contacts between adjacent DNA-SspC filaments are necessarily present because of the circularly closed conformation of bacterial chromosomes. Our geometric docking analysis indicates that the filaments pack in a particularly tight conformation when juxtaposed in an antiparallel geometry, whereas in the parallel configuration, lower spatial complementarity between adjacent filaments is observed (Fig. 3). The tight antiparallel contacts stabilize the assembly, thus compensating for the weaker parallel contacts. Due to the nonoptimal complementarity between adjacent DNA-SspC filaments that is imposed by the contribution of parallel contacts, it could be expected that DNA protection against small DNA-modifying agents would be less effective than that achieved against relatively bulky species such as nucleases. Indeed, the DNA-SspC complex has been shown to be as susceptible to small alkylating reagents as naked DNA (29, 30). The mechanism by which SspC protects DNA against formaldehyde (15) remains, however, enigmatic.
Our structural analysis further demonstrates that parallel interfilament contacts are less specific than antiparallel contacts, allowing for flexibility and a relatively large angular distribution (±4°) of packed DNA-SspC filaments. This larger conformational freedom may result in rearrangements of the filaments such that a higher spatial complementarity and hence a tighter, near-to-hexagonal packaging are obtained. These rearrangements, which are likely to entail winding of filaments around each other, would lead to DNA superhelical density within the complex by generating an overall twist. Notably, it has been previously shown that the supercoiling handedness exhibited by aggregates of closely packed chiral filaments is dictated by the chirality of these filaments (26). The unidirectional winding of DNA-SspC filaments, which is required to explain the SspC-induced effects on DNA supercoiling, is thus proposed to represent a direct outcome of the chiral nature of the filaments, combined with the drive to achieve the highest possible chromatin compactness.
We have previously shown that starved nonsporulating bacteria, as well as bacteria exposed to severe and prolonged DNA-damaging agents, protect their DNA complement through physical sequestration within tightly packed structures (7, 13, 37). In this study we demonstrate that the structure of sporal DNA-SspC filaments, in conjunction with their tight spatial organization, provides an effective means for a continuous protection of the spore chromosome. This protection is promoted by DNA saturation with SspC and the tight packaging of the resulting DNA-SspC filaments, as well as by the toroidal conformation of the spore genome. The toroidal structure may facilitate and promote DNA repair through NHEJ (36), by creating a relatively rigid matrix in which DNA fragments are kept together (12). The results support the notion that a protection and survival strategy that depends upon structural features is widespread among prokaryotes when exposed to prolonged periods of starvation and stress (17, 18).
Acknowledgments
This work was supported by the Israel Science Foundation, funded by the Academy of Sciences and Humanities, and by the Minerva Foundation, Germany.
We thank Ed Egelman for the IHRSR source code and David Morgan for his help in implementing the helical-reconstruction techniques.
REFERENCES
- 1.Adrian, M., J. Dubochet, S. D. Fuller, and J. R. Harris. 1998. Cryo-negative staining. Micron 29:145-160. [DOI] [PubMed] [Google Scholar]
- 2.Belliveau, B. H., T. C. Beaman, and P. Gerhardt. 1990. Heat-resistance correlated with DNA content in Bacillus megaterium spores. Appl. Environ. Microbiol. 56:2919-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egelman, E. H. 2000. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85:225-234. [DOI] [PubMed] [Google Scholar]
- 4.Eisenstein, M., I. Shariv, G. Koren, A. A. Friesem, and E. Katchalski-Katzir. 1997. Modeling supra-molecular helices: extension of the molecular surface recognition algorithm and application to the protein coat of the tobacco mosaic virus. J. Mol. Biol. 266:135-143. [DOI] [PubMed] [Google Scholar]
- 5.Francesconi, S. C., T. J. Macalister, B. Setlow, and P. Setlow. 1988. Immunoelectron microscopic localization of small, acid-soluble spore proteins in sporulating cells of Bacillus-Subtilis. J. Bacteriol. 170:5963-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 7.Frenkiel-Krispin, D., S. Levin-Zaidman, E. Shimoni, S. G. Wolf, E. J. Wachtel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 2001. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 20:1184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith, J., A. Makhov, L. Santiago-Lara, and P. Setlow. 1994. Electron microscopic studies of the interaction between a Bacillus subtilis α/β-type small, acid-soluble spore protein with DNA: protein binding is cooperative, stiffens the DNA, and induces negative supercoiling. Proc. Natl. Acad. Sci. USA 91:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes, C. S., E. Alarcon-Hernandez, and P. Setlow. 2001. N-terminal amino acid residues mediate protein-protein interactions between DNA-bound α/β-type small, acid-soluble spore proteins from Bacillus species. J. Biol. Chem. 276:2267-2275. [DOI] [PubMed] [Google Scholar]
- 10.Hayes, C. S., Z. Y. Peng, and P. Setlow. 2000. Equilibrium and kinetic binding interactions between DNA and a group of novel, nonspecific DNA-binding proteins from spores of Bacillus and Clostridium species. J. Biol. Chem. 275:35040-35050. [DOI] [PubMed] [Google Scholar]
- 11.Katchalski-Katzir, E., I. Shariv, M. Eisenstein, A. Friesem, C. Aflalo, and I. Vakser. 1992. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. USA 89:2195-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 13.Levin-Zaidman, S., D. Frenkiel-Krispin, E. Shimoni, I. Sabanay, S. G. Wolf, and A. Minsky. 2000. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc. Natl. Acad. Sci. USA 97:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levitt, M., and M. Gerstein. 1998. A unified statistical framework for sequence comparison and structure comparison. Proc. Natl. Acad. Sci. USA 95:5913-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loshon, C. A., P. C. Genest, B. Setlow, and P. Setlow. 1999. Formaldehyde kills spores of Bacillus subtilis by DNA damage and small, acid-soluble spore proteins of the α/β-type protect spores against this DNA damage. J. Appl. Microbiol. 87:8-14. [DOI] [PubMed] [Google Scholar]
- 16.Mason, J. M., and P. Setlow. 1986. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J. Bacteriol. 167:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minsky, A. 2003. Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50:367-376. [DOI] [PubMed] [Google Scholar]
- 18.Minsky, A., E. Shimoni, and D. Frenkiel-Krispin. 2002. Stress, order and survival. Nat. Rev. Mol. Cell Biol. 3:50-60. [DOI] [PubMed] [Google Scholar]
- 19.Mohr, S. C., N. V. Sokolov, C. M. He, and P. Setlow. 1991. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc. Natl. Acad. Sci. USA 88:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson, W. L., B. Setlow, and P. Setlow. 1990. Binding of DNA in vitro by a small, acid-soluble spore protein from Bacillus subtilis and the effect of this binding on DNA topology. J. Bacteriol. 172:6900-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, W. L., and P. Setlow. 1990. Dramatic increase in negative superhelicity of plasmid DNA in the forespore compartment of sporulating cells of Bacillus subtilis. J. Bacteriol. 172:7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen, C. H., D. G. Morgan, and D. J. DeRosier. 1996. Image analysis of helical objects: the Brandeis Helical Package. J. Struct. Biol. 116:167-175. [DOI] [PubMed] [Google Scholar]
- 23.Patrick, M. H., and D. M. Gray. 1976. Photochem. Photobiol. 24:507-513. [DOI] [PubMed] [Google Scholar]
- 24.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 25.Ragkousi, K., A. E. Cowan, M. A. Ross, and P. Setlow. 2000. Analysis of nucleoid morphology during germination and outgrowth of spores of Bacillus species. J. Bacteriol. 182:5556-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reich, Z., Z. S. Levin, S. B. Gutman, T. Arad, and A. Minsky. 1994. Supercoiling-regulated liquid-crystalline packaging of topologically-constrained, nucleosome-free DNA molecules. Biochemistry 33:14177-14184. [DOI] [PubMed] [Google Scholar]
- 27.Saxton, W. O., and W. Baumeister. 1982. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127:127-138. [DOI] [PubMed] [Google Scholar]
- 28.Setlow, B., and P. Setlow. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow, B., D. Sun, and P. Setlow. 1992. Interaction between DNA and α/β-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J. Bacteriol. 174:2312-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setlow, B., K. J. Tautvydas, and P. Setlow. 1998. Small, acid-soluble spore proteins of the α/β type do not protect the DNA in Bacillus subtilis spores against base alkylation. Appl. Environ. Microbiol. 64:1958-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setlow, P. 1992. DNA in dormant spores of Bacillus species is in an A-like conformation. Mol. Microbiol. 6:563-567. [DOI] [PubMed] [Google Scholar]
- 32.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 33.Setlow, P. 1988. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 42:319-338. [DOI] [PubMed] [Google Scholar]
- 34.Van Heel, M. 1987. Similarity measures between images. Ultramicroscopy 21:95-100. [Google Scholar]
- 35.Vreeland, R. H., W. D. Rosenzweig, and D. W. Powers. 2000. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407:897-900. [DOI] [PubMed] [Google Scholar]
- 36.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. D. di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, S. G., D. Frenkiel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 1999. DNA protection by stress-induced biocrystallization. Nature 400:83-85. [DOI] [PubMed] [Google Scholar]