Abstract
Introduction
Targeting activating oncogenic driver mutations in lung adenocarcinoma has led to prolonged survival in patients harboring these specific genetic alterations. The prognostic value of these mutations has not yet been elucidated. The prevalence of recently uncovered non-coding somatic mutation in promoter region of TERT gene is also to be validated in lung cancer. The purpose of this study is to show the prevalence, association with clinicalpathological features and prognostic value of these factors.
Methods
In a cohort of patients with non-small cell lung cancer (NSCLC) (n = 174, including 107 lung adenocarcinoma and 67 lung squamous cell carcinoma), EGFR, KRAS, HER2 and BRAF were directly sequenced in lung adeoncarcinoma, ALK fusions were screened using FISH (Fluorescence in situ Hybridization).TERT promoter region was sequenced in all of the 174 NSCLC samples. Associations of these somatic mutations and clinicopathological features, as well as prognostic factors were evaluated.
Results
EGFR, KRAS, HER2, BRAF mutation and ALK fusion were mutated in 25.2%, 6.5%, 1.9%, 0.9% and 3.7% of lung adenocarcinomas. No TERT promoter mutation was validated by reverse-sided sequencing. Lung adenocarcinoma with EGFR and KRAS mutations showed no significant difference in Disease-free Survival (DFS) and Overall Survival (OS). Cox Multi-variate analysis revealed that only N stage and HER2 mutation were independent predictors of worse overall survival (HR = 1.653, 95% CI 1.219–2.241, P = 0.001; HR = 12.344, 95% CI 2.615–58.275, P = 0.002).
Conclusions
We have further confirmed that TERT promoter mutation may only exist in a very small fraction of NSCLCs. These results indicate that dividing lung adenocarcinoma into molecular subtypes according to oncogenic driver mutations doesn't predict survival difference of the disease.
Introduction
Lung cancer is one of the most devastating diseases and the leading cause of cancer-related deaths worldwide [1]. Non-small cell lung cancer (NSCLC), which accounts for about 85% of lung cancer cases, is further divided to histological subtypes of adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma. Adenocarcinoma has become the most common lung cancer subtype [2]. The 5-year survival rate of NSCLC remains low as 15–16%, despite improvement of treatment in the past decades [1], [3]. Better understanding of the NSCLC tumorigenesis and factors associated with prognosis is needed.
Uncovering of activating mutations in tyrosine kinase domain of EGFR has led to developing and wide use of gefetinib and erlotinib, which has proven to be effective in the treatment for part of EGFR mutated lung adenocarcinomas [2], [4]. According to specific oncogenic driver mutations, lung adenocarcinoma is divided to molecular subtypes such as EGFR, KRAS, ALK, HER2, BRAF and so on [5], [6]. Most of these driver genes have specific targeted drugs in use or clinical trials. Compared to adenocarcinoma, knowledge on genetic alterations of lung squamous cell carcinoma is limited. Frequent mutation, amplification or loss of FGFR1, PTEN, PIK3CA, NFE2L2, NRF have been identified in comprehensive genomic study in lung squamous cell carcinoma [7]. Because the largely undefined mutational spectrum and low frequencies of these genes, targeted therapies in lung squamous cell carcinoma is lagging behind and constricted to clinical trials.
Recently, human cancer genome sequencing studies have revealed somatic mutaitons in the core promoter of human telomerase reverse transcriptase (hTERT).
Gene, which codes the catalytic subunit of telomerase [8]–[10]. Either C228T or C250T of hTERT promoter mutations elevated transcriptional activity of hTERT gene in the luciferase reporter assays [8]. The tumor types with frequent hTERT promoter mutations included Melanoma (71%), Glioma (51%), Myxoid liposarcoma (79%), Urothelial carcinoma of bladder (66%) [8], [9]. Evidences in above studies suggested that hTERT promoter mutations potentially act as a driver gene in melanoma. However, the prevalence of hTERT promoter mutation in lung cancer clinical samples is absent in these studies. It's necessary to show the prevalence of hTERT promoter mutation in NSCLC samples as lung cancer shares driver mutated genes with melanoma, such as BRAF and NRAS [8], [9], [11].
In this study, we aimed to show the comprehensive molecular and clinicopathological features in NSCLC, as well as the prognostic value of these characteristics.
Methods
Patients and tissues
Primary tumor samples were obtained from 174 patients who underwent potentially curative pulmonary resection at the Tianjin Medical University Cancer Hospital from Jan 2004 through Jan 2008. This study was approved by the Institutional Review Board of Tianjin Medical University Cancer Institute and Hospital. All participants gave written informed consent. Patients were enrolled in this specific study based upon the following criteria: they had a pathologic diagnosis of lung adenocarcinoma or squamous cell carcinoma, their tumor sample contained a minimum of 50% tumor cells as determined by study pathologists, they did not receive neoadjuvant chemotherapy, and they had sufficient tissue for molecular analysis. Clinicopathological features and prognosis information were collected.
DNA isolation and mutational analysis
Genomic DNA were isolated from Formalin-Fixed Paraffin-Embedded (FFPE) samples according to standard protocols (TIANquick FFPE DNA Kit, Tiangen Inc, China). EGFR (exons 18–22), HER2 (exon 20), KRAS (exons 2 to 3), BRAF (exon 15) and TERT promoter region were PCR amplified using DNA and directly sequenced. Multiplex PCR analysis was done with rTaq DNA polymerase (Toyobo, Osaka, Japan). Primers used in these experiments are listed in Table S1. ALK fusions were screened using FISH with FFPE slides.
Statistical analysis
Associations between mutations and clinical and biological characteristics were analyzed by χ2 or Fisher's exact test. Survival curves were drawn by the Kaplan–Meier method. The Cox proportional hazards regression (forward likelihood ratio model) was used for multivariate survival analyses. All data were analyzed using the Statistical Package for the Social Sciences Version 16.0 Software (SPSS Inc., Chicago, IL). The two-sided significance level was set at P<0.05.
Results
Patient Characteristics
One hundred and seven lung ADC and 67 SCC patients were enrolled in this study, including 105 men and 69 women (average 58 years; range, 39–77 years). There were 109 smokers and 65 non-smokers, accounting for 63% and 37%, respectively. The number of patients in stages I-III was 57, 46, and 69, respectively. Two samples were with undefined stage. The detailed information is listed in Table 1 .
Table 1. Demographics and clinicopathologic features of 174 patients with NSCLC.
| Variables | No. of patients (%) |
| Age | |
| ≤50 | 37(21.3) |
| 51–60 | 69(39.6) |
| 61–70 | 49(28.2) |
| >70 | 19(10.9) |
| Mean ± SD (range) | 58.17±9.44 |
| Histologic subtype | |
| Adenocarcinoma | 107(61.5) |
| Squamous cell carcinoma | 67(38.5) |
| Smoking status | |
| Smoker | 109(62.6) |
| Non-smoker | 65(37.4) |
| Stage | |
| IA | 24(13.8) |
| IB | 33(18.9) |
| IIA | 30(17.2) |
| IIB | 16(9.2) |
| IIIA | 68(39.1) |
| IIIb | 1(0.5) |
| NA | 2(1.1) |
| Total | 174 |
SD,standard deviation; NA,not applicable.
EGFR, KRAS, HER2 and BRAF Mutation Status and ALK fusions
Of the 107 lung adenocarcinoma samples, 25.2% (27/107) of tumors were found to harbor EGFR kinase domain mutations. Among these, 17 were deletions in exon 19 and 6 were L858R missense changes. Other alterations included 1 exon 20 insertion, exon 20 D807N and T790A mutations, 1 exom 18 R705K mutation. Six samples (5.6%) had a KRAS mutation, including five G12V mutations and an I36M missense mutation. Two samples harbored HER2 exon 20 mutation, including a 776 to 779 YVMA insertion and a S789P mutation. Only one BRAF L588F missense mutation was identified in these tumor samples ( Figure 1 ). ALK fusions were detected in 4 (3.7%) samples. Because mutations of EGFR, KRAS, HER2, BRAF, as well as ALK fusions have been well established to be existed in lung adenocarcinoma [5], [12]–[14], these mutation and fusion genes that ‘drives’ lung adenocarcinoma were not screened in lung squamous cell carcinoma.
Figure 1. Pie chart of EGFR, KRAS, HER2 and BRAF mutation spectrum from 107 lung adenocarcinomas.
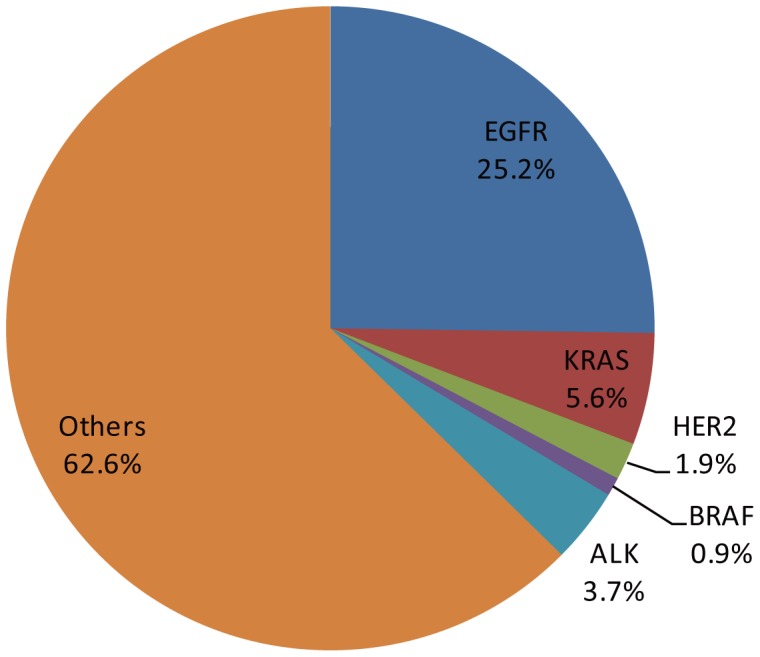
The mutation rates of EGFR, KRAS, HER2 and BRAF were 25.2, 5.6, 1.8 and 0.9%, respectively.
5′ non-coding region mutations of TERT gene in NSCLC
Neither C228T nor C250T mutations was validated in the 174 NSCLC samples after reverse sequencing. These results indicated that TERT promoter mutation may only exist in a very small fraction of primary NSCLC samples.
Survival Analysis
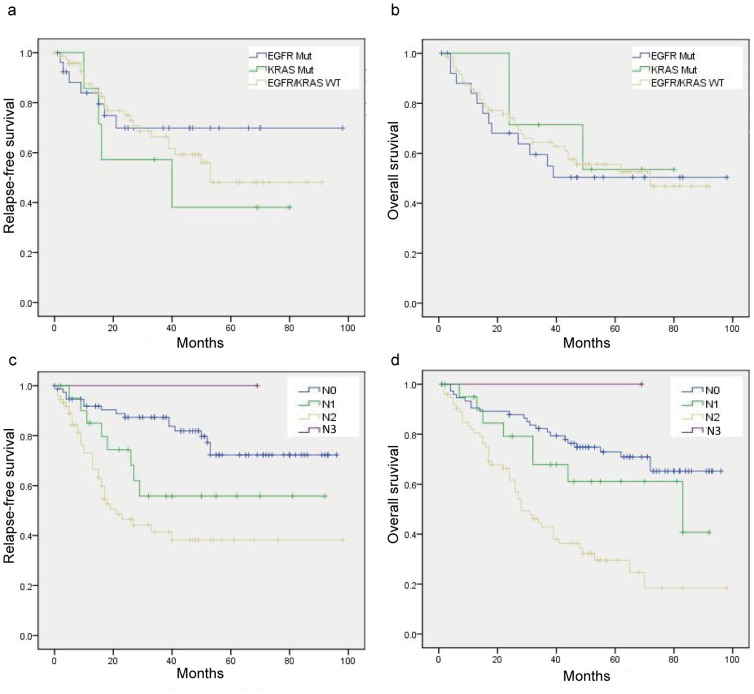
By univariate analysis, advanced clinical staging (P = 0.018), lymphnode metastasis (P<0.001), ≥2 station of metastatic lymphnode (P<0.001), received adjuvant chemotherapy (P = 0.004) were associated disease relapse ( Table 2 ). EGFR and KRAS mutation status were not associated with relapse-free survival (P = 0.600) or overall survival in lung adenocarcinoma (P = 0.873) ( Figure 2 ). Cox multivariate forward stepwise analysis (adjusting for age, gender, smoking history, clinical stage, histologic subtypes and mutational status) revealed that only lymphnode stage (N stage) and HER2 mutation was independent predictors of poorer overall survival (HR 1.653, 95% CI 1.219–2.241; P = 0.001; HR 12.344, 95% CI 2.615–58.275; P = 0.002). Detailed mutational and survival information is listed in Table S2.
Table 2. Association between clinical and molecular factors and disease relapse.
| Factors | Relapse | P | |
| Clinical Stage | Relapsed | Relapse-free | |
| I | 13 | 46 | 0.018 |
| II | 16 | 30 | |
| III | 32 | 37 | |
| EGFR status | |||
| Mutated | 7 | 20 | 0.326 |
| Wildtype | 29 | 51 | |
| KRAS status | |||
| Mutated | 4 | 3 | 0.188 |
| Wildtype | 32 | 68 | |
| Lymphnode metastasis | |||
| Positive | 44 | 52 | 0.001 |
| Negative | 17 | 61 | |
| No.of metastatic lymphnode station | |||
| <2 | 29 | 83 | 0.001 |
| ≥2 | 32 | 30 | |
| Adjuvent Chemotherapy | |||
| Received | 45 | 58 | 0.004 |
| Not-received | 16 | 55 | |
Figure 2. Relapse-free survival and overall survival in patients with NSCLC.
EGFR and KRAS mutation status were not associated with relapse-free survival (P = 0.600) or overall survival (P = 0.873) in lung adenocarcinoma; c–d. Lymphnode stage (N stage) was significantly associated with relapse-free survival (P = 0.001) or overall survival (P = 0.001) in NSCLC.
Discussion
NSCLC is a heterogeneous disease, outcome differs even in patients with identical clinicopathologic features. Identification of oncogenic driver mutations in lung adenocarcinoma has greatly promoted clinical use and development of targeted drugs [5], [6], [15], [16]. Though EGFR and KRAS mutations have been established as predictive and prognostic markers for patients who receive EGFR-TKIs [4], [17], [18], results on the prognostic value of these alterations at genomic level in patients who didn't receive EGFR-TKIs were rarely reported.
We have screened the prevalence of known driver mutations of EGFR, KRAS, HER2, BRAF and ALK in a cohort of 107 lung adenocarcinoma sample with complete prognostic information. The EGFR mutation, KRAS mutation and EGFR/KRAS wild type groups showed no significant difference in both relapse-free survival and overall survival in the present study. All the patients enrolled in this study didn't receive small molecular drugs targeting EGFR. Our results suggest that specific driver mutation of EGFR or KRAS gene doesn't predict survival advantage or disadvantage when these patients didn't receive specific target therapies. In the multivariate analysis, HER2 mutation was identified as independent predictor of unfavorable prognosis. Because only two samples harbored HER2 mutation, further study with larger number of samples are needed to confirm this result. Based on these facts, we conclude that to divide lung adenocarcinoma into molecular subtypes only predicts therapy response but not survival benefit in patients who didn't receive these drugs.
The emerging of somatic mutations of hTERT gene 5′ promoter region in melanoma has provided another promising candidate driver gene for lung cancer. Several studies have identified frequent hTERT 5′ promoter mutations in melanoma [8], [10], gliomas, as well as from relatively low rates of self renewal tissues like liposarcomas, hepatocellular carcinomas, urothelial carcinomas, squamous cell carcinomas of the tongue and medulloblastomas [9], [19]. We screened the hTERT 5′ promoter region in 174 NSCLC FFPE samples to explore the mutation rate and potential prognostic value. We identified only three C250T mutations from the 174 samples, but none of these were validated by reverse sequencing, which could be possibly caused by DNA damage in the process of formalin fixing and paraffin embedding. These results confirmed that hTERT 5′ promoter mutations may only exist in a very small fraction of NSCLC cases.
The limitation of the present study should be discussed. In order to explore the prognostic significance, we used FFPE samples which have complete survival information. However, the sensitivity of sequencing results is affected by the relatively low quality of FFPE samples, in which DNA is degraded and artificially mutated. The mutation rate of well known driver genes is lower than previous studies using high quality snap-frozen samples [20], [21], and the artificial mutations appeared in hTERT 5′ promoter region. Though consecutive samples with complete survival data are not always available, our study provided the mutation spectrum and prognostic association of these genetic alterations with available sample and techniques.
In conclusion, our work illustrated the mutation spectrum of EGFR, KRAS, HER2, BRAF and ALK in lung adenocarcinoma, as well as confirmed that hTERT 5′ promoter region mutation rarely occurred in NSCLC samples. We also confirmed that these driver mutations lack prognostic value in lung adenocarcinoma patients who didn't receive EGFR TKI treatment, and subtypes divided by these oncogenic driver mutations don't predict significant survival difference.
Supporting Information
List of primers used for polymerase chain reaction amplification of the EGFR, KRAS, HER2 and BRAF gene.
(DOCX)
Mutational and survival information of 174 NSCLC patients. Sex, 1 male, 2 female; Smoking, 0 non-smoker, 1 smoker; path, pathology; WT, wild-type; DEL, deletion; INS, insertion; NEG, negative; POS, positive.
(XLS)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The original data sheet of our experiment's results can be found in the supporting information.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81302001 to CG Li and 81201649 to BS Sun, http://www.nsfc.gov.cn), China Postdoctoral Science Foundation (2014M550147 to CG Li, http://res.chinapostdoctor.org.cn/BshWeb/index.shtml), Key Program for Anti-cancer Research of Tianjin Municipal Science and Technology Commission (12ZCDZSY15400 to CL Wang, http://www.tstc.gov.cn/) and Key Program of Application Foundation, Tianjin Municipal Science and Technology Commission (12JCYBJC17800 to CL Wang, http://www.tstc.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 4. Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, et al. (2003) Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin Oncol 21: 2237–2246. [DOI] [PubMed] [Google Scholar]
- 5. Pao W, Hutchinson KE (2012) Chipping away at the lung cancer genome. Nat Med 18: 349–351. [DOI] [PubMed] [Google Scholar]
- 6. Li C, Fang R, Sun Y, Han X, Li F, et al. (2011) Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 6: e28204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research N (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, et al. (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, et al. (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110: 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, et al. (2013) TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961. [DOI] [PubMed] [Google Scholar]
- 11. Bucheit AD, Syklawer E, Jakob JA, Bassett RL Jr, Curry JL, et al. (2013) Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. Cancer 119: 3821–3829. [DOI] [PubMed] [Google Scholar]
- 12. Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, et al. (2012) Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res 18: 1167–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perez-Moreno P, Brambilla E, Thomas R, Soria JC (2012) Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res 18: 2443–2451. [DOI] [PubMed] [Google Scholar]
- 14. Gold KA, Wistuba, II, Kim ES (2012) New strategies in squamous cell carcinoma of the lung: identification of tumor drivers to personalize therapy. Clin Cancer Res 18: 3002–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, et al. (2012) ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 30: 863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, et al. (2014) Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370: 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon JH, Kang CH, Kim HS, Seong YW, Park IK, et al. (2014) Prognostic and predictive role of epidermal growth factor receptor mutation in recurrent pulmonary adenocarcinoma after curative resection. Eur J Cardiothorac Surg. [DOI] [PubMed]
- 18. Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, et al. (2005) Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353: 123–132. [DOI] [PubMed] [Google Scholar]
- 19. Hurst CD, Platt FM, Knowles MA (2014) Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur Urol 65: 367–369. [DOI] [PubMed] [Google Scholar]
- 20. Sun Y, Ren Y, Fang Z, Li C, Fang R, et al. (2010) Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 28: 4616–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao B, Sun Y, Zhang J, Ren Y, Fang R, et al. (2010) Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol 5: 1130–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for polymerase chain reaction amplification of the EGFR, KRAS, HER2 and BRAF gene.
(DOCX)
Mutational and survival information of 174 NSCLC patients. Sex, 1 male, 2 female; Smoking, 0 non-smoker, 1 smoker; path, pathology; WT, wild-type; DEL, deletion; INS, insertion; NEG, negative; POS, positive.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The original data sheet of our experiment's results can be found in the supporting information.