Abstract
Biofilms have been implicated as an important reservoir for pathogens and commensal enteric bacteria such as Escherichia coli in natural and engineered water systems. However, the processes that regulate the survival of E. coli in aquatic biofilms have not been thoroughly studied. We examined the effects of hydrodynamic shear and nutrient concentrations on E. coli colonization of pre-established Pseudomonas aeruginosa biofilms, co-inoculation of E. coli and P. aeruginosa biofilms, and P. aeruginosa colonization of pre-established E. coli biofilms. In nutritionally-limited R2A medium, E. coli dominated biofilms when co-inoculated with P. aeruginosa, and successfully colonized and overgrew pre-established P. aeruginosa biofilms. In more enriched media, P. aeruginosa formed larger clusters, but E. coli still extensively overgrew and colonized the interior of P. aeruginosa clusters. In mono-culture, E. coli formed sparse and discontinuous biofilms. After P. aeruginosa was introduced to these biofilms, E. coli growth increased substantially, resulting in patterns of biofilm colonization similar to those observed under other sequences of organism introduction, i.e., E. coli overgrew P. aeruginosa and colonized the interior of P. aeruginosa clusters. These results demonstrate that E. coli not only persists in aquatic biofilms under depleted nutritional conditions, but interactions with P. aeruginosa can greatly increase E. coli growth in biofilms under these experimental conditions.
Introduction
In aquatic environments, microorganisms coexist in complex and heterogeneous biofilm communities that exhibit genotypes and phenotypes distinct from their planktonic counterparts [1]–[3]. Biofilms are of particular concern in engineered water distribution systems, as they have been found to harbour pathogens as diverse as enteric viruses, cysts of protozoa such as Cryptosporidium parvum and Giardia lamblia, and bacteria such as Pseudomonas aeruginosa, Escherichia coli, Campylobacter jejuni, Helicobacter pylori, Legionella pneumophila, and Aeromonas spp. [1], [3]–[5]. Residence within biofilms can provide microorganisms with access to higher concentrations of nutrients and protection from environmental stresses and chemical disinfectants [1], [2], [6], [7]. Biological interactions that occur between newly-associated pathogens and indigenous microbial populations have been shown to regulate pathogen persistence and growth [1], [6]–[8]. Prior studies on the survival of biofilm-associated pathogens [9]–[12] suggest that biofilms may play an especially important role in the persistence and dissemination of fastidious and stress-sensitive organisms in the environment [1]. A deeper understanding of the survival and growth potential of pathogens and fecal indicator organisms in aquatic biofilms is needed for the continued improvement of water treatment strategies and identification of potential sources of pathogen contamination.
Historically, E. coli has been used as an indicator of fecal contamination in the United States and elsewhere [13], [14]. For years, E. coli was believed to survive poorly outside of a living host and not grow in secondary habitats due to a constant exposure to environmental stresses [15]. However, recent reports have shown that E. coli can survive and potentially replicate in nutrient-rich aquatic environments [15]. Mounting evidence also suggests that E. coli and other enteric organisms can persist and potentially grow in drinking water distribution networks [16], [17]. Laboratory-scale studies have demonstrated that E. coli can become associated with pre-established indigenous biofilms of the type found in drinking water distribution systems [6], [18]. The prolonged survival of E. coli in aquatic biofilms may lead to the incorrect detection of fecal contamination and mask true breakthrough events in water distribution networks. Temperature, nutrient availability, concentration of disinfectants and antagonistic bacterial predation have all been shown to influence the persistence and growth of E. coli in aquatic environments [6], [7], [15], [19]. However, little is known of the conditions under which E. coli can colonize pre-existing aquatic biofilms, or the mechanisms of their interaction with typical aquatic biofilm-forming organisms.
Here, we investigated the effects of varying nutrient concentrations and fluid velocities on the ability of E. coli to form biofilms in co-culture with the robust and ubiquitous aquatic biofilm-forming bacterium Pseudomonas aeruginosa. To evaluate the effects of timing of organism introduction, we investigated E. coli colonization of pre-established P. aeruginosa biofilms, co-inoculation of E. coli and P. aeruginosa biofilms, and P. aeruginosa colonization of pre-established E. coli biofilms. Additionally, we examined the role of indole, an E. coli metabolite, in regulating E. coli and P. aeruginosa interactions in biofilms.
Experimental Procedures
Flow cells
Biofilm growth was observed using a 2-D planar flow cell, and a single-channel microfluidic flow cell [20]. The planar flow cell was used for the study of biofilms under a distribution of local velocities in a single experiment. The flow chamber is 35 mm×35 mm×0.6 mm in size, and consists of a transparent acrylic base and a glass coverslip. The glass coverslip allows for direct observations of biofilm development by microscopy. Controlled patterns of inflow can be imposed by means of inflow or outflow ports distributed around the periphery of the flow cell. Here, flow was introduced uniformly to the inflow ports on one side of the flow cell, producing a right-angle turning flow that imposes a distribution of velocities and medium flux rates over the biofilm. Nine regions (R1–9) at the center of the flow chamber were selected for biofilm imaging, forming a 10 mm×10 mm observation window. A detailed description of the flow cell design and performance can be found in Zhang et al. [20].
A polydimethylsiloxane (PDMS), single-channel microfluidic flow cell was used to study biofilm growth under a controlled, uniform velocity field. Each channel measures 35 mm×4 mm×1 mm in size, with a single inlet and outlet port. A glass coverslip at the base of each channel allows for in situ visualization of the biofilms. A detailed description of the flow cell design can be found in Song et al. [21].
Bacterial strains
Biofilm flow cell experiments were performed with several E. coli strains and a single typical P. aeruginosa biofilm-forming strain. An E. coli DH5α strain inserted with plasmid pUC encoding mCherry fluorescent proteins was used to study dual-species biofilm growth behavior. This strain is derived from E. coli K-12. The indole-deficient tnaA single deletion mutant E. coli JW3686 [22] and its parent strain E. coli BW25113 [22] were used to study the effects of indole on dual-species biofilm growth. Both E. coli JW3686 and E. coli BW25113 are derivatives of E. coli K-12. A P. aeruginosa PAO1 strain with a chromosomally expressed green fluorescent protein (GFP) was also used. P. aeruginosa PAO1 is the definitive laboratory model strain used for biofilm research. Stock cultures of P. aeruginosa and E. coli were streaked onto Luria-Bertani (LB) agar plates, and incubated for 24 hours at 37°C. Single colonies of each strain were transferred into separate tubes containing 3 mL of sterile LB broth, and grown overnight in a shaker at 37°C and 225 rpm for injection into the flow cells.
Flow cell experimental conditions
Mono- and mixed-culture biofilm experiments were run using R2A media at room temperature (24°C). R2A was chosen as a nutritionally depleted medium. R2A consists of 0.05 g/L yeast extract, 0.05 g/L proteose peptone No. 3, 0.05 g/L casamino acids, 0.05 g/L dextrose, 0.03 g/L sodium pyruvate, 0.03 g/L dipotassium phosphate, and 0.005 g/L magnesium sulfate [9]. This medium has most typically been used to grow and enumerate microorganisms from drinking water sources [23]. Gilson Miniplus 3 peristaltic pumps were used to circulate the R2A medium, as they produce minimal flow pulsations and are well suited for biofilm experiments. The flow was regulated to 0.8 mL/min (0.2 mL/min per inflow port) in planar flow cells. We chose this flow configuration because the resulting velocity gradient (0.96–1.74 mm/s) has been shown to produce distinct growth and detachment patterns in P. aeruginosa biofilms [20]. In the microfluidic flow cells, the flow was regulated to 0.2 mL/min. All experiments were replicated three times using independent flow cells run in parallel.
To investigate the colonization of pre-existing P. aeruginosa biofilms by E. coli in planar and microfluidic flow cells, 1 mL of a stationary-phase culture of P. aeruginosa (OD600 = 0.1) was first injected into each chamber and allowed to deposit on the flow cell coverslip under stagnant conditions for one hour. The flow of standard R2A was then initiated and maintained at a constant rate for 3 days. On day 3, the flow cells containing pre-established P. aeruginosa biofilms were inoculated using 1 mL of stationary-phase E. coli cultures (OD600 = 0.1), and the flow was halted for 30 min to facilitate E. coli deposition into the biofilm. The inflow of medium was then resumed and maintained at a constant rate for an additional 3 days. These experiments were also conducted using 4x and 8x concentrated R2A medium in microfluidic flow cells to investigate the effects of nutritional conditions on growth of biofilm co-cultures.
In co-inoculated experiments, 0.5 mL of stationary-phase cultures of E. coli and P. aeruginosa were diluted to an OD600 of 0.1 and mixed at a ratio of 1∶1 (1 mL total volume). The cell mixtures were then inoculated in both planar and microfluidic flow cells. Inoculation was followed by a 1-h stagnant period to facilitate attachment of cells, and the flow of standard R2A medium was then resumed for 3 days. In microfluidic flow cells, these experiments were also conducted using 4x and 8x concentrated R2A medium.
To investigate the colonization of pre-existing E. coli biofilms by P. aeruginosa, 1 mL of a stationary-phase culture of E. coli (OD600 = 0.1) was injected into a microfluidic flow cell with standard R2A medium and allowed to attach on the glass coverslip under stagnant conditions for one hour. The inflow of R2A medium was then initiated and maintained at a constant rate for a period of 3 days. On day 3, the flow chambers containing pre-established P. aeruginosa biofilms were inoculated using 1 mL of stationary-phase cultures of P. aeruginosa (OD600 = 0.1), and the flow was halted for 30 min to facilitate P. aeruginosa deposition. The flow was then resumed and maintained at a constant rate for 3 additional days.
Batch experiments
Batch cultures were grown for comparison with biofilm results. Pure cultures of P. aeruginosa and E. coli were grown in LB medium overnight at 24°C, and subsequently diluted to an OD600 of 0.1 (1.6×108 CFU/mL). Erlenmeyer flasks containing 20 mL of sterile R2A medium were inoculated using 150 µL of each culture. The mixed cultures were incubated in a shaker (150 rpm) at 24°C overnight. Each day for 3 days, 300 µL of the mixed culture was transferred into a new Erlenmeyer flask containing 20 mL of fresh R2A. The batch experiments were repeated using an incubation temperature of 37°C. These mixed batch cultures were sampled daily for CFU using selective media in order to differentiate P. aeruginosa and E. coli. Ampicillin (100 µg/mL) was used to select for P. aeruginosa, and cefsulodin (20 µg/mL) was used to select for E. coli [24]. Batch cultures were repeated in triplicate.
Imaging procedures
Biofilm micrographs were obtained using a Leica SP5 confocal laser scanning microscope, and collected with Leica Confocal Software. The three-dimensional biofilm images were generated from the planar image stacks using the image processing software VOLOCITY (Improvision, Inc.) Quantitative analysis of the biofilm structures was conducted using the COMSTAT image processing software [25].
Antibiotic selection for mCherry plasmid-encoding E. coli was not performed in flow cell experiments to avoid negative effects on P. aeruginosa in mixed biofilms. As a result, mCherry fluorescence was lost after a period of 18–24 hours. Imaging of E. coli using mCherry fluorescence was therefore only used during the initial cell deposition and attachment phase. E. coli was subsequently imaged by counterstaining the mixed biofilms using SYTO 62, a cell-permeant nucleic acid stain (Life Technologies). P. aeruginosa was imaged by constitutively expressed gfp fluorescence. Biofilms counterstaining was conducted at the end of each experiment using a 50 µM solution of SYTO 62 for a period of 30 minutes in the dark. The flow was then resumed for 20 min in order to wash out unbound stain.
Results
Colonization of pre-established P. aeruginosa biofilms by E. coli
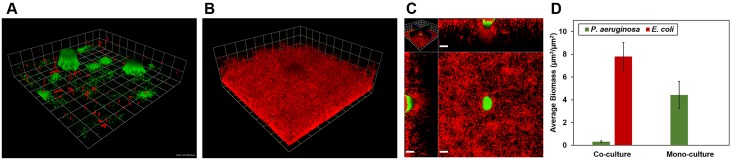
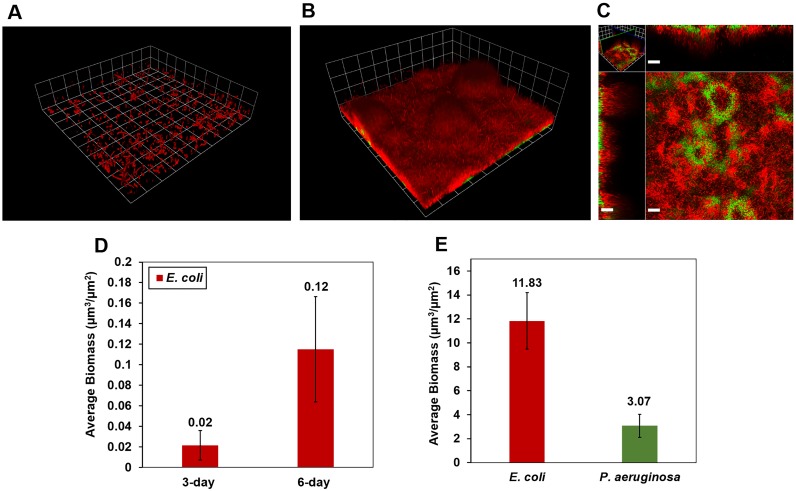
E. coli colonization of pre-existing, 3-day-old P. aeruginosa biofilms is shown in Fig. 1. P. aeruginosa biofilms grew in clusters with scattered cells present between colonies (Fig. 1A). Within three days of introduction, E. coli was able to extensively colonize the biofilm (Fig. 1B, C). E. coli overgrew P. aeruginosa clusters and colonized the interior of clusters as well (Fig. 1C). The average E. coli biomass was consistently greater than that of P. aeruginosa (Fig. 1D). Moreover, we found substantially less P. aeruginosa in mixed culture compared to mono-species biofilms on day 6 (Fig. 1D). This indicates that E. coli not only dominated in mixed culture, but also hindered P. aeruginosa growth. These results show that E. coli can successfully colonize, outcompete, and outgrow established P. aeruginosa biofilms in the nutritionally limited R2A medium.
Figure 1. Colonization of P. aeruginosa biofilms by E. coli in R2A medium in a microfluidic flow cell.
E. coli appears red and P. aeruginosa appears green or yellow. A) E. coli deposited on 3-day old P. aeruginosa biofilm (grid unit is 23.8 µm). B) Resulting mixed E. coli – P. aeruginosa biofilm on day 6 (grid unit is 23.8 µm). C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 20 µM). D) P. aeruginosa and E. coli biomass in mixed biofilms on day 6 compared with biomass of 6-day old mono-cultured P. aeruginosa biofilms.
Co-inoculation of P. aeruginosa and E. coli
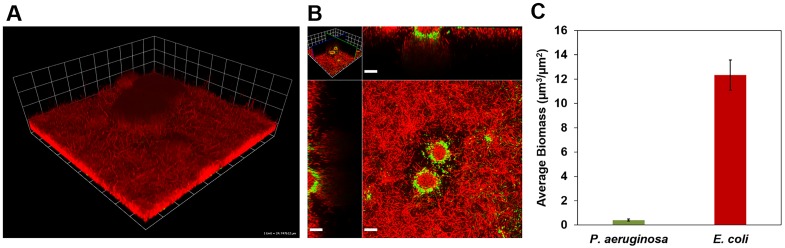
Growth of co-inoculated, 3-day-old biofilms in microfluidic flow cells is shown in Fig. 2. E. coli grew extensively in a lawn-like fashion (Fig. 2A), and consistently over-grew P. aeruginosa, which was present primarily in isolated clusters (Fig. 2B). E. coli also successfully colonized the interior of the P. aeruginosa clusters, restricting P. aeruginosa to a thin shell near the exterior of each cluster. The average biomass data presented in Fig. 2C highlights the overwhelming success of E. coli in colonizing the landscape compared to P. aeruginosa. Conversely, P. aeruginosa consistently achieved greater population density than E. coli in batch co-cultures (Fig. S1, S2 in File S1). These differences emphasize that population growth dynamics and competition for resources change substantially in biofilms.
Figure 2. Co-development of E. coli and P. aeruginosa biofilms in a microfluidic flow cell and R2A medium.
E. coli appears red and P. aeruginosa appears green or yellow. A) 3-day old co-inoculated P. aeruginosa - E. coli biofilm (grid unit is 23.8 µm). B) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel A (scale bar = 20 µM). C) P. aeruginosa and E. coli biofilm biomass on day 3.
Effects of nutrient concentrations on the colonization of pre-established P. aeruginosa biofilms by E. coli
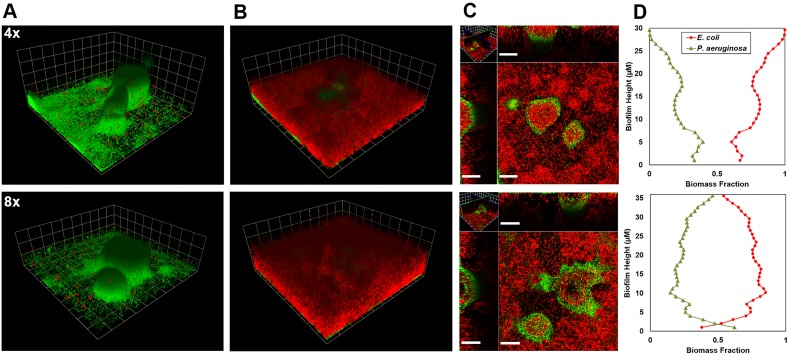
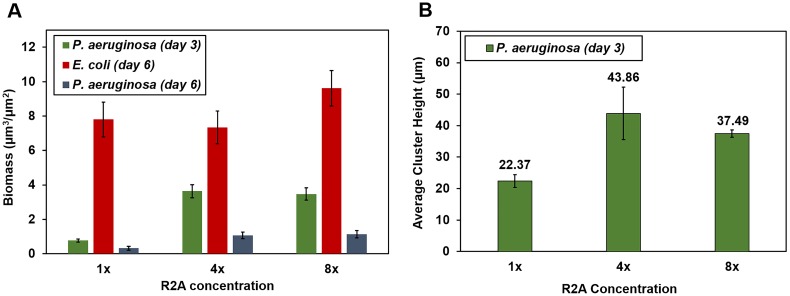
Larger and morphologically distinct P. aeruginosa biofilms grew in enriched R2A medium (Fig. 3A). Significantly more P. aeruginosa biomass and taller clusters grew under enriched 4x and 8x R2A media relative to the standard (depleted) R2A medium (t-test P<0.01) (Fig. 4). However, differences in biomass between 4x and 8x R2A media were not significant (P>0.60). P. aeruginosa biomass decreased after E. coli inoculation. P. aeruginosa biomass in mixed culture on day 6 (3 days after E. coli inoculation) was consistently lower than on day 3 (at the time of E. coli inoculation) for all nutritional conditions.
Figure 3. Colonization of P. aeruginosa biofilms by E. coli under varying R2A medium concentrations in a microfluidic flow cell.
E. coli appears red and P. aeruginosa appears green. Column A) E. coli deposited on 3-day old mono-species P. aeruginosa biofilms under 4x and 8x R2A concentrations (grid unit is 23.8 µm). B) P. aerugiosa and E. coli biofilms on day 6, 3 days after introduction of E. coli (grid unit is 23.8 µm). C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 40 µm). D) Biomass fraction vs. biofilm height graph. P. aeruginosa biofilm biomass increased with R2A concentration. E. coli dominated the community under all medium concentrations regardless of the P. aeruginosa cluster morphology. Biofilms were counter-stained by SYTO 62.
Figure 4. E. coli colonization of pre-established P. aeruginosa biofilms under varying R2A medium concentrations.
A) Average biofilm biomass vs. R2A concentration for 1) P. aeruginosa on day 3, 2) E. coli on day 6, and 3) P. aeruginosa on day 6. B) Average P. aeruginosa cluster height vs. R2A concentration on day 3.
E. coli grew to substantially greater biomass than P. aeruginosa within 3 days of inoculation (Fig. 4B). Unexpectedly, E. coli biomass was not significantly different under enriched conditions (4x and 8x R2A) compared to standard R2A (P>0.50 and P>0.09 respectively). The biofilm morphology of E. coli did not vary, with lawn-like growth throughout the chamber and colonization of the interior of P. aeruginosa clusters under all conditions tested (Fig. 3B, C). P. aeruginosa clusters were readily colonized by E. coli, with substantial intergrowth occurring even in the largest and tallest P. aeruginosa clusters (Fig. 3C). Despite greater P. aeruginosa growth under more nutritionally rich conditions, mixed biofilms were still dominated by E. coli throughout the entire biofilm height (Fig. 3D). Similar patterns of colonization were observed in co-cultured biofilms as well (Fig. S3, S4 in File S1).
Effects of varying fluid velocities on colonization of pre-established P. aeruginosa biofilms by E. coli
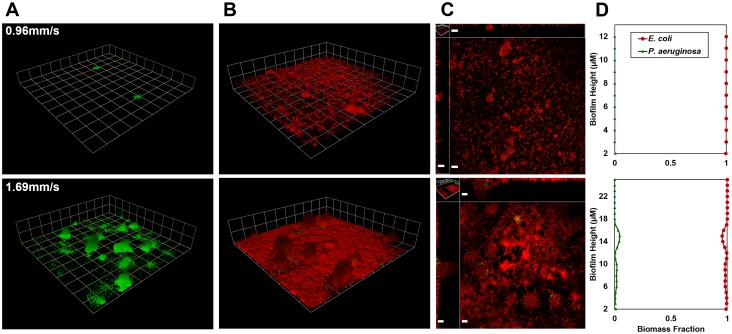
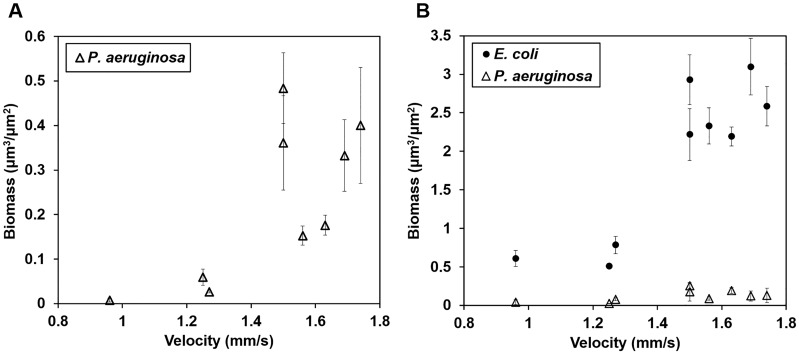
Higher local fluid velocities increased the growth of both P. aeruginosa and E. coli in biofilms (Fig. 5, 6). P. aeruginosa grew to significantly greater biomass in regions of higher velocity (1.50–1.74 mm/s) than in regions of lower velocity (0.96–1.27 mm/s), both on day 3 (P<0.001) and day 6 (P<0.01). E. coli strongly dominated biofilm biomass on day 6, 3 days after its inoculation, in all regions of the flow cell regardless of local fluid velocities (Fig. 5B, 6B). E. coli grew consistently in a lawn-like fashion, overgrowing P. aeruginosa clusters and also colonizing the interior of clusters. E. coli was the dominant species over the full height of the biofilm (Fig. 5D). E. coli also developed greater biomass in regions of higher velocity (P<0.001). Similar trends with velocity were also observed in co-inoculation experiments (Fig. S5, S6 in File S1).
Figure 5. Colonization of P. aeruginosa biofilms by E. coli under a controlled flow gradient in a planar flow cell and in R2A medium.
E. coli appears red and P. aeruginosa appears green. Results are presented for two local fluid velocities, 0.96 mm/s (top row) and 1.69 mm/s (bottom row). Column A) E. coli deposited on 3-day old P. aeruginosa biofilms (grid unit is 23.8 µm). B) Mixed P. aerugiosa and E. coli biofilms on day 6, 3 days after E. coli inoculation (grid unit in B is 23.8 µm). C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 15 µm). D) Distribution of P. aerugiosa and E. coli biomass as function of height, indicating that E. coli was the dominant species throughout the biofilm. E. coli appears red and P. aeruginosa-GFP appears green.
Figure 6. Biomass of P. aeruginosa and E. coli biofilms subjected to different local velocities in R2A medium.
A) Average biomass vs. fluid velocity for mono-cultured, 3-day old P. aeruginosa biofilms. B) Average biomass vs. fluid velocity on day 6 of experiments on colonization of P. aeruginosa by E. coli (after 3 days of mono-culture P. aeruginosa growth plus an additional 3 days after introduction of E. coli).
Colonization of E. coli biofilms by P. aeruginosa
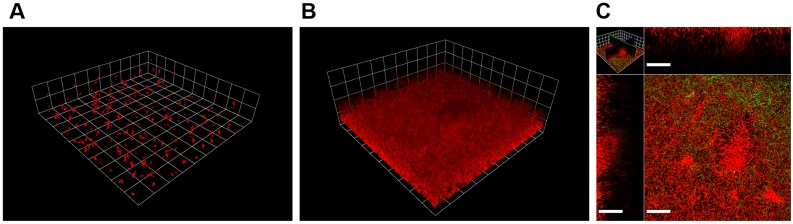
The E. coli strain used in this study formed biofilms poorly in mono-culture. After 3 days of growth, E. coli only formed sparse, discontinuous biofilms (Fig. 7A, D). Similarly limited E. coli growth occurred after 6 days of monospecies culturing (Fig. 7D). However, the introduction of P. aeruginosa caused a dramatic increase in E. coli growth (Fig. 7B, C). On day 6 of the colonization experiments, 3 days after the inoculation of P. aeruginosa on 3-day old E. coli, P. aeruginosa grew into discrete clusters, and E. coli again grew into an extensive lawn and colonized the interior and exterior of P. aeruginosa clusters (Fig. 7C). As a result, E. coli biofilm biomass after 3 days of mono-culture growth plus 3 days after introduction of P. aeruginosa was substantially greater than the E. coli biofilm biomass after 6 days of mono-culture growth (P<0.001) (Fig. 7E, F). These findings demonstrate that the introduction of P. aeruginosa can enhance the growth of E. coli and facilitate the formation of biofilms.
Figure 7. Colonization of E. coli biofilms by P. aeruginosa in R2A medium.
E. coli appears red and P. aeruginosa-GFP appears green or yellow. A) After 3-days of mono-species growth, E. coli formed sparse biofilms composed of small, isolated cell clusters (grid unit is 23.8 µm). B) Mixed P. aeruginosa and E. coli biofilm on day 6, after 3 days of mono-species E. coli growth plus 3 additional days of multi-species growth after inoculation of P. aeruginosa. (grid unit in B is 23.8 µm). Following introduction of P. aeruginosa, E. coli grew prolifically and adopted a configuration similar to that observed in other mixed-species experiments. C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 20 µm). D) Biomass of 3-day- and 6-day-old E. coli mono-cultured biofilms. E) Biomass of co-developed biofilms after 3 days of E. coli growth plus 3 additional days of multi-species growth after inoculation of P. aeruginosa.
Colonization of indole-deficient E. coli biofilms by P. aeruginosa
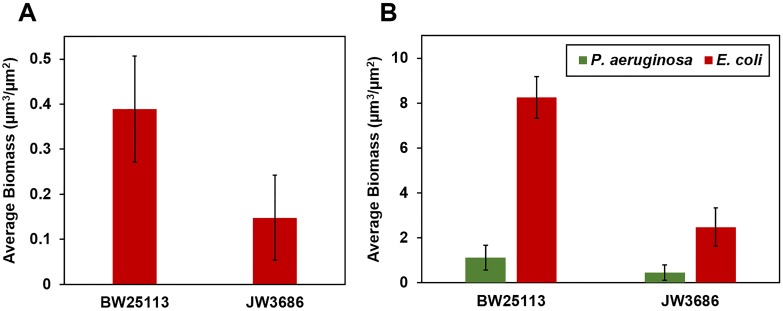
Mono-cultured E. coli JW3686 formed sparse and discontinuous biofilms over a 3-day growth period (Fig. 8A, 9A). After the introduction of P. aeruginosa however, E. coli grew rapidly in the mixed-species biofilms. By day 6 of the experiment, 3 days after P. aeruginosa inoculation, E. coli formed lawn-like biofilms with isolated larger colonies interspersed with P. aeruginosa (Fig. 8B, C, 9B). Control experiments using the indole-positive parent strain, E. coli BW25113, also resulted in very similar mono- and dual-species biofilm growth (Fig. 9A, B, and Fig. S7 in File S1). Mono-cultured E. coli BW25113 formed biofilms poorly over a 3-day growth period (Fig. S7A in File S1), but the introduction of P. aeruginosa triggered a rapid growth response that enabled E. coli to outcompete and colonize the P. aeruginosa biofilms (Fig. 9B, and Fig. S7B, C in File S1).
Figure 8. Colonization of indole-deficient E. coli JW3686 biofilms by P. aeruginosa in R2A medium.
E. coli appears red and P. aeruginosa-GFP appears green or yellow. A) Mono-cultured, 3-day old E. coli formed sparse biofilms composed of small, isolated cell clusters (grid unit is 23.8 µm). B) Mixed P. aeruginosa and E. coli biofilm on day 6, after 3 days of mono-species E. coli growth plus 3 additional days of multi-species growth after inoculation of P. aeruginosa. (grid unit in B is 23.8 µm). Following introduction of P. aeruginosa, E. coli grew extensively and adopted a configuration similar to that observed in other mixed-species experiments. C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 40 µm).
Figure 9. Biomass of E. coli JW3686 and BW25113 biofilms in mono-culture and after colonization by P. aeruginosa in R2A medium.
A) Average biomass of mono-cultured E. coli on day 3. B) Average biomass of E. coli and P. aeruginosa in mixed biofilms on day 6, 3 days after introduction of P. aeruginosa.
Discussion
We investigated the effects of hydrodynamic shear and nutrient concentrations on E. coli colonization of pre-established Pseudomonas aeruginosa biofilms, co-inoculation of E. coli and P. aeruginosa biofilms, and P. aeruginosa colonization of pre-established E. coli biofilms. In R2A medium, P. aeruginosa formed robust biofilms with continuous surface coverage and mound-shaped clusters, as is typical of this organism under a wide range of environmental conditions [26], [27]. Conversely, E. coli only formed sparse biofilms with very small, discontinuous microcolonies in mono-culture. Many E. coli strains are known to be poor biofilm formers [28], [29]. In particular, E. coli K-12 strains lacking the F episome, a plasmid responsible for conjugative pili [26], and strains with low cell motility exhibit poor biofilm growth [28], [30], [31]. Because the E. coli DH5α strain used in this study does not carry the F episome and is not highly motile [28], [32], we expected minimal E. coli biofilm growth. However, the introduction of P. aeruginosa triggered a growth response that enabled extensive lawn-like biofilm formation by E. coli.
Deposited E. coli consistently overgrew pre-established P. aeruginosa biofilms and also colonized the interior of P. aeruginosa clusters under nutrient-limited conditions. We also observed this behavior in co-inoculated experiments, and in experiments where P. aeruginosa was introduced to pre-established E. coli biofilms. The colonization of the cluster interiors by E. coli is likely associated with the process of coordinated cell dispersal in P. aeruginosa biofilms. During these dispersal events, cells actively evacuate the interior of P. aeruginosa clusters through breaches in the cluster wall [33] and leave behind hollow, shell-like structures [33]–[36]. The “hollowing” of bacterial microcolonies from coordinated cell evacuation has been previously documented, though the suspected mechanisms responsible for these events differ from species to species [33], [36]–[39]. The transfer of exogenous solutes into the interior regions of the biofilm clusters has also been suggested to regulate dispersal events [33]. Because we observed void formation in P. aeruginosa biofilms under all experimental conditions tested, this phenomenon appears to generally facilitate E. coli colonization by providing access to the interior of biofilm cell clusters.
P. aeruginosa grew substantially less in mixed culture than in mono-culture under identical nutritional and flow conditions. Moreover, P. aeruginosa biofilm biomass decreased following introduction of E. coli. The reduction of P. aeruginosa biomass in dual-species biofilms suggests strong antagonistic behavior by E. coli towards P. aeruginosa. These results were surprising, as P. aeruginosa is known to be a robust biofilm-forming microorganism in a wide range of aquatic environments, and can produce a variety of antimicrobial agents that adversely affect the growth of other organisms in biofilms [40]–[42]. However, similar overgrowth of P. aeruginosa in dual-species biofilms with Flavobacterium has also been reported by Zhang et al. [43]. By comparison, P. aeruginosa consistently outgrew E. coli in batch experiments, demonstrating that biofilm growth can modify inter-species competition and the relative growth rates of individual species in mixed culture.
Accumulation of extracellular indole has been previously reported to decrease E. coli biofilm formation by hindering cell motility [45], [47]. Indole is an E. coli metabolite that is produced from the amino acid tryptophan by the enzyme tryptophanase [46]. P. aeruginosa degrades indole [24], and can potentially enhance E. coli biofilms by eliminating the growth inhibition caused by extracellular indole [45], [48]. However, under the conditions imposed in this study, we found that indole-producing and indole-deficient strains of E. coli grew similarly in biofilms. The indole-deficient E. coli mutant JW3686 (tnaA) grew poorly in mono-culture, but was able to form extensive lawn-like biofilms after the introduction of P. aeruginosa. We observed the same growth behavior in experiments using the indole-producing strains E. coli BW25113 and E. coli DH5α. The most prominent effects of indole on E. coli and P. aeruginosa have been observed at physiologically relevant concentrations between 500 µM and 1 mM, in media that is rich with exogenous tryptophan; e.g., LB medium [24], [44], [45], [47]. However, the tryptophan content in R2A medium (2.3–2.5 µM) is substantially less than that of LB medium (510 µM) (BD Bionutrients Technical Manual Third Edition). This suggests that indole may play a larger role in the growth behavior of E. coli – P. aeruginosa biofilms only in media that contains greater concentrations of tryptophan. We also show that the triggered E. coli biofilm growth phenomenon after introduction of P. aeruginosa is not limited to E. coli DH5α, but can also occur in other E. coli strains including BW25113 and the indole-deficient JW3686 mutant.
Biofilm growth was more extensive under faster local velocities and in more nutrient-rich R2A media (4x and 8x concentrations). In regions of faster flow, increased biofilm growth occurred for both P. aeruginosa and E. coli. This behavior has been previously attributed to more rapid delivery of substrates and nutrients to biofilms under higher local velocities [20]. In more concentrated R2A media, increased growth occurred primarily in P. aeruginosa biofilms, but not E. coli. This suggests that, unlike P. aeruginosa, the growth of E. coli was not constrained by nutrient availability in the R2A medium. However, E. coli remained the dominant species in co-culture under all velocities and media tested regardless of P. aeruginosa biofilm growth and morphology. E. coli consistently colonized outer regions of P. aeruginosa clusters and extensively colonized the larger and taller clusters. The results demonstrate that E. coli can outcompete P. aeruginosa in dual-species biofilms under nutrient-depleted conditions.
In conclusion, we demonstrated that E. coli is capable of colonizing and overgrowing P. aeruginosa biofilms under both nutrient-limited and enriched conditions, and over a range of fluid velocities. We further showed that the introduction of P. aeruginosa triggered a dramatic growth response in E. coli that enabled extensive biofilm formation. By comparison, P. aeruginosa outgrew E. coli in co-cultured planktonic batch experiments, illustrating that biofilm growth substantially modifies inter-species competition and growth rates. These results show that interactions with pre-established bacterial biofilms such as P. aeruginosa can greatly increase the ability of E. coli to persist and grow in aquatic environments.
Supporting Information
Figures S1–S7. Figure S1. Mean population density of E. coli and P. aeruginosa grown in R2A medium over a 3-day period at 37°C. Both populations remained steady for the duration of the experiment, with P. aeruginosa exhibiting a higher population concentration. Figure S2. Mean population density of E. coli and P. aeruginosa grown in R2A medium over a 3-day period at 24°C. Both populations remained relatively steady for the duration of the experiment, with P. aeruginosa exhibiting a higher population concentration. Figure S3. Co-development of 3-day old E. coli and P. aeruginosa biofilms under varying R2A medium concentrations in a microfluidic flow cell. Column: A) 3-day old co-inoculated P. aeruginosa – E. coli biofilms with under 4x and 8x R2A concentrations (grid unit is 23.8 µm). B) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel A (scale bar = 40 µm). C) Biomass fraction vs. biofilm height graph. Biofilms were counter-stained by SYTO 62. E. coli appears red and P. aeruginosa-GFP appears green. Figure S4. Average biofilm biomass vs. R2A concentration for co-development of 3-day old E. coli and P. aeruginosa biofilms. Figure S5. Co-development of 3-day old E. coli and P. aeruginosa biofilms under a controlled flow gradient in a planar flow cell and in R2A medium. Results are presented for two local fluid velocities, 0.96 mm/s (top row) and 1.69 mm/s (bottom row). Column: A) 3-day old co-inoculated P. aeruginosa – E. coli biofilms (grid unit is 23.8 µm). B) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel A (scale bar = 20 µm). C) Distribution of P. aerugiosa and E. coli biomass as function of height, indicating that E. coli was the dominant species throughout a majority of the biofilm. E. coli appears red and P. aeruginosa-GFP appears green. Figure S6. Biofilm biomass on day 3 for the co-development of P. aeruginosa and E. coli biofilms subjected to different local velocities in R2A medium. Figure S7. Colonization of E. coli BW25113 biofilms by P. aeruginosa in R2A medium. E. coli appears red and P. aeruginosa-GFP appears green or yellow. A) After 3-days of mono-species growth, E. coli formed sparse biofilms composed of small, isolated cell clusters (grid unit is 23.8 µm). B) Mixed P. aeruginosa and E. coli biofilm on day 6, after 3 days of mono-species E. coli growth plus 3 additional days of multi-species growth after inoculation of P. aeruginosa. (grid unit in B is 23.8 µm). Following introduction of P. aeruginosa, E. coli grew prolifically and adopted a configuration similar to that observed in other mixed-species experiments. C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 40 µm).
(DOCX)
Acknowledgments
The authors would like to thank Dr. Bradley Borlee and Matthew Parsek (University of Washington) for providing the E. coli DH5α test strain. The authors would also like to thank the Coli Genetic Stock Center (Yale University) for providing the E. coli JW3686 and BW25113 strains. Imaging work was performed at the Northwestern University Biological Imaging Facility generously supported by the Northwestern University Office for Research.
Funding Statement
This work was supported by the Water Research Foundation Project 4259 (website: http://www.waterrf.org/Pages/Index.aspx). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parsek MR, Singh PK (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57: 677–701 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- 2. Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 3. Szewzyk U, Szewzyk R, Manz W, Schleifer K (2000) Microbiological Safety of Drinking Water. Annu Rev Microbiol 54: 81–127. [DOI] [PubMed] [Google Scholar]
- 4. Berry D, Xi C, Raskin L (2006) Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol 17: 297–302 10.1016/j.copbio.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 5. Park S, Mackay W, Reid D (2001) Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res 35: 1624–1626 10.1016/S0043-1354(00)00582-0 [DOI] [PubMed] [Google Scholar]
- 6. Williams MM, Braun-howland EB (2003) Growth of Escherichia coli in Model Distribution System Biofilms Exposed to Hypochlorous Acid or Monochloramine. Appl Environ Microbiol 69: 5463–5471 10.1128/AEM.69.9.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banning N, Toze S, Mee B (2003) Persistence of biofilm-associated Escherichia coli and Pseudomonas aeruginosa in groundwater and treated effluent in a laboratory model system. Microbiology 149: 47–55 10.1099/mic.0.25938-0 [DOI] [PubMed] [Google Scholar]
- 8. Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, et al. (2006) Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72: 3916–3923 10.1128/AEM.03022-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murga R, Forster TS, Brown E, Pruckler JM, Fields BS, et al. (2001) Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147: 3121–3126. [DOI] [PubMed] [Google Scholar]
- 10. Lehtola MJ, Torvinen E, Kusnetsov J, Pitkänen T, Maunula L, et al. (2007) Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl Environ Microbiol 73: 2854–2859 10.1128/AEM.02916-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stout JE, Yu VL, Best M (1985) Ecology of Legionella pneumophila within water distribution systems. Environ Microbiol 49: 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ica T, Caner V, Istanbullu O, Nguyen HD, Ahmed B, et al. (2012) Characterization of mono- and mixed-culture Campylobacter jejuni biofilms. Appl Environ Microbiol 78: 1033–1038 10.1128/AEM.07364-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leclerc H, Mossel DA, Edberg SC, Struijk CB (2001) Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol 55: 201–234 10.1146/annurev.micro.55.1.201 [DOI] [PubMed] [Google Scholar]
- 14.USEPA (1986) Ambient Water Quality Criteria for Bacteria - 1986. Washington DC.
- 15. Ishii S, Sadowsky MJ (2008) Escherichia coli in the Environment: Implications for Water Quality and Human Health. Microbes Environ 23: 101–108 10.1264/jsme2.23.101 [DOI] [PubMed] [Google Scholar]
- 16. LeChevallier MW, Babcock TM, Lee RG (1987) Examination and characterization of distribution system biofilms. Appl Environ Microbiol 53: 2714–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LeChevallier MW, Welch NJ, Smith DB (1996) Full-scale studies of factors related to coliform regrowth in drinking water. Appl Environ Microbiol 62: 2201–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fass S, Dincher ML, Reasoner DJ, Gatel D, Block J (1996) Fate of Escherichia coli experimentally injected in a drinking water distribution pilot system. Water Res 30: 2215–2221. [Google Scholar]
- 19. Juhna T, Birzniece D, Rubulis J (2007) Effect of Phosphorus on survival of Escherichia coli in drinking water biofilms. Appl Environ Microbiol 73: 3755–3758 10.1128/AEM.00313-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang W, Sileika TS, Chen C, Liu Y, Lee J, et al. (2011) A novel planar flow cell for studies of biofilm heterogeneity and flow-biofilm interactions. Biotechnol Bioeng 108: 2571–2582 10.1002/bit.23234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song JL, Au KH, Huynh KT, Packman AI (2014) Biofilm responses to smooth flow fields and chemical gradients in novel microfluidic flow cells. Biotechnol Bioeng 111: 597–607 10.1002/bit.25107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 1–11 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chu W, Zere TR, Weber MM, Wood TK, Whiteley M, et al. (2012) Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl Environ Microbiol 78: 411–419 10.1128/AEM.06396-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heydorn A, Nielsen A, Hentzer M, Sternberg C, Givskov M, et al. (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407. [DOI] [PubMed] [Google Scholar]
- 26. Lyczak JB, Cannon CL, Pier GB (2000) Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2: 1051–1060. [DOI] [PubMed] [Google Scholar]
- 27. Donlan RM, Costerton JW (2002) Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin Microbiol Rev 15: 167–193 10.1128/CMR.15.2.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood TK, González Barrios AF, Herzberg M, Lee J (2006) Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72: 361–367 10.1007/s00253-005-0263-8 [DOI] [PubMed] [Google Scholar]
- 29. Beloin C, Roux A, Ghigo J (2008) Escherichia coli biofilms. Curr Top Microbiol Immunol 322: 249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30: 285–293. [DOI] [PubMed] [Google Scholar]
- 31. Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S (2003) Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48: 933–946. [DOI] [PubMed] [Google Scholar]
- 32. Zhuang W, Yuan D, Li J-R, Luo Z, Zhou H-C, et al. (2012) Highly potent bactericidal activity of porous metal-organic frameworks. Adv Healthc Mater 1: 225–238 10.1002/adhm.201100043 [DOI] [PubMed] [Google Scholar]
- 33. Purevdorj-Gage B (2005) Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151: 1569–1576 10.1099/mic.0.27536-0 [DOI] [PubMed] [Google Scholar]
- 34. Hunt SM, Werner EM, Huang B, Hamilton A, Stewart PS, et al. (2004) Hypothesis for the Role of Nutrient Starvation in Biofilm Detachment Hypothesis for the Role of Nutrient Starvation in Biofilm Detachment. Appl Environ Microbiol 70: 7418–7425 10.1128/AEM.70.12.7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barraud N, Hassett DJ, Hwang S-H, Rice S a, Kjelleberg S, et al. (2006) Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188: 7344–7353 10.1128/JB.00779-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sauer K, Camper AK, Ehrlich GD, William J, Davies DG, et al. (2002) Pseudomonas aeruginosa Displays Multiple Phenotypes during Development as a Biofilm. J Bacteriol 184: 1140–1154 10.1128/JB.184.4.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, et al. (2000) Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol 182: 6482–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaplan JB, Ragunath C, Ramasubbu N, Fine DH (2003) Detachment of Actinobacillus actinomycetemcomitans Biofilm Cells by an Endogenous β-Hexosaminidase Activity. J Bacteriol 185: 4693–4698 10.1128/JB.185.16.4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaplan JB, Meyenhofer MF, Fine DH (2003) Biofilm Growth and Detachment of Actinobacillus actinomycetemcomitans Biofilm Growth and Detachment of Actinobacillus actinomycetemcomitans. J Bacteriol 185: 1399–1404 10.1128/JB.185.4.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuhard J, Nygaard P (1987) Purines and pyrimidines. Escherichia coli and Salmonella: cellular and molecular biology. Washington DC: American Society for Microbiology. 445–473.
- 41. Heo Y-J, Chung I-Y, Choi KB, Cho Y-H (2007) R-type pyocin is required for competitive growth advantage between Pseudomonas aeruginosa strains. J Microbiol Biotechnol 17: 180–185. [PubMed] [Google Scholar]
- 42. Waite RD, Curtis MA (2009) Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J Bacteriol 191: 1349–1354 10.1128/JB.01458-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei Zhang, Tadas Sileika AIP (2013) Effects of fluid flow conditions on interactions between species in biofilms. FEMS Microbiol Ecol 84: 344–354 10.1111/1574-6941.12066.Effects [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee J, Attila C, Cirillo SLG, Cirillo JD, Wood TK (2009) Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol 2: 75–90 10.1111/j.1751-7915.2008.00061.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J, Jayaraman A, Wood TK (2007) Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 15: 1–15 10.1186/1471-2180-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isaacs H, Chao D, Yanofsky C, Saier MH (1994) Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli. Microbiology: 2125–2134. [DOI] [PubMed]
- 47. Bansal T, Englert D, Lee J, Hegde M, Wood TK, et al. (2007) Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75: 4597–4607 10.1128/IAI.00630-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Domka J, Lee J, Wood TK (2006) YliH (BssR) and YceP (BssS) Regulate Escherichia coli K-12 Biofilm Formation by Influencing Cell Signaling. Appl Environ Microbiol 72: 2449–2459 10.1128/AEM.72.4.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S7. Figure S1. Mean population density of E. coli and P. aeruginosa grown in R2A medium over a 3-day period at 37°C. Both populations remained steady for the duration of the experiment, with P. aeruginosa exhibiting a higher population concentration. Figure S2. Mean population density of E. coli and P. aeruginosa grown in R2A medium over a 3-day period at 24°C. Both populations remained relatively steady for the duration of the experiment, with P. aeruginosa exhibiting a higher population concentration. Figure S3. Co-development of 3-day old E. coli and P. aeruginosa biofilms under varying R2A medium concentrations in a microfluidic flow cell. Column: A) 3-day old co-inoculated P. aeruginosa – E. coli biofilms with under 4x and 8x R2A concentrations (grid unit is 23.8 µm). B) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel A (scale bar = 40 µm). C) Biomass fraction vs. biofilm height graph. Biofilms were counter-stained by SYTO 62. E. coli appears red and P. aeruginosa-GFP appears green. Figure S4. Average biofilm biomass vs. R2A concentration for co-development of 3-day old E. coli and P. aeruginosa biofilms. Figure S5. Co-development of 3-day old E. coli and P. aeruginosa biofilms under a controlled flow gradient in a planar flow cell and in R2A medium. Results are presented for two local fluid velocities, 0.96 mm/s (top row) and 1.69 mm/s (bottom row). Column: A) 3-day old co-inoculated P. aeruginosa – E. coli biofilms (grid unit is 23.8 µm). B) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel A (scale bar = 20 µm). C) Distribution of P. aerugiosa and E. coli biomass as function of height, indicating that E. coli was the dominant species throughout a majority of the biofilm. E. coli appears red and P. aeruginosa-GFP appears green. Figure S6. Biofilm biomass on day 3 for the co-development of P. aeruginosa and E. coli biofilms subjected to different local velocities in R2A medium. Figure S7. Colonization of E. coli BW25113 biofilms by P. aeruginosa in R2A medium. E. coli appears red and P. aeruginosa-GFP appears green or yellow. A) After 3-days of mono-species growth, E. coli formed sparse biofilms composed of small, isolated cell clusters (grid unit is 23.8 µm). B) Mixed P. aeruginosa and E. coli biofilm on day 6, after 3 days of mono-species E. coli growth plus 3 additional days of multi-species growth after inoculation of P. aeruginosa. (grid unit in B is 23.8 µm). Following introduction of P. aeruginosa, E. coli grew prolifically and adopted a configuration similar to that observed in other mixed-species experiments. C) Horizontal section near the base of the biofilm and vertical sections of the biofilm shown in panel B (scale bar = 40 µm).
(DOCX)