Abstract
The hypertrophic growth of cardiac myocytes is a highly dynamic process that underlies physiological and pathological adaptation of the heart. Accordingly, a better understanding of the molecular underpinnings of cardiac myocyte hypertrophy is required in order to fully appreciate the causes and functional consequences of the changes in the size of the healthy and diseased heart. Hypertrophy is driven by increases in cardiacmyocyte protein, which must be balanced by cellular ability to maintain protein quality in order to avoid maladaptive accumulation of toxic misfolded proteins. Recent studies have shown that the endoplasmic reticulum (ER), which, in cardiac myocytes, comprises the sarco/endoplasmic reticulum (SR/ER), is the site of most protein synthesis. Thus, the protein quality control machinery located at the SR/ER is likely to be an important determinant of whether the heart responds adaptively to hypertrophic growth stimuli. The SR/ER-transmembrane protein, ATF6, serves a critical protein quality control function as a first responder to the accumulation of potentially toxic, misfolded proteins. Misfolded proteins transform ATF6 into a transcription factor that regulates a gene program that is partly responsible for enhancing protein quality control. Two ATF6-inducible genes that have been studied in the heart and shown to be adaptive are RCAN1 and Derl3, which encode proteins that decrease protein-folding demand, and enhance degradation of misfolded proteins, respectively. Thus, the ATF6-regulated SR/ER protein quality control system is important for maintaining protein quality during growth, making ATF6, and other components of the system, potentially attractive targets for the therapeutic management pathological cardiac hypertrophy. This article is part of a Special Issue entitled “Protein Quality Control, the Ubiquitin Proteasome System, and Autophagy”.
Keywords: ATF6, Cardiac hypertrophy, Cardiac myocyte, Sarcoplasmic reticulum, Endoplasmic reticulum, Protein quality control
1. Introduction
The heart plays a critical role as a pump, propelling blood to all parts of the body in precisely the quantities necessary to match the needs of the organism. Maintaining efficient cardiac function under physiological, as well as pathological conditions, is intimately linked to the heart’s ability to change size, which, in the adult, is driven mainly by cardiac myocyte atrophy or hypertrophy [1]. For example many cardiac pathologies are associated with hypertrophic growth of cardiac myocytes. Although potentially compensatory at first, pathological hypertrophy often leads to an eventual failure of the heart to perform its function as a pump, and is therefore considered maladaptive [2]. Since pathological cardiac hypertrophy often precedes heart failure, a potentially powerful therapeutic approach would be to intervene with the pathologic hypertrophic growth process, which has been shown to avert the life-threatening heart failure [3].
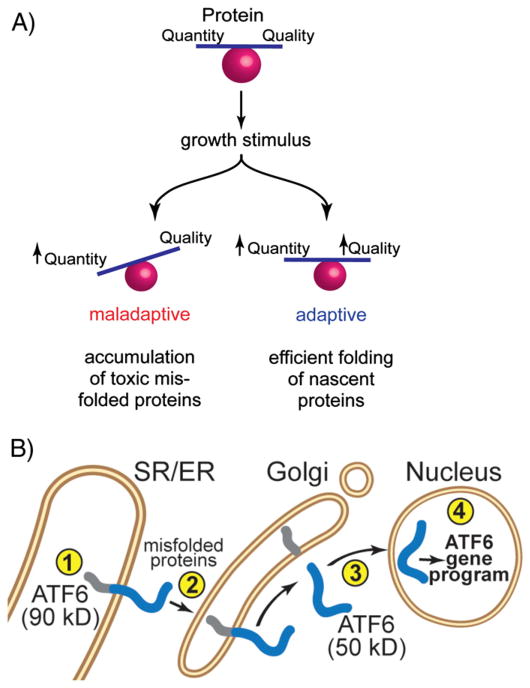
The critical nature of cardiac myocyte hypertrophy under both physiological and pathological conditions has driven numerous studies aimed at gaining a better understanding of the molecular mechanisms by which cardiac myocytes exhibit such dynamic growth responses [2,4]. In part, and not surprisingly, such studies have demonstrated that increases in cardiac myocyte size require increases in the quantity of cardiac myocyte protein [5–9]. Moreover, in order to avert cell death, increases in cardiac myocyte growth are associated with increases in protein quantity that must be balanced by cellular ability to supervise and manage the quality of new protein [10] (Fig. 1A), which requires correct protein folding, as well as the degradation of potentially toxic terminally misfolded proteins [11].
Fig. 1.
Balancing protein quantity and quality at the sarco/endoplasmic reticulum of cardiac myocytes: Panel A — Balancing protein quantity and quality is required for adaptive responses to growth stimuli: Upon a growth stimulus, if protein synthesis increases without a coordinate increase in the ability to monitor and control protein quality (left), the response is maladaptive due to the accumulation of toxic misfolded proteins. However, when increases in protein synthesis are balanced with the cellular ability to control protein quality (right), the response is adaptive. Panel B — ATF6 is a component of the SR/ER protein quality control network: ATF6 resides in the SR/ER of cardiac myocytes (➀). Accumulation of misfolded proteins in the ischemic or hypertrophic heart results in the translocation of ATF6 to the Golgi apparatus (➁), where it is cleaved by transmembrane proteases (➂), which liberates a cytosolic, 50 kD fragment. The 50 kD fragment of ATF6 has a nuclear localization sequence that facilitates the translocation of ATF6 to the nucleus, where it binds to specific elements in the regulatory regions of ATF6-responsive genes and regulates the ATF6 gene program (➃).
2. The SR/ER as a major site of protein synthesis
It has long been believed that about 2/3 of proteins are translated on cytosolic ribosomes, while the remaining 1/3, comprising secreted and membrane proteins, are translated on SR/ER-associated ribosomes [12–14]. In terms of the protein synthesis that underlies cardiacmyocyte hypertrophy, this concept has resulted in emphasis being focused mainly on the protein quantity and quality control machinery located in the cytosol [15–19]. However, recent paradigm-shifting studies have shown that, in addition to secreted and membrane proteins, ribosomes associated with the ER are also responsible for translation of many cytosolic and nuclear proteins [20]. For example, while not studied in cardiac myocytes, in model cell lines, as much as 96% of the transcripts encoding cytosolic proteins are associated with, and translated by ER-bound ribosomes [21]. Furthermore, the rate of protein synthesis by ER-bound ribosomes exceeds that of cytosolic ribosomes by up to 4-fold, indicating that the ER is a privileged site for protein synthesis [22]. Thus, ER-bound ribosomes play a global role in cellular protein synthesis. Consequently, the protein quality control machinery associated with the SR/ER is likely to play an important part in determining whether hypertrophic cardiac myocyte growth is adaptive or maladaptive (Fig. 1A) [23].
The SR/ER in cardiacmyocytes has an elaborate protein quality control system, many of the details of which were elucidated during studies of secreted and membrane protein synthesis, ER stress, and the unfolded protein response, reviewed in [11,24]. Most secreted and membrane proteins are co-translationally translocated across the SR/ER membrane, after which they are folded and further modified [25–27]. Properly folded proteins are then transported to the Golgi, where they are sorted to their final destinations; however, misfolded proteins are not sorted, but are instead degraded before they can enter the Golgi apparatus [28–30]. Since misfolded proteins can be toxic, their degradation is considered adaptive. Thus, in response to growth stimuli, increased protein synthesis at the SR/ER must be balanced by the ability to manage protein quality in order to achieve adaptive growth [31] (Fig. 1A).
3. ATF6 and protein quality control in the SR/ER
An elaborate regulatory network associated with the SR/ER is responsible for managing protein quality at this location; essential elements of the network are three trans-ER membrane proteins, activating transcription factor 6 (ATF6), protein kinase R [PKR]-like ER kinase (PERK), and inositol requiring enzyme (IRE)-1, which serve as the proximal effectors of ER stress [32]. The three proximal effectors of ER stress are activated with different strengths and time courses by various ER stresses [33]. In response, ATF6, IRE-1, and PERK regulate the expression of common, and unique genes, the products of which can be oriented toward ER stress relief, or, if the stress cannot be resolved, toward cell death. Although all of the effectors can serve adaptive, as well as maladaptive roles, among the three, ATF6 exerts mainly adaptive functions in most cell types examined to date [34,32, 35,36]. The 90 kD form of ATF6 is a single-pass SR/ER-transmembrane protein (Fig. 1B, ➀) that monitors the folding status of proteins made in the SR/ER. If protein synthesis outpaces protein-folding capacity, ATF6 senses the accumulation of misfolded proteins, a situation sometimes called ER stress, then translocates to the Golgi (Fig. 1B, ➁), where it is cleaved by the Golgi-localized proteases, SP1 and SP2 [37]. The resulting 50 kD cytosolic fragment translocates to the nucleus (Fig. 1B, ➂), binds to elements in ATF6-responsive genes, and regulates their expression (Fig. 1B, ➃). Thus, activated ATF6 is an important element of the unfolded protein response.
The 50 kD form of ATF6 is, itself, subject to regulation by proteasome-dependent protein quality control, most likely, in the nucleus. Prior to its activation, ATF6 is a relatively stable protein that is anchored in the SR/ER [38]. However, after its activation, and translocation to the nucleus, the 50 kD form of ATF6 is rapidly degraded. In fact, the rapid degradation of activated ATF6, which can be slowed by proteasome inhibitors, is coupled to the engagement of ATF6 in the transcriptional process [39]. Mapping studies have revealed an 8-amino acid region residing in the transcriptional activation domain of ATF6 that is responsible for ATF6-mediated gene induction, as well as its rapid degradation [40]. Thus, the ability of ATF6 to induce genes is tightly temporally regulated by virtue of an obligate coupling of its ability to activate transcription with its rapid degradation.
To examine the function of ATF6 in the heart, a transgenic (TG) mouse line in which ATF6 can be activated conditionally in cardiac myocytes, was generated [41]. Using this mouse model it was shown that, upon ATF6 activation, many prototypical ER stress response genes were induced, even though the half-life of transgene-encoded ATF6 in the mouse heart decreased, consistent with the coupling of ATF6 transcriptional induction with degradation. Upon activation, ATF6 protected the heart from ischemic damage, in vivo. ATF6 has also been shown to be adaptive in a mouse model of pressure overload hypertrophy [42]. Transcript profiling showed that among the numerous genes induced by ATF6 in the heart were some that were not previously known to be ATF6-inducible, but which could contribute to the adaptive effects of ATF6 in the heart [43]. One such gene encodes regulator of calcineurin 1 (RCAN1), which is a moderator of nuclear factor of activated T cells (NFAT) activity in most eukaryotic cells [44].
4. ATF6 as a regulator of calcineurin/NFAT signaling in cardiac myocytes
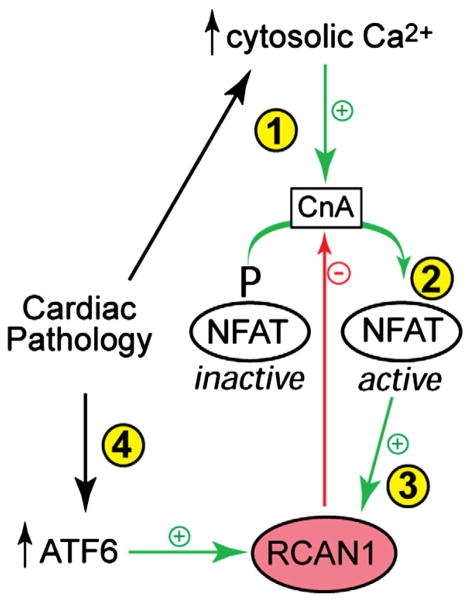
Cardiac pathologies, including cardiac hypertrophy and myocardial ischemia, can increase cytosolic calcium in cardiac myocytes, which activates the cytosolic phosphatase, calcineurin A (CnA) (Fig. 2, ➀). CnA then dephosphorylates NFAT (Fig. 2, ➁), which translocates to the nucleus and activates genes, some of which contribute to hypertrophic growth [45]. NFAT also increases the expression of RCAN1, which can bind to, and inhibit CnA (Fig. 2, ➂). Thus, RACN1 can decrease NFAT activity and moderate the hypertrophic growth of cardiac myocytes. Therefore, as an NFAT-inducible feedback inhibitor of NFAT-mediated gene expression and cardiac myocyte growth, RCAN1 participates in an autoregulatory signaling circuit that contributes to determining the extent of cardiac hypertrophy [46]. Consistent with this concept was the finding that overexpression of RCAN1 in transgenic mouse hearts inhibited pressure overload-induced cardiac hypertrophy by inhibiting CnA/NFAT signaling [47]. Moreover, when subjected to myocardial infarction, the hearts of RCAN1 transgenic mice exhibited less cardiac hypertrophy and reduced heart failure, consistent with growth modulating, adaptive roles for RCAN1 [48]. Cardiac pathology can also activate ATF6 [42,49] (Fig. 2, ➂). The finding that RCAN1 was ATF6-inducible in the mouse heart suggested a mechanism by which ATF6 and the unfolded protein response, which are both activated in the pathologic heart, might contribute to reducing the demands on the protein quality machinery to ensure balanced adaptive responses to growth stimuli. In this way, by inducing RCAN1, ATF6 can reduce the amount of protein that must be properly folded. Thus, via RCAN1 induction, ATF6 indirectly enhances protein quality by decreasing protein-folding demands [43].
Fig. 2.
RCAN1 is an ATF6-inducible gene in the heart: Cardiac pathologies lead to increases in cytosolic calcium in cardiac myocytes, which activates the calcineurin A (CnA) (➀), a phosphatase that removes a critical phosphate from inactive NFAT. Dephosphorylated NFAT translocates to the nucleus (➁), where it controls genes, some of which are responsible for pathological hypertrophic cardiacmyocyte growth, which can lead to heart failure. NFAT also induces RCAN1 (➂), which inhibits CnA and, in so doing, inhibits the dephosphorylation of NFAT, thus shifting NFAT more toward inactivation. ATF6, which is activated during certain pathological conditions in the heart, can also transcriptionally induce RCAN1 (➃), which provides a mechanism by which SR/ER protein quality control integrates functionally with CnA/NFAT-mediated hypertrophic signaling.
5. ATF6 as a regulator of ER associated protein degradation (ERAD) in cardiac myocytes
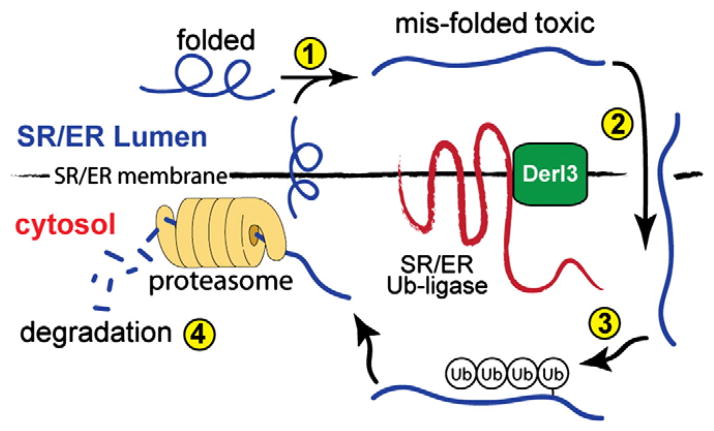
Another gene that is induced by ATF6 in the heart is degradation in endoplasmic reticulum protein 3, or derlin-3 (Derl3), a member of a family comprised of Derl1, Derl2, and Derl3. The derlin family has been extensively studied in yeast, where the homologue of human Derl1, Der1p, was found to be involved in protein degradation at the ER [50], a process known as ER associated degradation, or ERAD, reviewed in [51,52]. ERAD begins with translocation of misfolded proteins from the SR/ER membrane, or lumen (Fig. 3, ➀ & ➁) to the cytosol (Fig. 3, ➁), where they are ubiquitinated on the cytosolic side of the SR/ER [30] by SR/ER-transmembrane ubiquitin (Ub) ligases (Fig. 3, ➂). The resulting poly-ubiquitinated proteins are then degraded by proteasomes located on the cytosolic face of the SR/ER (Fig. 3, ➃). ERAD reduces toxicity by degrading terminally misfolded proteins and is, therefore, considered adaptive [29].
Fig. 3.
ER associated degradation: Misfolded luminal and membrane proteins in the SR/ER (➀) are retrotranslocated from the SR/ER into the cytosol (➁), where they are ubiquitinated by one of several SR/ER-transmembrane ubiquitin ligases (➂). These ligases work in conjunction with Derlin-3 (Derl3), which facilitates retrotranslocation, as well as misfolded protein substrate recognition by the ubiquitin ligases. Following ubiquitination, misfolded proteins are degraded by proteasomes associated with the cytosolic face of the SR/ER (➃).
Although the exact function of Derl3 in ERAD is not known, it is believed that derlin family members facilitate the retro-translocation of misfolded proteins from the ER lumen to the cytosolic face of the ER, and that they work in concert with SR/ER-transmembrane ubiquitin ligases to recognize misfolded protein [53,54]. Among the Derlin family members, only Derl3 was induced by ATF6 in cardiac myocytes, in vivo [55]. In fact, in the mouse heart, ATF6 induced Derl3 expression by more than 400-fold over control. Moreover, Derl3 was the only Derlin family member that was induced in the infarcted mouse hearts, and in cultured cardiacmyocytes subjected to simulated ischemia. Thus, it was possible that Derl3 overexpression might enhance ERAD, and in so doing, might serve an adaptive role in the ischemic heart by decreasing the accumulation of potentially toxic misfolded proteins, which is known to occur during ischemia. Indeed, Derl3 overexpression decreased cardiac myocyte death in response to simulated ischemia, while inhibiting endogenous Derl3 augmented cardiac myocyte death. Moreover, Derl3 was shown to enhance ERAD in cardiac myocytes, which contributed to the observed adaptive protection from cell death upon ischemia/reperfusion. Thus, overexpression of Derl3 enhanced ERAD in cardiac myocytes, which contributed to protection from cell death. This adaptive response was, at least partly, due to the efficient removal of potentially toxic misfolded proteins that accumulate in the ischemic and hypertrophic heart, which would contribute to balancing protein quantity and quality in cardiac myocytes.
6. Conclusions
The importance of protein synthesis in the SR/ER of cardiacmyocytes is just beginning to be understood. The proteins synthesized in the SR/ER are likely to constitute a major proportion of proteins that comprise cardiac myocytes. Dynamic changes in cardiac myocyte protein require responses in protein synthesis that are balanced with protein quality control machinery. The protein quality control machinery at the ER, which, in large part, is managed by ATF6, is responsible for regulating the expression of genes that can help balance protein quality with protein quantity. Thus, since SR/ER protein synthesis plays a major role in the plasticity with which cardiac myocytes can respond to environmental cues, the SR/ER protein quality control machinery must also play a major role in ensuring a balance between the amount of protein being synthesis, and the ability of cardiac myocytes to fold newly synthesized proteins. Therefore, in the heart, genes under the control of ATF6, such as those that encode RCAN1 and Derlin-3, are likely to be involved in determining whether cardiac myocytes survive responses to growth stimuli. Accordingly, since the SR/ER protein quality control system is likely to be a convergence point for signaling pathways that regulate protein quantity and quality in cardiac myocytes, it is also a potentially fertile ground for the development of new strategies aimed at managing cardiac diseases that hinge on the regulation of protein quantity and quality, such as pathological cardiac hypertrophy and heart failure.
Acknowledgments
Research in the Glembotski lab is supported by National Institutes of Health, grants HL-075573, HL-085577, and HL-104535.
The author wishes to thank Dr. Christopher Nicchitta, from the Duke University School of Medicine, for enlightening discussions concerning the major role played by SR/ER-bound ribosomes in protein synthesis. The author also thanks Dr. Shirin Doroudgar, from the San Diego State University Heart Institute, for insightful and thought provoking discussions about functions for the SR/ER protein quality control system in the heart, as well as critical reading of the manuscript.
Footnotes
Disclosures
None.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123:37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–9. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 4.Maillet M, van Berlo JH, Molkentin JD. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan HE, Beinlich CJ. Contributions of increased efficiency and capacity of protein synthesis to rapid cardiac growth. Mol Cell Biochem. 1997;176:145–51. [PubMed] [Google Scholar]
- 6.Razeghi P, Baskin KK, Sharma S, Young ME, Stepkowski S, Essop MF, et al. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun. 2006;342:361–4. doi: 10.1016/j.bbrc.2006.01.163. [DOI] [PubMed] [Google Scholar]
- 7.Taegtmeyer H, Harinstein ME, Gheorghiade M. More than bricks and mortar: comments on protein and amino acid metabolism in the heart. Am J Cardiol. 2008;101:3E–7E. doi: 10.1016/j.amjcard.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 8.Zak R, Martin AF, Reddy MK, Rabinowitz M. Control of protein balance in hypertrophied cardiac muscle. Circ Res. 1976;38:I145–50. [PubMed] [Google Scholar]
- 9.Simpson P, McGrath A, Savion S. Myocyte hypertrophy in neonatal rat heart cultures and its regulation by serum and by catecholamines. Circ Res. 1982;51:787–801. doi: 10.1161/01.res.51.6.787. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. Am J Physiol Heart Circ Physiol. 2011;301:H2207–19. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–84. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 12.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–58. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 13.Kelly RB. Pathways of protein secretion in eukaryotes. Science. 1985;230:25–32. doi: 10.1126/science.2994224. [DOI] [PubMed] [Google Scholar]
- 14.Simon SM, Blobel G. Mechanisms of translocation of proteins across membranes. Subcell Biochem. 1993;21:1–15. doi: 10.1007/978-1-4615-2912-5_1. [DOI] [PubMed] [Google Scholar]
- 15.Pagan J, Seto T, Pagano M, Cittadini A. Role of the ubiquitin proteasome system in the heart. Circ Res. 2013;112:1046–58. doi: 10.1161/CIRCRESAHA.112.300521. [DOI] [PubMed] [Google Scholar]
- 16.Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol. 2013;304:H1283–93. doi: 10.1152/ajpheart.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. J Mol Cell Cardiol. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlossarek S, Carrier L. The ubiquitin–proteasome system in cardiomyopathies. Curr Opin Cardiol. 2011;26:190–5. doi: 10.1097/HCO.0b013e32834598fe. [DOI] [PubMed] [Google Scholar]
- 19.Glembotski CC. Clarifying the cardiac proteasome paradox: protein quality control. Circ Res. 2012;111:509–12. doi: 10.1161/CIRCRESAHA.112.275917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–27. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–37. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell. 2008;19:623–32. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glembotski CC. Roles for the sarco-/endoplasmic reticulum in cardiac myocyte contraction, protein synthesis, and protein quality control. Physiology (Bethesda) 2012;27:343–50. doi: 10.1152/physiol.00034.2012. [DOI] [PubMed] [Google Scholar]
- 24.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44:453–9. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 1808;2011:912–24. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Brodsky JL. Translocation of proteins across the endoplasmic reticulum membrane. Int Rev Cytol. 1998;178:277–328. doi: 10.1016/s0074-7696(08)62139-7. [DOI] [PubMed] [Google Scholar]
- 27.Doroudgar S, Glembotski CC. The cardiokine story unfolds: ischemic stress-induced protein secretion in the heart. Trends Mol Med. 2011;17:207–14. doi: 10.1016/j.molmed.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci U S A. 1996;93:13797–801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537–76. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Needham PG, Brodsky JL. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim Biophys Acta. 2013;1833(11):2447–57. doi: 10.1016/j.bbamcr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 32.Doroudgar S, Glembotski CC. New concepts of endoplasmic reticulum function in the heart: programmed to conserve. J Mol Cell Cardiol. 2012;55:85–91. doi: 10.1016/j.yjmcc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DuRose JB, Tam AB, Niwa M. Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell. 2006;17:3095–107. doi: 10.1091/mbc.E06-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K. Divest yourself of a preconceived idea: transcription factor ATF6 is not a soluble protein! Mol Biol Cell. 2010;21:1435–8. doi: 10.1091/mbc.E09-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 38.Doroudgar S, Glembotski CC. ATF6 [corrected] and thrombospondin 4: the dynamic duo of the adaptive endoplasmic reticulum stress response. Circ Res. 2013;112:9–12. doi: 10.1161/CIRCRESAHA.112.280560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thuerauf DJ, Morrison L, Glembotski CC. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004;279:21078–84. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- 40.Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–9. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- 41.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–93. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 42.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, et al. A thrombospondin-dependent pathway for a protective ER stress response. Cell. 2012;149:1257–68. doi: 10.1016/j.cell.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belmont PJ, Tadimalla A, Chen WJ, Martindale JJ, Thuerauf DJ, Marcinko M, et al. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J Biol Chem. 2008;283:14012–21. doi: 10.1074/jbc.M709776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 45.Vega RB, Bassel-Duby R, Olson EN. Control of cardiac growth and function by calcineurin signaling. J Biol Chem. 2003;278:36981–4. doi: 10.1074/jbc.R300023200. [DOI] [PubMed] [Google Scholar]
- 46.Rothermel BA, Vega RB, Williams RS. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med. 2003;13:15–21. doi: 10.1016/s1050-1738(02)00188-3. [DOI] [PubMed] [Google Scholar]
- 47.Rothermel BA, McKinsey TA, Vega RB, Nicol RL, Mammen P, Yang J, et al. Myocyte-enriched calcineurin-interacting protein, MCIP1, inhibits cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2001;98:3328–33. doi: 10.1073/pnas.041614798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rooij E, Doevendans PA, Crijns HJ, Heeneman S, Lips DJ, van Bilsen M, et al. MCIP1 overexpression suppresses left ventricular remodeling and sustains cardiac function after myocardial infarction. Circ Res. 2004;94:e18–26. doi: 10.1161/01.RES.0000118597.54416.00. [DOI] [PubMed] [Google Scholar]
- 49.Doroudgar S, Thuerauf DJ, Marcinko MC, Belmont PJ, Glembotski CC. Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J Biol Chem. 2009;284:29735–45. doi: 10.1074/jbc.M109.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taxis C, Vogel F, Wolf DH. ER-Golgi traffic is a prerequisite for efficient ER degradation. Mol Biol Cell. 2002;13:1806–18. doi: 10.1091/mbc.01-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2011;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–7. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14296–301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–93. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belmont PJ, Chen WJ, San Pedro MN, Thuerauf DJ, Gellings Lowe N, Gude N, et al. Roles for endoplasmic reticulum-associated degradation and the novel endoplasmic reticulum stress response gene Derlin-3 in the ischemic heart. Circ Res. 2010;106:307–16. doi: 10.1161/CIRCRESAHA.109.203901. [DOI] [PMC free article] [PubMed] [Google Scholar]