Abstract
In previous reports it was demonstrated that the Nipah virus V and W proteins have interferon (IFN) antagonist activity due to their ability to block signaling from the IFN-α/β receptor (J. J. Rodriguez, J. P. Parisien, and C. M. Horvath, J. Virol. 76:11476-11483, 2002; M. S. Park et al., J. Virol. 77:1501-1511, 2003). The V, W, and P proteins are all encoded by the same viral gene and share an identical 407-amino-acid N-terminal region but have distinct C-terminal sequences. We now show that the P protein also has anti-IFN function, confirming that the common N-terminal domain is responsible for the antagonist activity. Truncation of this N-terminal domain revealed that amino acids 50 to 150 retain the ability to block IFN and to bind STAT1, a key component of the IFN signaling pathway. Subcellular localization studies demonstrate that the V and P proteins are predominantly cytoplasmic whereas the W protein is localized to the nucleus. In all cases, STAT1 colocalizes with the corresponding Nipah virus protein. These interactions are sufficient to inhibit STAT1 activation, as demonstrated by the lack of STAT1 phosphorylation on tyrosine 701 in IFN-stimulated cells expressing P, V, or W. Therefore, despite their common STAT1-binding domain, the Nipah virus V and P proteins act by retaining STAT1 in the cytoplasm while the W protein sequesters STAT1 in the nucleus, creating both a cytoplasmic and a nuclear block for STAT1. We also show that the IFN antagonist activity of the P protein is not as strong as that of V or W, perhaps explaining why Nipah virus has evolved to express these two edited products.
The interferon (IFN) response is one of the host's primary defense mechanisms against viral infection. IFN-α/β is synthesized and secreted by cells in direct response to viral infection, specifically to viral products including double-stranded RNA, which triggers a cascade of kinase reactions (13, 49) and leads to the activation of specific cellular transcription factors (e.g., IFN regulatory factor 3 [IRF-3] and NF-κB) required for IFN synthesis. The antiviral effects of IFN are mediated by the products of IFN-stimulated genes (ISG), whose promoters are activated through a signal transduction pathway following binding of IFN to the IFN receptor. Receptor binding activates the JAK tyrosine kinases, JAK1 and TYK2, which in turn phosphorylate STAT1 and STAT2 on tyrosines 701 and 689, respectively (11). The phosphorylated STATs heterodimerize, associate with IRF-9, and translocate to the nucleus. This complex is known as ISG factor 3 (ISGF3) and binds to IFN-sensitive response elements (ISRE) within the promoters of ISGs, resulting in transcriptional upregulation of these genes and establishment of an antiviral state (11).
Viruses have devised various strategies to circumvent this IFN-induced antiviral state through the expression of viral proteins that inhibit specific components of the IFN system (14, 29, 31). Several viral IFN antagonists have been identified in viruses with negative-sense RNA genomes, such as the NS1 protein of influenza A virus (15), the VP35 protein of Ebola virus (4), the NSs protein of Bunyamwera virus (58), and the ML protein of Thogoto virus (23). Within the family Paramyxoviridae, there is a large number of viruses that encode IFN antagonists (22). Respiratory syncytial virus (RSV), a member of the subfamily Pneumovirinae, expresses two nonstructural proteins, NS1 and NS2, which for bovine RSV have been shown to act cooperatively to block the IFN response (48). The IFN antagonist functions of members of the subfamily Paramyxovirinae are all encoded by the phosphoprotein (P) gene and in most cases are carried out by one of the accessory proteins V and C. These proteins all target the IFN signaling, or JAK/STAT, pathway but do so through distinct mechanisms. The Sendai virus C proteins (a nested set of four proteins) interact with STAT1 and prevent pY701-STAT1 formation in response to IFN stimulation (16, 18, 52). There is also evidence that they can affect the dephosphorylation and stability of STAT1 and that the different C proteins may have differential effects (19, 20, 46). The V protein of simian virus 5 (SV5) targets STAT1 for degradation through the assembly of a degradation complex that also requires the presence of STAT2, damaged DNA binding protein 1, and cullin 4A (1, 12, 40, 54). The SV5 V protein also inhibits IFN-β synthesis, so this single protein has the ability to target multiple pathways (26, 44). The human parainfluenza virus 2 V protein targets STAT2 for degradation through a mechanism analogous to that described for SV5 V (35, 39, 54, 61). Similarly, the mumps virus V protein causes loss of STAT1 (30, 59) as well as STAT3 (55). Thus, it appears that the V proteins of these three rubulaviruses all exert their effects through a degradation mechanism, although they have distinct targets. The V protein of measles virus, a morbillivirus, prevents nuclear translocation of STAT1 and STAT2 following IFN treatment but does not affect tyrosine phosphorylation (37). The Newcastle disease virus (NDV) V protein has also been shown to have IFN antagonist activity (28, 42), and there is evidence that it is able to block IFN production (M.-S. Park, unpublished data) and induce loss of STAT1 (28).
Many of the IFN antagonist proteins mentioned above have also been shown to act as virulence factors and therefore have direct effects on viral pathogenicity (6, 15, 17, 32, 43). There is also growing evidence that viral IFN antagonists play a role in determining the host range of a virus, presumably due to a species-specific restriction on their cellular target protein (3, 5, 38, 41, 60).
Nipah virus is a newly emergent paramyxovirus and has been identified as the etiological agent responsible for an outbreak of disease among pigs and humans which occurred in Malaysia and Singapore in 1998 to 1999 (7). Infection in humans was associated with severe febrile encephalitis with a high mortality rate (21), indicating that Nipah virus is a highly virulent human pathogen. In contrast, the virus caused a predominantly respiratory illness in pigs, which were identified as the source of infection for humans. Recently, the fruit bat has been identified as the natural reservoir of Nipah virus (8), and in addition to humans and pigs, there is evidence of infection in dogs, cats, and horses (27). Sequence analysis shows that Nipah virus is closely related to Hendra virus, another zoonotic paramyxovirus, and together these two viruses make up the Henipavirus genus within the subfamily Paramyxovirinae (24, 56). It has been shown that Nipah virus encodes IFN antagonist activity in its V, W, and C proteins (42, 45). The mode of action for C is unclear, but the V protein is able to block IFN signaling by preventing IFN-induced STAT phosphorylation and nuclear translocation (45). The W protein also inhibits IFN signaling, which led to the discovery that it is the common N-terminal portion of V and W that encodes the anti-IFN function (42).
We show here that the Nipah virus P protein also blocks IFN signaling and that the STAT1-binding domain of P, V, and W lies within amino acids 50 to 150 in the common N-terminal domain. We further demonstrate differential activity for the Nipah virus P, V, and W proteins, and we show that while P and V inhibit STAT1 in the cytoplasm, W acts from the nuclear compartment.
MATERIALS AND METHODS
Cells, virus, and expression plasmids.
Vero, 293T, A549, and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS). The green fluorescent protein (GFP)-expressing NDV has been described previously (42). The Nipah virus V and W protein expression plasmids have been described previously (42). An expression construct containing the cDNA for STAT1α under the control of a cytomegalovirus promoter (47) was provided by Jim Darnell (Rockefeller University, New York, N.Y.).
Cloning of the Nipah virus P ORF and truncations thereof.
Nipah virus cDNA of the P open reading frame (ORF) was constructed by PCR without a template by using overlapping deoxyoligonucleotides corresponding to GenBank accession number NC_002728. The P ORF corresponds to nucleotides 2406 to 4535, but nucleotides 2429, 2432, and 2438 have been mutated to C, C, and A, respectively, to eliminate the first two AUG start codons in the C ORF and to create an early stop codon. The PCR product was cloned into the mammalian expression plasmid pCAGGS (36). The domains encoding all truncated versions of the V protein were amplified by PCR, by using the full-length V construct as a template, and cloned into pCAGGS. Where indicated, a hemagglutinin (HA) epitope tag was added at the N terminus or a V5 epitope tag was added at the C terminus.
NDV-GFP complementation assay with A549 cells.
A549 cells were seeded onto 24-well plates, and the monolayer was transfected the following day with 2 μg of a plasmid and 2 μl of Lipofectamine 2000 reagent (Invitrogen) per well. Twenty-four hours later, the cells were washed briefly with phosphate-buffered saline (PBS) and infected with NDV-GFP (multiplicity of infection, 1) at room temperature for 1 h. The infected cells were incubated for 24 h at 37°C under 5% CO2 and analyzed for levels of GFP expression by fluorescence microscopy and fluorescence-activated cell sorter (FACS) analysis, as described previously (42). A549 cells were used for this assay because they are susceptible to infection with NDV-GFP and are relevant for studying the ability of human viral proteins to overcome the antiviral state in human cells.
Reporter gene assays with Vero cells.
Vero cells (which are defective in IFN production) were transfected with a reporter gene plasmid containing the chloramphenicol acetyltransferase (CAT) gene under the control of an ISRE promoter (1 μg), a Renilla luciferase plasmid (1 μg), and the expression plasmid of interest (5 μg), as described previously (42). Cells were treated 24 h later with 1,000 IU of human IFN-β (Calbiochem, La Jolla, Calif.)/ml. Incubation was continued for a further 24 h before the cells were harvested and analyzed for CAT and luciferase activities.
Immunoblotting and immunoprecipitation.
For analysis of the levels of STAT1 phosphorylation, 293T cells were transfected in suspension with 5 μg of a plasmid by using Lipofectamine 2000 (Invitrogen). Transfected cells were incubated for 20 to 24 h, fresh medium without FBS was added, and incubation was continued for 2 to 4 h. Fresh medium (without FBS) containing 1,000 IU of human IFN-β/ml was added, and the cells were incubated for a further 30 min. Cells were then washed in ice-cold PBS and lysed in 100 μl of extract buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% IGEPAL, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 0.1 mM sodium vanadate, and protease inhibitors [Complete; Roche]). Total-cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to a nitrocellulose membrane and probed with antibodies against STAT1 (catalog no. 610185; Transduction Laboratories) or the tyrosine 701-phosphorylated form of STAT1 (catalog no. 9171; Cell Signaling). After incubation with a peroxidase-labeled secondary antibody, blots were analyzed by chemiluminescence.
For coimmunoprecipitation of STAT1, 293T cells were transfected with 5 μg of the HA or V5 epitope-tagged Nipah virus expression plasmid or an empty vector and 5 μg of the STAT1 plasmid. Following a 20- to 24-h incubation, cells were washed in ice-cold PBS and lysed in 500 μl of extract buffer for 30 min on ice. Extracts were centrifuged at 13,000 rpm for 15 min in a Heraeus Biofuge Pico microcentrifuge, the supernatant was collected, and 2 μl of an anti-HA antibody (catalog no. H6908; Sigma) or 2 μl of an anti-V5 antibody (Invitrogen) was added and incubated overnight at 4°C with rotation. Protein A-agarose (Roche) was added, and incubation at 4°C was continued for 1 to 2 h. The agarose was washed three times with extract buffer and boiled in 2× SDS-PAGE sample buffer. Proteins were analyzed by SDS-PAGE and immunoblotting with an antibody against STAT1 as described above. 293T cells were used for these assays so as to achieve the highest possible transfection efficiency.
Immunofluorescence.
HeLa cells were transfected in suspension with 1 μg of the HA-tagged Nipah virus expression plasmid and 2 μg of the STAT1 expression plasmid and were seeded onto glass coverslips. Following a 20- to 24-h incubation, they were fixed and permeabilized in ice-cold methanol. For analysis of the presence of phosphorylated STAT1, the cells were first serum starved (as described above) and then treated with 100 ng of human IFN-α (catalog no. 9906; Cell Signaling)/ml for 30 min prior to fixation. Fixed cells were probed with antibodies against the HA epitope and either STAT1 or phosphorylated STAT1 and were analyzed by fluorescence microscopy following incubation with fluorescein isothiocyanate- or Texas red-conjugated secondary antibodies. HeLa cells were used, because they allow easy distinction of the cytoplasmic and nuclear compartments.
RESULTS
The Nipah virus P protein has IFN antagonist activity.
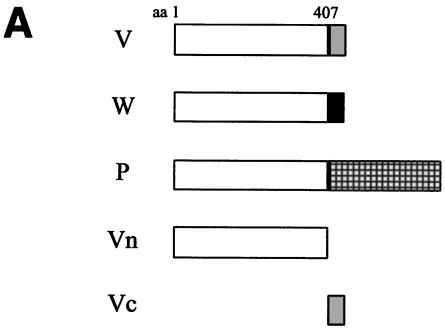
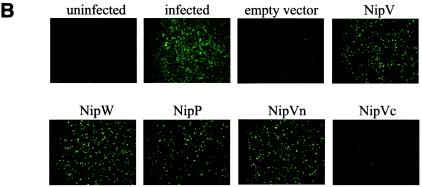
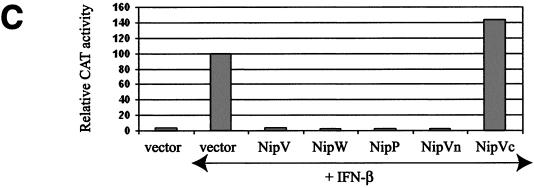
The Nipah virus P gene is predicted to encode four proteins: the phosphoprotein (P), V, W, and C. The V and W proteins are predicted to be expressed from edited transcripts of the P gene where either 1 or 2 additional G residues are inserted at the editing site, respectively (24). This creates a frameshift such that the V, W, and P proteins (the latter expressed from the unedited transcript) have a common 407-amino-acid N-terminal domain but unique C-terminal domains (Fig. 1A). A fourth protein, C, is predicted to be expressed from an alternate ORF in the P gene; however, in all the constructs described here, this ORF has been eliminated by mutation of the start codon and insertion of a stop codon at position 4 to prevent potential effects due to C protein expression. We have previously shown that the common N-terminal domain alone (referred to here as Vn) has the ability to inhibit IFN signaling, as do full-length V and W (42). The P protein has a much longer C-terminal portion than V and W. To examine whether the N-terminal domain retains the ability to block IFN within the context of the full-length P protein, IFN antagonist function was tested in an NDV-GFP assay (42). This assay is based on the observation that plasmid transfection can induce an IFN response, and if the transfected cells are subsequently infected with a recombinant GFP-expressing NDV, the virus fails to replicate efficiently and little GFP expression is observed (Fig. 1B, empty vector). However, if the plasmid encodes an IFN antagonist protein, the virus can replicate and GFP expression is restored. When the V, W, P, or Vn protein was expressed, there was an increase in GFP expression relative to that in cells transfected with the empty vector, whereas with the C-terminal portion of V (Vc), no effect was observed (Fig. 1B). This result indicates that, like the V and W proteins, the P protein can prevent the establishment of an IFN-induced antiviral state. The P protein was further tested for its ability to block IFN-α/β-induced gene expression, as previously demonstrated for V (42, 45) and W (42). Expression of V, W, P, or Vn dramatically reduced activation of an IFN-α/β-inducible reporter gene relative to that with the empty vector control or to expression of Vc (Fig. 1C). These results confirm those obtained by the NDV-GFP assay and show that the V, W, and P proteins all target the IFN signaling pathway and most likely do so through their shared N-terminal portion.
FIG. 1.
The Nipah virus P protein has IFN antagonist activity. (A) Schematic representation of the Nipah virus V, W, and P proteins, their common N-terminal domain (Vn), and the unique C-terminal domain of V (Vc). Open box, N-terminal domain shared by all three proteins; shaded, solid, and checked boxes, unique C-terminal domains of V, W, and P, respectively, which arise due to the insertion of 1, 2, or 0 extra nontemplate G residues. (B) GFP expression in A549 cells following NDV-GFP infection. First panel, untransfected, uninfected cells; second panel, untransfected, NDV-GFP-infected cells. For the remaining panels, cells were transfected with the indicated expression plasmid and subsequently infected with NDV-GFP. (C) Inhibition of IFN-β-induced reporter gene expression in the presence of V, W, P, or Vn. Vero cells were transfected with an ISRE-CAT reporter plasmid and the indicated expression plasmid and were then either mock treated or treated with 1,000 IU of IFN-β/ml (+IFN-β) for 24 h. Data are expressed as the relative CAT activity determined 1 day posttreatment, normalized to a constitutively expressed Renilla luciferase control.
The 50-150 amino acid domain common to the P, V, and W proteins has IFN antagonist activity and inhibits activation of an ISRE promoter.
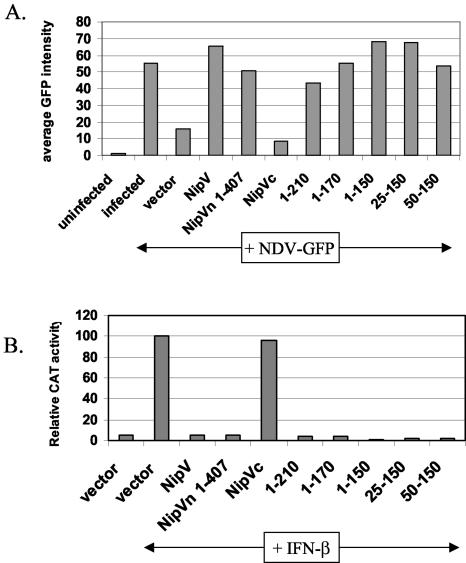
The identification of IFN antagonist activity for the V, W, and P proteins is unique for Nipah virus, since activity has been associated only with the V (and C) proteins of paramyxoviruses. For some V proteins, it has been shown that the conserved Vc is required to block IFN (30, 35, 42), but despite sharing approximately 50% amino acid conservation with the Vc's of other paramyxoviruses, Nipah virus Vc does not display IFN antagonist properties by either the NDV-GFP or the ISRE-CAT assay. When one compares the N-terminal domain of Nipah virus V with those of other paramyxoviruses, the most striking difference is that Nipah and Hendra viruses have a roughly 210 amino acid N-terminal extension. To investigate the possibility that this unique domain encodes the IFN antagonist activity, a construct encoding amino acids 1 to 210 was tested by both the NDV-GFP and ISRE-CAT assays. In the NDV-GFP assay (Fig. 2A), expression of the domain comprising amino acids 1 to 210 (the 1-210 domain) resulted in increased GFP expression, indicating that this region has IFN antagonist activity. Likewise, in the ISRE-CAT assay, there was an almost complete loss of CAT activity in the presence of the 1-210 domain (Fig. 2B). Therefore, in both of these tests, the 1-210 domain behaves similarly to the full-length V protein and Vn. To further map the minimal domain that retains IFN antagonist activity, both C- and N-terminal deletions were made. It was found that expression of amino acids 50 to 150 still resulted in increased GFP expression (Fig. 2A) and decreased CAT activity (Fig. 2B), indicating that the minimal domain required for IFN antagonist activity lies within amino acids 50 to 150.
FIG. 2.
The minimal domain that retains IFN antagonist activity is contained within amino acids 50 to 150. (A) Amino acids 50 to 150 constitute the smallest domain that rescues GFP expression in the NDV-GFP complementation assay. A549 cells were transfected with the indicated plasmid followed by NDV-GFP infection. Average GFP expression, as determined by FACS analysis, is shown. IFN antagonist activity is indicated by an increase in GFP expression compared to that with the vector control. (B) Amino acids 50 to 150 constitute the smallest domain that shows complete inhibition of IFN-β-inducible reporter gene expression. Vero cells were transfected with an ISRE-CAT reporter construct and the indicated expression construct and were then either mock treated or treated with 1,000 IU of IFN-β/ml (+IFN-β) for 24 h. Data are expressed as the relative CAT activity determined 1 day posttreatment, normalized to a constitutively expressed Renilla luciferase control.
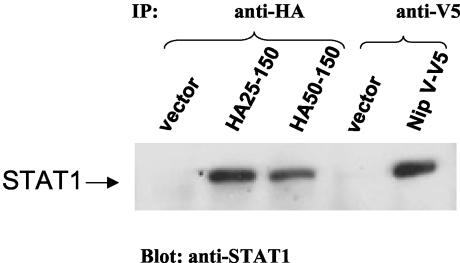
The 50-150 amino acid domain retains the ability to interact with STAT1.
It has been shown previously that expression of Nipah virus V does not result in degradation of either STAT1 or STAT2; rather, V acts by sequestering STAT1 and STAT2 into high-molecular-weight complexes (45). To investigate whether the minimal domain identified as having IFN antagonist properties (50-150) was also able to bind STAT1, HA-tagged constructs of the 25-150 and 50-150 truncations were tested for their abilities to coimmunoprecipitate STAT1 (Fig. 3). A full-length V construct tagged with a V5 epitope was used as a control. Full-length V and the 25-150 and 50-150 truncations were all able to bind STAT1. These results suggest that the STAT1-binding domain lies between amino acids 50 and 150. This finding correlates with the ability of these domains to block the effects of IFN in the NDV-GFP assay (Fig. 2A) and to inhibit the activation of promoters of ISGs (Fig. 2B).
FIG. 3.
Amino acids 50 to 150 constitute the smallest domain that is able to interact with STAT1. 293T cells were transfected with a STAT1 expression construct and the indicated Nipah virus construct. Protein complexes were coimmunoprecipitated with an antibody against the HA or V5 epitope, and the interacting protein was analyzed by SDS-PAGE and immunoblotting with an anti-STAT1 antibody. The position of STAT1 is indicated.
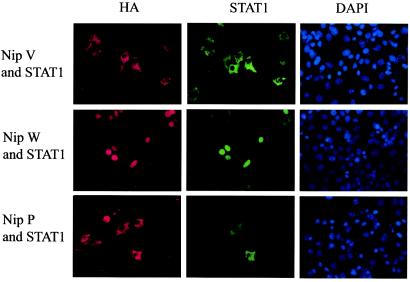
Nipah virus W localizes to the nucleus.
The STAT1-binding domain is present in the P, V, and W proteins, due to its location in the shared N-terminal domain. This begs the question of why the virus produces three proteins that appear to have the same activity. To address this question, we compared the subcellular localization of HA-tagged P, V, and W proteins by indirect immunofluorescence, using an antibody against the HA tag (Fig. 4, left panels). The V protein has previously been shown to have a cytoplasmic distribution (45), a finding confirmed in the present study. The P protein was also found to localize to the cytoplasm; however, the W protein showed a distinctly different localization, with staining exclusively in the nucleus. Experiments performed with untagged versions of the same proteins, using antisera raised against the V protein, yielded the same localization results (data not shown).
FIG. 4.
Nipah virus W is localized to the nucleus and causes redistribution of STAT1 to the nucleus, whereas V and P localize to the cytoplasm and retain STAT1 in the cytoplasm. HeLa cells were transfected with the STAT1 expression plasmid and HA-tagged V, W, or P. Cells were fixed and permeabilized 1 day later. Proteins were stained with antibodies against the HA epitope (left panels) or STAT1 (center panels), and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (right panels).
STAT1 colocalizes with W in the nucleus and with P and V in the cytoplasm.
To examine what effect the nuclear localization of W has on the localization of STAT1, which in its inactive state is both cytoplasmic and nuclear (33), cells were transfected with HA-tagged V, W, or P constructs and a plasmid encoding STAT1 and were costained for HA and STAT1 (Fig. 4, left and center panels). In cells expressing W, STAT1 also displayed nuclear localization, whereas in cells expressing P or V, STAT1 was entirely cytoplasmic. Therefore, STAT1 colocalizes with P and V in the cytoplasm and with W in the nucleus.
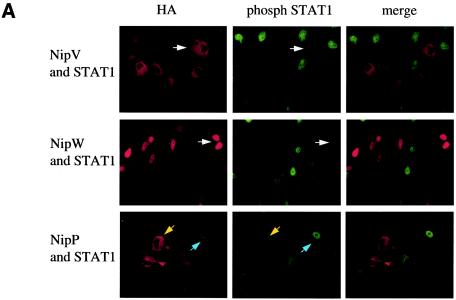
The P, V, and W proteins all inhibit phosphorylation of STAT1 in response to IFN.
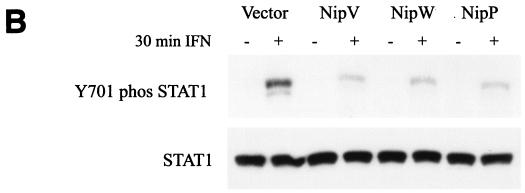
Having demonstrated that expression of P, V, or W results in redistribution of STAT1 to either the cytoplasmic or the nuclear compartment, we next examined the effect of IFN-α/β on STAT1 activation in cells expressing P, V, or W. Upon IFN treatment, STAT1 becomes phosphorylated on tyrosine 701, dimerizes, and translocates to the nucleus (11). To monitor the formation of activated STAT1, cells transfected with STAT1 and either P, V, or W were treated with IFN for 30 min, fixed, permeabilized, and stained with an anti-HA antibody (Fig. 5A, left panels) and an antibody that specifically recognizes the nuclear, phosphorylated form of STAT1 (Fig. 5A, center panels). In those cells not expressing V or W, activated STAT1 was clearly visible in the nucleus; however, if either the V or the W protein was expressed in the cell, there was no evidence of phosphorylated STAT1 in the nucleus (Fig. 5A). Among cells expressing the P protein, some had activated STAT1 in the nucleus while others did not (Fig. 5A), indicating that P is not as capable of blocking STAT1 activation as V or W in this system. However, these first experiments involved overexpression of STAT1. To compare the effects of P, V, and W on the activation of endogenous STAT1, cells were transfected to a high efficiency with the indicated plasmid, either treated with IFN for 30 min or left untreated, and lysed, and the cell extracts were analyzed by Western blotting for the presence of phosphorylated STAT1 (Fig. 5B, upper panel) and bulk STAT1 (lower panel). Levels of bulk STAT1 remained unchanged in the presence or absence of IFN. As expected, the tyrosine-phosphorylated form of STAT1 was seen only in those cells treated with IFN; however, in the presence of either V, W, or P, the amount of phosphorylated STAT1 was smaller than that in cells transfected with the empty vector. The small amounts of phosphorylated STAT1 seen in the V, W, or P samples are likely derived from the small percentage of untransfected cells.
FIG. 5.
Nipah virus V, W, and P all inhibit IFN-induced phosphorylation of STAT1. (A) HeLa cells were transfected with a STAT1 plasmid and an HA-tagged V, W, or P plasmid. STAT1 phosphorylation was induced by treatment with 100 ng of IFN-α/ml for 30 min. Proteins were stained with antibodies against the HA epitope (left panels) or pY701-STAT1 (center panels). In cells expressing V or W, there was no evidence of phosphorylated STAT1 in the nucleus (white arrows). Among cells expressing P, some had activated STAT1 in the nucleus (blue arrows), while others did not (yellow arrows). Merged images are shown in the panels on the right. (B) 293T cells were transfected with the indicated plasmids. One day later, STAT1 phosphorylation was induced by treatment with 1,000 IU of IFN-β/ml for 30 min. Cells were lysed, and levels of pY701-STAT1 (upper panel) and bulk STAT1 (lower panel) were determined by Western blot analysis.
The P protein has a weaker ability to block IFN signaling than V or W.
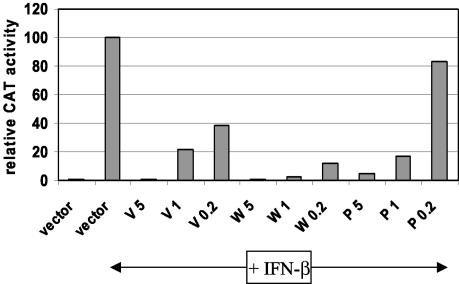
Given that P is unable to inhibit STAT1 phosphorylation to the same extent as V and W when STAT1 is overexpressed (Fig. 5A), we explored whether P may be less effective than V or W at blocking IFN signaling. The ISRE-CAT assay was repeated, this time using limiting amounts of the P, V, and W expression plasmids (Fig. 6). When 5 μg of plasmid was used, all three proteins inhibited reporter gene activation to similar extents. However, when 0.2 μg was used, P displayed an almost complete loss of activity, whereas V and W were still able to block reporter gene activation to a significant extent, with W showing stronger activity than V. To exclude the possibility that this result was due to different levels of protein expression, a portion of the cell extract was analyzed by Western blotting with an anti-HA antibody. Comparable levels of expression were seen with 5 μg of plasmid, but no Western blot signal was detectable at the lower concentrations for any of the constructs (data not shown). These results indicate that the IFN antagonist activity of W is strongest, that of V is intermediate, and that of P is weakest.
FIG. 6.
At limiting concentrations, Nipah virus P has a weaker ability to block IFN signaling than the V and W proteins. Vero cells were transfected with an ISRE-CAT reporter plasmid and either an empty vector or 5, 1, or 0.2 μg of an HA-tagged V, W, or P plasmid. In all cases, the total DNA was made up to 5 μg with the empty vector. Cells were either mock treated or treated with 1,000 IU of IFN-β/ml for 24 h, lysed, and analyzed for CAT activity. Data are expressed as relative levels of CAT activity, normalized to a constitutively expressed luciferase control.
DISCUSSION
There is a precedent for viruses of the subfamily Paramyxovirinae to express proteins that specifically target the IFN signaling pathway and inhibit the cellular IFN response. These antagonist proteins are all encoded by the viral phosphoprotein (P) gene either from alternate ORFs or as a result of RNA editing. For Sendai virus (a respirovirus), the C proteins mediate this effect, whereas for members of the Rubulavirus, Morbillivirus, and Avulavirus genera, the antagonist function is encoded by the V protein. Nipah virus belongs to the newly described Henipavirus genus, and in addition to the P protein, its P gene has the ability to encode V, W, and C proteins. All three of the latter proteins have been shown to have IFN antagonist activity (42, 45). While the target of Nipah virus C remains to be determined, V and W specifically inhibit the IFN signaling pathway. This is the first W protein of a paramyxovirus (here defined as the protein translated from a +2G edited transcript) to be ascribed a function. The observation that both V and W were IFN antagonists led to the finding that it is the common amino-terminal domain of Nipah virus V and W that has IFN antagonist activity (42). This situation differs remarkably from that of the V proteins of human parainfluenza virus 2, mumps virus, and NDV, for which it has been shown that expression of the C-terminal domain alone is sufficient to block the IFN response; for these viruses, expression of the P protein, which has an N-terminal domain identical to that of V, is inactive in the same assay (30, 35, 42).
In the present report we show that the Nipah virus P protein also has IFN antagonist activity, confirming that this function is encoded in the common N-terminal domain of the P, V, and W proteins. The ability of the P protein to block IFN is another unusual feature of Nipah virus (and perhaps Hendra virus), because in all other members of the Paramyxovirinae, the anti-IFN function has been associated only with the accessory proteins C and V. However, the Ebola virus VP35 protein is a functional counterpart of the paramyxovirus phosphoproteins (34) and has also been identified as an IFN antagonist (2, 4). Thus, it would appear that the P genes of negative-strand RNA viruses, although not highly conserved, are multifunctional. It would appear, too, that in the majority of paramyxoviruses the IFN antagonist function has been taken over by the accessory proteins, but in Nipah virus it has been maintained in the P protein (and in V and W) by virtue of its placement in the shared N-terminal domain.
One of the reasons for the placement of Nipah and Hendra viruses in a separate genus is that their P proteins are much larger than those of other paramyxoviruses (24). The majority of this extra length (approximately 210 amino acids) is due to an extended N-terminal domain (57), which is also present in the V and W proteins. Alignment of the Nipah and Hendra V proteins with the other paramyxovirus V proteins shows a high degree of conservation in the C-terminal domain but hardly any over the N-terminal domain. We found that this extended domain, expressed alone, has the ability to inhibit IFN signaling, as does a smaller truncation containing residues 50 to 150. Furthermore, this minimal domain is also capable of interacting with STAT1, which is the proposed mechanism of action for Nipah virus V (45).
The STAT1-binding domain identified in the N terminus is present in the P, V, and W proteins, so the question of why the virus produces three proteins that have identical IFN antagonist functions remains. The differential localization of V and W in the cytoplasm and nucleus, respectively, may provide an answer to this question. Nipah virus W is analogous to the W proteins described for Sendai virus and NDV (9, 51), but the ORFs for the latter two proteins are terminated by a stop codon soon after the editing site, whereas the Nipah virus W ORF extends for 43 amino acids beyond the editing site. Within this unique C-terminal portion are several basic amino acid residues which are strong candidates for a nuclear localization signal. We have shown here that expression of Nipah virus W results in a redistribution of STAT1 to the nucleus in the absence of IFN stimulation. In unstimulated cells STAT1 shuttles between the nucleus and cytoplasm; therefore, W could bind STAT1 through its N terminus soon after it is expressed, and then the complex could translocate to the nucleus, or, alternatively, W could bind STAT1 once it enters the nucleus and prevent it from shuttling back to the cytoplasm. In contrast, V retains STAT1 in the cytoplasm, and there is evidence that, like STAT1, V can shuttle between the cytoplasm and nucleus (45). Ultimately, however, V is predominantly cytoplasmic, suggesting that it has a strong export signal. The Nipah virus P protein is also cytoplasmic and, like the V protein, causes redistribution of STAT1 to the cytoplasm. The result is that in cells expressing W, STAT1 is nuclear, whereas in cells expressing V or P, STAT1 is cytoplasmic. In response to IFN stimulation, STAT1 becomes phosphorylated on tyrosine 701, dimerizes, and translocates to the nucleus (11). It has been shown here and by Rodriguez et al. (45) that expression of V inhibits IFN-induced phosphorylation of STAT1. In cells expressing W, STAT1 is already in the nucleus, and therefore one possibility was that this nuclear STAT is already in its phosphorylated form. However, this possibility was excluded by our demonstration that IFN-treated cells expressing W do not contain tyrosine-phosphorylated forms of STAT1, presumably because STAT1 is excluded from the cytoplasm and thus is not accessible for phosphorylation by tyrosine kinases. Because both V and P retain STAT1 in the cytoplasm and also inhibit its phosphorylation, the mere act of binding appears to be sufficient to block STAT1 activation.
It will be important to determine the cellular distribution of STAT1 in infected cells, but because Nipah virus is classified as a BSL-4 agent and live virus experiments therefore require high-containment facilities, this project is not immediately feasible. One would predict, however, that STAT1 localization would be determined by the relative levels of P, V, and W expression in the cell. In this regard it has been shown that plasmid clones of the P gene, amplified from mRNA of infected cells, contain either 0, 1, or 2 extra G residues at the editing site, which confirms that both edited transcripts (i.e., V and W) are produced (24). Moreover, there is a report which indicates that the frequency of editing in Nipah and Hendra viruses is higher than that in measles and Sendai viruses and that this increase is due to the production of more W transcripts, while the proportions of V transcripts are similar (B. H. Harcourt, A. Tamin, B. Newton, A. Sanchez, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota, Abstr. 11th Int. Conf. Negative-Strand Viruses, abstr. 70, 2000).
Apart from expression levels, another consideration is how possible interactions with other viral proteins may affect the abilities of P, V, and W to function as IFN antagonists. As a phosphoprotein, the P protein functions as a cofactor for the polymerase; its C-terminal domain is predicted to be involved in oligomerization (25, 53) and in interactions with both L (50) and the assembled form of N (25), while the N terminus has a role in chaperoning unassembled N protein (10, 25). It is therefore quite conceivable that one or more of these interactions, which are required for viral RNA synthesis, interfere with the ability of P to bind STAT1 and therefore suppress its IFN antagonist activity. In fact, we have shown here that even when expressed alone, P (which has a long C-terminal domain) is inherently less able to inhibit IFN signaling than equivalent amounts of V and W (which have shorter C-terminal domains). This finding offers an explanation for the continued generation of two additional proteins that share the same STAT1-binding domain but localize to different compartments of the cell and therefore provide the virus with a dual strategy for blocking both cytoplasmic and nuclear forms of STAT1.
The present study and those of Rodriguez et al. (45) and Park et al. (42) provide evidence that all four proteins predicted to be encoded by the Nipah virus P gene (i.e., P, V, W, and C) are able to inhibit the host IFN response. At first glance it may appear that P, V, and W are redundant, but evidence presented here indicates that P has weaker activity, and its role during viral RNA synthesis may interfere with this activity even further. V and W appear to differ only in terms of their cellular distribution, and it remains to be seen what effect elimination of one or the other has on the virus's ability to counteract the IFN response. Several viral IFN antagonists have been identified as virulence factors (6, 15, 17, 32, 43), and therefore it is tempting to speculate that the high level of virulence of the Nipah virus in humans is due to the expression of these multiple IFN antagonist proteins. In addition to the simple presence of more protein, the unique mechanism whereby different proteins act from different compartments of the cell may contribute to severe impairment of the host innate immune response. Nipah and Hendra viruses also have a broad host range (e.g., humans, pigs, bats, horses, dogs, cats) (27). Multiple IFN antagonist proteins may be needed because some act in a species-specific manner, as described for the NS1 protein of influenza A virus (3) and for several paramyxovirus V proteins (38, 41, 60), facilitating the virus's ability to replicate in multiple hosts.
Acknowledgments
This work was supported by NIH grants to A.G.-S., P.P., and C.F.B. C.F.B. is an Ellison Medical Foundation New Scholar in Global Infectious Diseases.
We thank Neva Morales for expert technical assistance. We thank Jim Darnell (Rockefeller University) for providing the STAT1 expression plasmid.
REFERENCES
- 1.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 8.Chua, K. B., C. L. Koh, P. S. Hooi, K. F. Wee, J. H. Khong, B. H. Chua, Y. P. Chan, M. E. Lim, and S. K. Lam. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 9.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran, J., J. B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 18.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcin, D., J. B. Marq, S. Goodbourn, and D. Kolakofsky. 2003. The amino-terminal extensions of the longer Sendai virus C proteins modulate pY701-Stat1 and bulk Stat1 levels independently of interferon signaling. J. Virol. 77:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 21.Goh, K. J., C. T. Tan, N. K. Chew, P. S. Tan, A. Kamarulzaman, S. A. Sarji, K. T. Wong, B. J. Abdullah, K. B. Chua, and S. K. Lam. 2000. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N. Engl. J. Med. 342:1229-1235. [DOI] [PubMed] [Google Scholar]
- 22.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 23.Hagmaier, K., S. Jennings, J. Buse, F. Weber, and G. Kochs. 2003. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J. Virol. 77:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 25.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 26.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 27.Hooper, P., S. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 28.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 77:8676-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 30.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C-terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-α. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 31.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 32.Mebatsion, T., S. Verstegen, L. T. De Vaan, A. Romer-Oberdorfer, and C. C. Schrier. 2001. A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J. Virol. 75:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer, T., K. Gavenis, and U. Vinkemeier. 2002. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res. 272:45-55. [DOI] [PubMed] [Google Scholar]
- 34.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 37.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 40.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 44.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito, S., T. Ogino, N. Miyajima, A. Kato, and M. Kohase. 2002. Dephosphorylation failure of tyrosine-phosphorylated STAT1 in IFN-stimulated Sendai virus C protein-expressing cells. Virology 293:205-209. [DOI] [PubMed] [Google Scholar]
- 47.Schindler, C., X. Y. Fu, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA 89:7836-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 50.Smallwood, S., K. W. Ryan, and S. A. Moyer. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154-163. [DOI] [PubMed] [Google Scholar]
- 51.Steward, M., I. B. Vipond, N. S. Millar, and P. T. Emmerson. 1993. RNA editing in Newcastle disease virus. J. Gen. Virol. 74:2539-2547. [DOI] [PubMed] [Google Scholar]
- 52.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 53.Tarbouriech, N., J. Curran, C. Ebel, R. W. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]
- 54.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 55.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L. F., W. P. Michalski, M. Yu, L. I. Pritchard, G. Crameri, B. Shiell, and B. T. Eaton. 1998. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 72:1482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1α is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]