Abstract
Precursors entering the T-cell developmental pathway traverse a progression of states characterized by distinctive patterns of gene expression. Of particular interest are regulatory genes, which ultimately control the dwell time of cells in each state and establish the mechanisms that propel them forward to subsequent states. Under particular genetic and developmental circumstances, the transitions between these states occur with different timing, and environmental feedbacks may shift the steady-state accumulations of cells in each state. The fetal transit through pro-T cell stages is faster than in the adult, and subject to somewhat different genetic requirements. To explore causes of such variation, this review presents previously unpublished data on differentiation gene activation in pro-T cells of pre-TCR deficient mutant mice, and a quantitative comparison of the profiles of transcription factor gene expression in pro-T cell subsets of fetal and adult wildtype mice. Against a background of consistent gene expression, several regulatory genes show marked differences between fetal and adult expression profiles, including those encoding two bHLH antagonist Id factors, the Ets family factor SpiB, and the Notch target gene Deltex1. The results also reveal global differences in regulatory alterations triggered by the first TCR-dependent selection events in fetal and adult thymopoiesis.
SEPARATE STAGES OF THE T-LINEAGE COMMITMENT PROCESS
Regulatory requirements: the basic checklist
As hematopoietic multipotent progenitors work their way towards T cell differentiation, they shed other developmental options in a gradual process of T-lineage commitment. Accompanying the commitment process is what can be called the “specification” process, that is, the positive regulation of T-cell gene expression as the cells begin to adopt T-lineage characteristics. Both the positive events that promote T-lineage gene expression and the negative processes that block alternative fates are driven by shifting combinations of transcription factors that propagate new gene-expression and developmental states through successive cell cycles.
There has been a great deal of progress in the past few years identifying the regulatory inputs that guide T-lineage differentiation (rev. by (1;2); and see M. K. Anderson review, this volume). In general, pro-T cell emergence depends on at least nine different kinds of regulatory contributions, schematically shown in Fig. 1. These come from E2A and HEB bHLH transcription factors, Runx family transcription factors, transcription factor GATA-3, transcription factor c-Myb, Ikaros-type zinc finger family transcription factors, and TCF-1/LEF-1 HMG box transcription factors activated by β- (or γ-) catenin. An early, hit and run function provided by the Ets family transcription factor PU.1 is also important for establishing the pro-T cell compartment. While most of these factors are thought to work as activators, crucial inputs also come from dedicated repressors, including the growth-promoting Gfi1 (Growth Factor Independence) zinc finger repressor. Finally, a crucial input comes from Notch/Delta signaling interactions, which are mediated through RBPSuh (CSL, RBP-Jκ) transcription factor with MAML as a cofactor (3-5).
Figure 1.
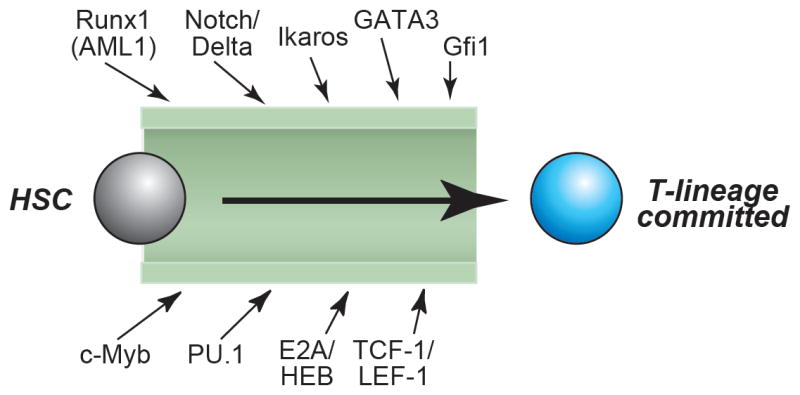
Crucial regulatory inputs into the T-cell commitment process.
Genetic evidence shows that eight transcription factors or transcription factor families plus the Notch/Delta signaling system have essential roles in the generation of committed pro-T cells from hematopoietic stem cells (HSC). The figure summarizes data reviewed in (1).
None of these factors is a lineage-specific “master regulator”, and despite much effort, none has yet been identified for T-cell specification. Individually, most of these inputs are shared between the T-cell program and at least one other hematopoietic developmental program(s). It is only in the correct combination that they specifically promote T-cell development, and correct levels of expression are needed for their positive effects as well (2;6). Thus a critical feature of the pro-T cell developmental program is precise regulation of expression of the regulators themselves in precursor cells. Importantly, in the T-cell developmental process the “correct levels” for each of these factors are not fixed, but need to undergo dynamic alterations from one developmental stage to another (1).
Notch signaling, commitment, and pro-T cell maintenance
Of the critical T-cell regulators, the role of Notch and its mediators has drawn special attention. This is because Notch action appears switch-like, suggesting a tight linkage between commitment and specification. Notch signaling not only promotes the acquisition of T cell characteristics but also blocks precursor entry into the B lymphoid lineage pathway. Extra Notch signaling (e.g. in ectopic sites) leads to more T cell development, while any loss of Notch signaling in the thymus can impair T-cell development and unleash B cell development at that site (rev. by (7-9)). It has been tempting to suppose that the B-lineage inhibiting features of Notch signaling are equivalent to its T-lineage promoting features, and that B-lineage inhibition by Notch is equivalent to T-lineage commitment. However, this is not the way most precursors appear to become committed in vivo. B lineage inhibition is not the conclusion of T-lineage commitment, and it is not the last role that Notch signaling needs to serve in pro-T cell development.
A multitude of studies have now shown, both in fetal and in adult thymocytes, that B-cell potential is lost before or immediately after precursors arrive in the thymus, whereas individual intrathymic precursors remain capable of giving rise to natural killer (NK) cells, dendritic cells (DC), and/or even macrophages as well as T cells (10-22). Meanwhile, Notch signaling from the thymic microenvironment is needed for additional, continuing roles in pro-T cell development and viability extending long after B-lineage potential is suppressed (14;23;24). This lineage-specific survival role extends right up to the point when the first productive rearrangement of the T-cell receptor (TCR) genes has occurred. We can therefore define at least three different stages in the pro-T cell commitment process: loss of B-lineage potential, Notch-dependent expansion while DC and NK potential is preserved, and loss of DC and NK potential. The temporal split between the loss of B cell potential and the loss of other alternative potentials confirms that different mechanisms must regulate different components of the commitment process, as previously suggested (25), and gives a lower bound to the duration of the process as a whole. T-lineage commitment may actually begin even earlier, with a separate loss of erythro-megakaryocytic potential before the loss of B-cell potential (22;26-28). If so, the full process must be even more protracted and mechanistically segmented.
T-cell development is therefore a composite of distinct regulatory subroutines. One subroutine presumably represses the B-cell fate, others repress the NK fate and the DC fate, and others cause onset of pro-T cell gene expression. Meanwhile, the proliferation and checkpoint arrest timing that accompanies these events may be regulated by yet other regulatory subcircuits. Because these subroutines are not active simultaneously, they need to be accounted for by time-dependent changes in regulatory factor activities.
Stages and success criteria for pro-T cell differentiation
The various phases of pro-T cell differentiation within the thymus can be mapped against the changing surface expression patterns of CD44, CD25, HSA (CD24), and c-kit on cells that have not yet expressed TCR complexes or the coreceptors CD4 or CD8. These define the DN1-DN4 stages (18;20;29)(see legend to Fig. 2). Fig. 2 summarizes the stages at which major changes in developmental potential occur. Changes from the DN1 stage through the DN3 stage are all T-cell receptor independent, and these encompass the pro-T cell stages. Throughout the DN1 and DN2 stages, commitment is still incomplete. These cells can still give rise to natural killer cells, dendritic cells, or even macrophages, if moved to a new signaling microenvironment (10;19;24;30-33). Commitment is only complete by the DN3 stage. Subsequently, transit from the DN3 stage to a DN4 stage then depends strictly on TCR-dependent selection, either β-selection or γδ-selection, and marks the end of pro-T cell development.
Figure 2.
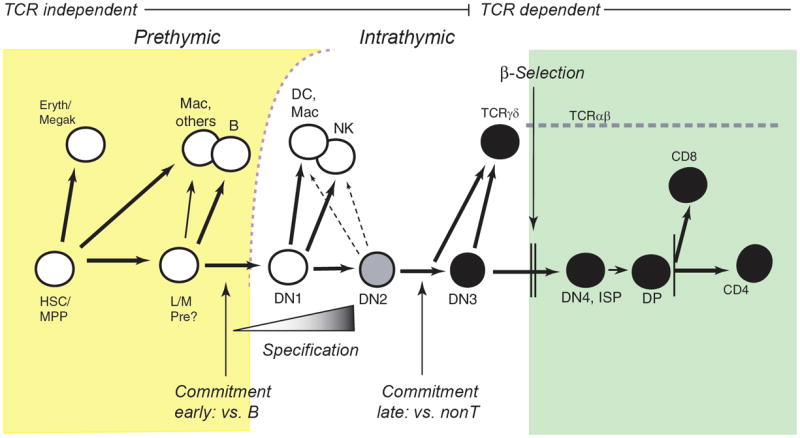
Summary of T-cell development: stages, branchpoints, and checkpoints.
Prethymic (yellow background) and intrathymic (white, green backgrounds) stages of T-cell development are diagrammed, in the context of the developmental options open to cells at each stage. White circles depict multipotent progenitors, black circles depict T-lineage committed stages, and the gray circle depicts the intermediate state of specified but uncommitted DN2 pro-T cells. “Commitment early” is the loss of B-cell potential, described in the text, and “commitment late” is the conclusion of T-lineage commitment with the loss of NK, DC, and macrophage (Mac) potentials. “Specification” denotes the gradual and asynchronous onset of T-lineage differentiation gene expression as described in the text. “Commitment late” concludes the TCR-independent phase of T cell development. Although cells from the DN3 stage onward can continue to give rise to different types of T cells, they can no longer give rise non-T cells unless genetically perturbed. “β-selection” is the strong proliferative and differentiative stimulus given only to those cells that successfully undergo productive TCRβ rearrangement; this marks the end of the pro-T cell stages. TCRγδ cells in general emerge when both γ and δ TCR chains are successfully rearranged before successful TCRβ rearrangement.
Abbreviations: HSC/MPP: hematopoietic stem cell/multipotent progenitor; L/M Pre, putative lympho-myeloid restricted progenitor (26); Eryth/megak, erythroid or megakaryocytic. For all intrathymic stages, DN= CD4- CD8- (and surface TCR- here). DN1 = c-kit++ CD44+ CD25-; DN2 = c-kit++ CD44+ CD25+ HSA+; DN3 = c-kit- CD44- CD25+ HSA+; DN4 = c-kit- CD44-CD25- HSA+ intracellular TCRβ+. After DN stages, ISP = immature single positive, CD4- CD8+ intracellular TCRβ+; DP = double positive, TCRlow CD4+ CD8+ ; CD8 = mature TCRαβ+ CD4- CD8+; CD4 = mature TCRαβ+ CD4+ CD8-. DN1, DN2, and DN4 are the main stages of proliferation in the thymus. Most TCR gene rearrangement occurs in the DN3 and DP stages.
The pro-T cell stages of development, which precede TCR expression, could be considered peripheral to the immunological functions of T cells, but they are important to accomplish several “goals”. One is extensive proliferation, amplifying a small number of precursors to a very large number of cells auditioning for selection (34). Another goal is to lead a precursor from multipotentiality to T-lineage commitment. A third goal is to arm the cell with the signaling apparatus that will enable it to detect successful TCR gene rearrangement. Fourth, of course, is the assembly of at least one good TCR coding sequence by RAG-mediated recombination of the TCRβ locus or the TCRγ and δ loci. The fifth goal is to transform the survival requirements of these hematopoietic cells to make them completely dependent on TCR expression, either a pre-TCR (pTα + TCRβ) or a γδ TCR. Thus, this fifth goal of the pro-T cell stage effectively sets a deadline for TCR gene rearrangement and enforces it with a death penalty. Of course this last “goal” must be delayed with respect to proliferation. Indeed, the interval from precursor entry to β-selection has been estimated as about two weeks in the adult thymus, encompassing at least 10-12 rounds of cell proliferation (and probably more), and with at least 7-10 rounds of division in the fetal thymus (34-36). Correct timing of the regulatory change concluding lineage commitment may be critical for generation of an adequate number of T-cell precursors. This is particularly the case in the adult thymus, where after the proliferative burst at β-selection, further selection and differentiation are carried out essentially without additional cell division (37;38).
HOW MUCH PLASTICITY IS INTRINSIC TO PRO-T CELL DEVELOPMENT?
Fast or slow timing of state transitions
Diagrams like Fig. 2 imply that T-cell development is a kind of pipeline through which the cells inexorably move forward in a fixed timecourse. However, several systems provide evidence that the flow of cells through the pro-T cell stages can be subject to regulatory variations.
Passage from DN1 precursor stage to DN2 and DN3 stages may be regulated by homeostatic feedbacks in some cases. In Rag-knockout animals and other mutants that cannot assemble a pre-TCR, there is no successful differentiation beyond the DN3 stage, and this appears to alter the passage through the stages leading up to the DN3 stage. The DN2 compartment is generally much reduced and canonical c-kithi DN1 cells are scarce or absent (39-41). Thus, precursors may not be allowed to enter the thymus (40), or their development may be arrested immediately within the thymus with downregulation of c-kit, or both.
Additional evidence for flexibility within the pro-T period has emerged recently through the use of the new OP9-DL1 stromal coculture system, which supports T-cell development with a combination of Notch ligand and cytokine-mediated signals (42;43). While the cells progress to β-selection and generate DP cells nicely in this system, there is often a distortion of the distribution of cells among subsets as they pass from the DN1 to the DN3 stage, suggesting that the DN2 compartment is substantially expanded under these conditions (e.g., (44)). Environmental factors such as IL-7 concentrations as well as differences in precursor sources (adult vs. fetal) can influence the differentiative “pausing” in vitro (45)(T. Taghon & EVR, unpublished data). This is most interesting because in this situation the proliferation continues, decoupled from differentiation to some extent. More work is needed to determine exactly how well the gene expression in these cultured DN2 cells matches that of DN2 cells in vivo, or exactly how many additional cell cycles, if any, they can undergo before moving on to the DN3 state; the evidence is still mostly suggestive. Nevertheless, the pattern seen in the OP9-DL1 system is a hint that the DN2 state can be maintained through more cell divisions than it normally occupies in the thymus, where the cells are forced to migrate between domains with differing levels of IL-7 and Notch ligand presentation (36;46). Conceivably the regulatory cascade within DN2 cells does not necessarily cause them to “fall forward” to the DN3 state unless environmental conditions help to “push” them there.
Possibilities of detours or shortcuts in thymocyte development
Most surprisingly, recent evidence has suggested that individual cells can make their ways through the pro-T cell stages by noncanonical routes that appear to miss some normal stages altogether. None of these cases are fully analyzed yet, but they suggest that under some circumstances the dwell time in the DN2 and/or DN3 stages can be greatly reduced. One case is the phenotype of DN thymocytes in a newly described mutant which has abnormal thymic epithelium as a result of a splicing mutation in the Foxn1 gene (47). After birth, the thymus in these mice becomes generally hypocellular, but relative to other subsets the DN3 thymocyte subset appears to be most severely depleted, even under conditions where DP cells are still readily detectable. The suggestion is that defects in the microenvironment force those DP cells that do emerge to take a route that shortcuts past the DN3 stage. A second, striking case is that of the development of certain uncommitted precursors in the thymus that do not fit neatly into the standard DN1-DN2-DN3 classification. While the most potent early T-lineage precursors are the c-kit+ subset(s) of CD25- CD44+ cells (18;29;48), for which we have reserved the term “DN1”in this review, there are also certain c-kitlow HSA+ subsets of CD25- CD44+ cells that contain other kinds of precursors showing some T-lineage potential. These can give rise to DP cells, with reduced efficiency, but they do so without apparently generating any detectable DN2/3-like intermediates at earlier timepoints (18). A third, final case of a possible detour is the effect observed in murine or human T-cell development when the transcription factor GATA-3 is overexpressed by retroviral transduction. Once again, there are hints that a modest number of cells can succeed in reaching the DN4 and/or DP state, but with a notable depletion of any DN3-stage intermediates or their equivalent in human thymocytes (49;50)(T. Taghon and E. V. R., unpublished data). This suggests that the cells may skip to β-selection directly from the DN1 or DN2 stage.
Taken together, these cases raise the possibility that T-cell development could be less like a pipeline and more like the path of a river through a valley, with varying degrees of “eddying” and “meandering” in the normal case which can be expanded or bypassed in abnormal conditions (Fig. 3). Variability in such paths can be understood in terms of a varying balance among the “self-renewal” and “progression” functions at each pro-T cell stage that respectively amplify and cause exit of cells from that stage. However, the question remains: how much of this potential variation is used physiologically? One natural comparison is between the pro-T cell differentiation process in adult and fetal thymus.
Figure 3.
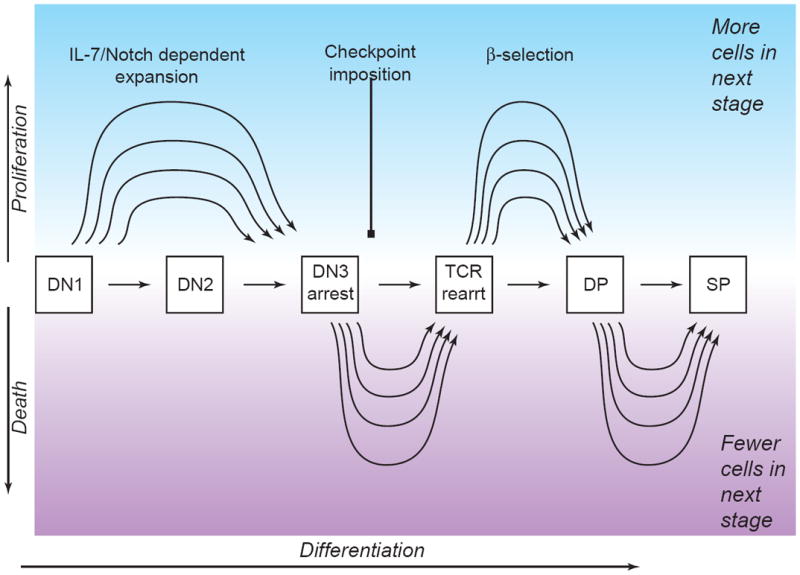
Developmental progression vs. population dynamics mechanisms: sources of potential flexibility in the subset distributions of developing T-lineage cells.
Schematic depiction of the flexibility, in principle, that could enable the same number of DN1 cells to give rise to more or less DN2 and later derivatives in a steady state thymus without affecting the differentiation program as such. Evidence suggests that proliferation, selection criteria, and timing of cell death may be based on separable mechanisms from acquisition of T-cell characteristics (see text).
Behavioral and lineage-choice differences among fetal, postnatal, and adult T-cell development
There are a number of striking differences between fetal and adult hematopoiesis and lymphoid development. First and foremost of these is the difference in time required for fetal mouse thymocyte maturation as compared to postnatal mouse thymocyte maturation. In the fetus, the first lymphoblastic immigrants to the thymus arrive at E13.5 or so and the first DP thymocytes emerge by E16.5, a total span of 3 days. In the postnatal thymus, the immigrants are estimated to take at least ten days to reach the DN3 stage, and about two weeks to differentiate into DP cells (35). This suggests a difference of at least fourfold in the overall length of time spent traversing the pro-T cell stages. In part, the difference in transit time may be due to distinct properties of the hematopoietic stem cells that are made at different stages of ontogeny (rev. by (51;52)). The earliest intraembryonic stem cells (from the aortic-gonadal-mesonephric region), fetal liver stem cells, and adult bone marrow stem cells are all subtly different in their self-renewal potentials and genetic requirements, and all markedly different from the first hematopoietic precursors in the extraembryonic yolk sacs. Accordingly, the timing, cell fate assignments, and genetic requirements of early murine T-cell development from these precursors are somewhat different in three different stages of life: fetal, young postnatal (up to 3-4 weeks), and adult.
The fetal thymus is unique for harboring at least one developmental pathway that is not supported in the postnatal thymus at all. The first cells to seed the fetal thymus, around E12.5-13.5, are also the one group of precursors that can generate specialized lineages of TCRγδ cells that have largely invariant receptors (53;54)(rev. by (55-57)). These γδ cells are the first TCR+ cells to be produced by the fetal thymus (around E15.5). They have distinctive homing properties, as they are targeted to epithelia for their function, and they have distinctive cytokine receptor usage even before leaving the thymus, as they depend on IL-2/IL-15Rβ (CD122) for proliferation as well as on IL-7R signaling for inducing their TCRγ gene recombination (58;59) (60). The production of these γδ cells appears to depend on unique properties both of the hematopoietic precursors in the first wave of fetal thymocyte development and of the fetal thymic microenvironment (54). It is notable that the rearrangement events that generate the specific fetal types of TCRγ and TCRδ chains are facilitated by lower levels of activity of the bHLH protein E2A, and disfavored by higher levels (61). Complementary to the fetal γδ T-cell lineages, there is another important T-cell lineage, the CD1-restricted invariant NKT cells, that is produced only by the postnatal thymus, and not in the fetus (62;63).
Several aspects of the ordering of developmental lineage choice appear to be different between fetal and adult precursors. Prethymically, a major difference is that all fetal precursors that still retain B as well as T potential also retain myeloid potential (22;27;64;65); truly lymphoid-restricted “common lymphoid progenitors” are reported only after birth (66;67). Within the fetal thymus, a major population of DN1 precursors shows T/NK potential, and a wave of NK cells matures in the thymus before any T-cell maturation (32;60;68-70). NK cells are dramatically expanded in fetal thymic organ culture when IL-2 is added (60). This is much more prominent than NK development in the normal adult thymus.
Different genetic requirements among fetal, postnatal, and adult pro-T cells
Fetal thymocytes differ from postnatal thymocytes in their dependence on two transcription factors, the Hunchback-type zinc finger transcription factor Ikaros (Znfn1a1) and the Ets-family transcription factor PU.1 (Sfpi1, Spi-1). Mutants with null Ikaros function have no fetal thymopoiesis, but immediately after birth a fairly normal wave of T cell development begins and eventually generates a substantially well-populated thymus (71). It has been more difficult to study the role of PU.1 in fetal vs. adult thymocytes because true null mutants of PU.1 do not permit survival beyond birth (72-74), at least in part because of a complete lack of macrophages as well as acute hematopoietic progenitor defects. However, a severely hypomorphic PU.1 mutant exists that allows mice to survive for a week or two after birth if kept on antibiotics (75). This mutation, also, completely blocks fetal thymocyte development, but after birth, T cell development begins in the thymus and continues as long as the animals can be kept alive, even while macrophage and B-cell development remains absent (75). Neither the Ikaros case nor the PU.1 case should be interpreted to mean that postnatal T-cell development is really “independent” of the factor’s activity. In Ikaros-null mutants, the ability of postnatal T-cell development to proceed is almost certainly due to the compensatory activity of closely related Ikaros-family transcription factors, such as Helios and/or Aiolos, because in mice transgenic for a dominant-negative allele of Ikaros there is no T-cell development at all (71;76). In the case of PU.1, most recently, conditional knockout alleles have been made that enable the PU.1 gene to be disrupted in adult bone marrow. These studies show that even in the adult, a complete deletion of PU.1 eliminates the ability of the bone marrow to contribute to T-cell development in an adoptive transfer host (74;77). Thus the emergence of some T cells in the postnatal hypomorphic PU.1 mutant may reflect some residual activity and the absence of competition in the mutant environment. Still, the striking difference in impact of these mutations on the thymus before and after birth implies that the fetal and postnatal T-cell development programs use these factors in quantitatively or qualitatively different ways.
Fetal and postnatal T-cell development differ in the ways they use IL-7 receptors for survival and population expansion. IL-7R signaling works through activation of PI3K and STAT5 for proliferative, antiapoptotic, and differentiative effects (78). Postnatal T-cell development is acutely dependent on canonical IL-7 receptors, i.e. heterodimers of IL-7Rα (CD127) and γc (CD132). Mutants lacking either chain are severely disabled in postnatal T-cell development, specifically through the pro-T cell stages (40;79-89). This pro-T cell-specific role of IL-7R signaling is partly based on its ability to maintain anti-apoptotic Bcl-2 levels, but Bcl-2 transgene expression is not sufficient to replace all roles of IL-7R signaling in pro-T cell proliferation (82;85;90). IL-7R signaling has another role as well, a very specific requirement to activate STAT5, which is needed to open the TCRγ loci for recombination and make γδ T-cell development possible (91-98). Beyond the DN3 stage, IL-7 receptor signaling must be downregulated to allow adult pro-T cells to generate DP cells (99). Thus, any IL-7R-deficient mutant cells that do manage to reach the β-selection checkpoint can then undergo efficient proliferation and differentiation to the DP stage and beyond.
In view of these diverse roles in postnatal T-cell development, it is remarkable that fetal T-cell development appears to begin almost normally in IL-7R mutants. Fetal TCRγδ rearrangement is blocked in the mutants, but proliferation apparently is not. Thus the first wave of fetal precursors appears to traverse the DN1-DN3 stages comparably in wildtype animals and in at least one IL-7R mutant strain (100). This is not because fetal thymocytes are cytokine-independent: fetal thymocyte generation is reported to be more, not less, dependent on STAT5 than postnatal thymocyte development (98). However, there appears to be a general division of labor between the IL-7R and IL-2/IL-15R complexes for maintaining survival of fetal-lineage cells (59;60;101). Analysis of IL-7-deficient mutants shows that of the surviving γδ T cells, fetally derived γδ cells which populate the skin are less dependent on environmental IL-7 for their maintenance in the periphery than adult-generated γδ cells (102). To the extent that the IL-2/IL-15R complex transduces slightly different signals than the IL-7R complex, with tighter coupling to Grb2/Ras pathways (103-105), fetal thymocytes (and especially fetal-lineage γδ cells) may also develop in a somewhat different activation state than adult thymocytes. Accompanying their reduced dependence on IL-7R signaling, fetal pro-T cells are also less sensitive than adult pro-T cells to inhibition of DP generation by IL-7R signaling at β-selection (45).
A three-way split between the fetal, immediate postnatal, and fully adult thymocyte developmental requirements is revealed through the functions of the Wnt/ β-catenin/ TCF pathway, with a reversal of the pattern seen for PU.1 and Ikaros mutants. The TCF-1 transcription factor (encoded by the Tcf7 gene) is expressed very highly in thymocytes (see Fig. 4). When mutated, there is little effect on fetal thymocyte development, but postnatal development is strongly impaired at β-selection (DN4 to DP), and as the animals age beyond a month old, an increasingly severe defect is seen in the transit of pro-T cells from the DN1 to the DN2 stage (106;107). The relatively mild effect in fetal development is due to the masking of TCF-1 deficiency by the activity of the closely related LEF-1 gene. Thus, in double mutant fetal thymocytes a strong block is revealed very similar to the block in single TCF-1 mutant postnatal thymocytes (108). The double mutant phenotypes show that TCF-1 and LEF-1 are needed specifically for TCRα expression, for αβ-lineage T cell development immediately after β-selection, and for replenishment of the DN1/2 precursor compartments in fetal thymocytes. The same general functions can be ascribed to TCF-1 in postnatal T-cell development (109-111), so the difference is that after birth LEF-1 does not seem to be able to compensate for TCF-1 deficiency.
Figure 4.
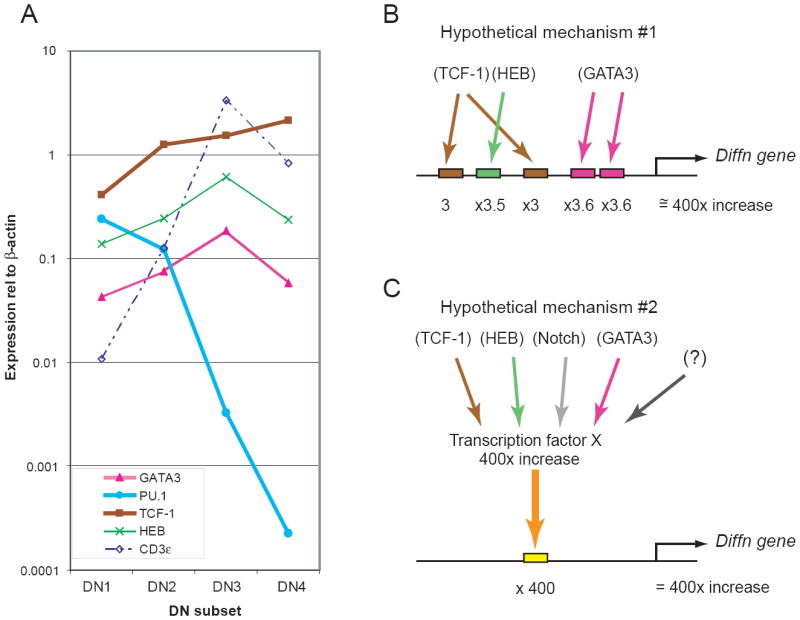
Expression patterns of T-lineage transcription factor and differentiation genes in pro-T cells: disparate rates of developmental change.
(A) Q-PCR analysis of the expression of four critical T-lineage transcription factors and the differentiation gene CD3ε in the DN1-3 stages and through β-selection. Expression levels of these genes were determined in two independent series of sorted DN1-DN4 thymocytes, normalized to β-actin and averaged, and are shown on a log scale. Results for all genes were obtained from the same samples. See text for discussion. (B) and (C) Two models to explain why the increase in “differentiation gene” expression from before specification to commitment should be so much steeper than the increase in T-lineage transcription factor gene expression. (B) Expression of the differentiation gene may depend on combinatorial interactions between transcription factors, so that individual cis-regulatory site occupancies must be multiplied to generate the output expression level. (C) Expression of the differentiation gene may depend not on the known factors, but on an unknown transcription factor that undergoes a steeper increase in its own expression from DN1 to DN3 stage. In this version of the model the unknown transcription factor (X) is itself proposed to be activated by an unspecified combination of other T-lineage regulators. A third possible type of model, not shown, could include steeply downregulated factors like PU.1 as possible rate-limiting antagonists of differentiation gene expression. Unfortunately, the regulatory sequences for CD3ε itself are still uncharacterized and cannot help to distinguish yet between these types of models.
To summarize, even though both the adult and fetal thymus produce recognizable T cells, they do so by somewhat different pathways that depend on different viability and proliferation functions and may also permit access to different lineage alternatives. Thus, while certain aspects of T-lineage specification are fundamentally constant, they are embedded in processes with significant regulatory flexibility.
MAJOR MOLECULAR BENCHMARKS FOR PRO-T CELL DEVELOPMENTAL PROGRESSION
Differentiation gene expression patterns and regulatory gene expression patterns
The most obvious activity that the regulators of T-cell development must carry out is to turn on the “differentiation genes” that confer critical functional properties on the differentiating hematopoietic cells. By this criterion, the T-cell program is under way by the DN2 stage: this is what is meant by “specification” in Fig. 2. For example, Fig. 4A shows quantitative real-time PCR analysis of the expression of the differentiation gene CD3ε in DN1-4 thymocyte subsets, and patterns similar to that of CD3ε here have also been reported for pTα (112;113). Additional data for Rag1 are shown in Fig. 5 and reported elsewhere (114)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. Rothenberg, manuscript submitted). All of these genes are activated detectably by the DN2 stage with a strong increase between DN2 and DN3. Presumably at least some of the temporal control of genes like CD3ε, pTα, and the Rag genes reflects their requirements for the T-lineage transcription factors that are also specifically upregulated during the specification process. The T-lineage-specific transcription factors GATA-3 and TCF-1 and the critical T-lineage bHLH family member HEB also increase in expression from DN1 to DN3 (Fig. 4A). However, transcripts of these regulatory genes increase in expression much more gradually than the CD3ε gene. While the general upward trends are timed similarly, there is a striking difference in slope. At the same time, the early-acting factor PU.1 drops in expression by up to four orders of magnitude between the DN1 and DN3 stages.
Figure 5.
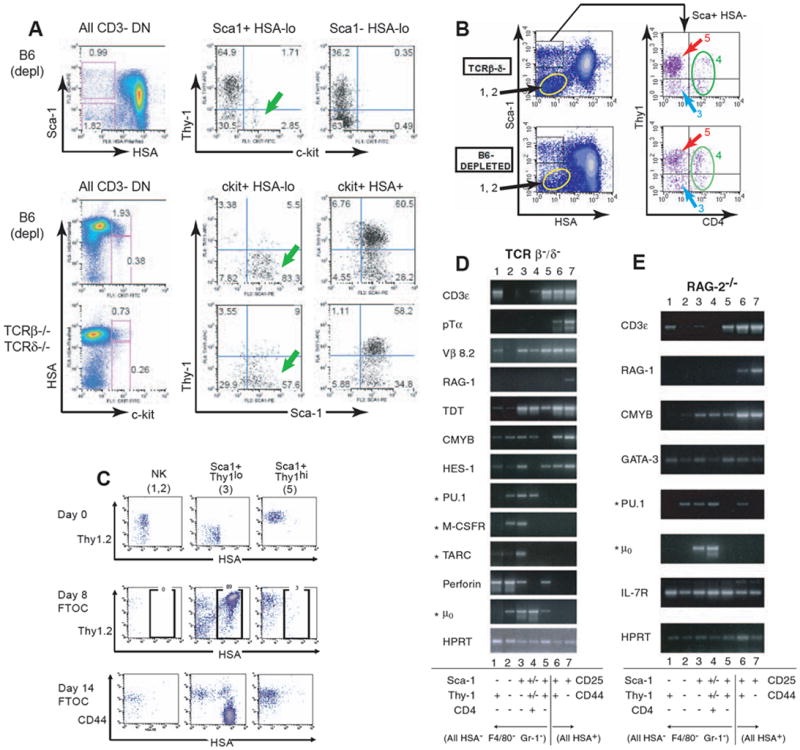
Asynchronous and noncoordinate onset of T-lineage gene expression during T-lineage specification: alternative programs of gene activation?
This figure compares gene expression in definitively specified pro-T cells (DN2, DN3) with gene expression in diverse populations of pre-specification thymocytes [all CD44+ CD25-, mostly also HSA (CD24) low] that differ in T-lineage potential.
(A) The most primitive c-kit+ DN1 cells are found in the Sca-1+ Thy-1lo HSAlo population (green arrows). Data are shown from a five-color flow cytometric analysis of wildtype DN thymocytes (B6 depl = B6 depleted of CD8+, TCR+, and non-lymphoid cells). A parallel analysis is shown of TCRβ-/- δ-/- mutant thymocytes which cannot progress beyond the DN3 stage, confirming that all the subsets shown are generated through TCR-independent processes. HSA is strongly expressed by all CD25+ DN cells (DN2 and DN3), by DN4 cells, and by any B-lineage precursors (39). The HSA-low/negative subsets can be subdivided by Thy-1, Sca-1, NK1.1, c-kit, and other markers to provide sensitive discrimination of DN1 cells (“DN1a”, (18)) and other immature minority populations (39;115;120;124;192). Top panels: c-kit+ DN1 cells are found in the Sca-1+ population of HSAlo cells, not the Sca-1- HSAlo cells. Middle panels: HSAlo c-kit+ cells (“DN1a”, (18)) are predominantly Sca-1+ Thy-1lo, but as they acquire HSA (“DN1b”, (18)) they also become Thy-1hi.
(B) Definition of thymocyte subsets for gene expression analysis. In both wildtype thymocytes and mutants blocked in pre-TCR assembly, Sca-1hi and Sca-1lo subsets of HSAlo thymocytes are further subdivided by Thy-1 and CD4 staining. TCRβ-/- δ-/- results shown in upper panel are representative of Rag-2-/- and SCID subpopulations as well (M. A. Y., unpublished data). In all these mutants, populations 1 and 2, Sca-1lo Thy-1+ and Thy-1- cells, are shown elsewhere to express NK1.1 and CD122 almost uniformly and represent NK lineage cells (115;120;124). Sca-1+ HSAlo cells were separated into CD4- Thy-1lo (population 3) and CD4- Thy-1+ (population 5) subsets. CD4+ cells were sorted separately (population 4) (118); but in mutant thymocytes that cannot undergo β-selection, this population does not contain any T-cell precursors (122). Numbering of subsets in this panel corresponds to numbers in panel C and lane numbers in panels D & E.
(C) TCR-independent differentiation potential in TCRβ-/-δ-/- HSA-low DN cells is confined to cells in HSA-low DN population 3. Mutant thymocytes unable to go through β-selection were used to eliminate the possibility of more mature cell contaminants in any HSA-low DN fractions. Thy-1, CD44, and HSA staining profiles are shown for populations 1+2, population 3, and population 5, isolated as in panel B, both initially and after reaggregation into thymic epithelial lobes and culture for 8-14 days in fetal thymus organ culture. Note that only descendents of population 3 generated HSA+ Thy-1+ CD44- DN3 cells. Descendents of populations 1+2 remained unchanged and also retained predominant NK1.1+ phenotype after fetal thymic organ culture (M. A. Y., unpublished data). Descendents of population 5 failed to proliferate or generate DN3 cells in spite of their high Thy-1+ phenotype and expression of certain other T-lineage genes (panels E & E).
(D) and (E) Expression of T-lineage and non-T-lineage genes in subsets of pro-T cells and other HSA-low DN thymocytes. Thymocytes from two different mutant mouse strains that are arrested at the β-selection checkpoint were used as starting material to exclude any possible contamination with mature TCR+ cells. Populations 1-5 were sorted as indicated (cf. panel B) and compared with DN2 (CD25+ CD44+ HSA+, pop. 6) and DN3 (CD25+ CD44- HSA+, pop. 7) cells from the same thymus. Semiquantititative RT-PCR analyses were performed as previously described (115;119;124;192) using primers for T-lineage genes and “lineage inappropriate” genes (indicated by asterisks: genes expressed preferentially in B-lineage, monocytic, and dendritic cells). The results shown are representative of over six independent experiments with Rag-2-/-, TCRβ-/- δ-/-, and SCID thymocytes (H. W., R. A. D., and E. V. R., unpublished data). Note that all the T-lineage genes and none of the “inappropriate” genes are expressed in both DN2 and DN3 stages, whereas the gene expression patterns in populations 3, 4, and 5 reveal substantial diversity and suggest some early cross-lineage gene activation.
The difference in steepness of induction between T-lineage differentiation genes and T-lineage regulatory genes raises the question of exactly how the transcription factor activity brings about specification, and two non-exclusive possibilities can be suggested (Fig. 4B, C). One is that the regulation of targets like CD3ε depends on combinatorial action of multiple T-lineage transcription factors, such that the differences in the factor concentrations act multiplicatively (Fig. 4B). Another is that GATA-3, TCF-1, and HEB are not the right transcription factors to be looking at in this comparison, and that some other regulatory input, which itself increases more steeply, is the one that truly accounts for the magnitude of differentiation gene induction (Fig. 4C). Data shown below will suggest a limited number of candidates for factors that could provide qualitatively specific positive functions with a sharper slope of developmental increase. However, such candidates are a small minority. We have made a systematic attempt to identify most of the transcription factors that are actively expressed during the pro-T cell stage (E.-S. David, G. Buzi, L. Rowen, R. A. Diamond, M. K. Anderson, and E. V. R., in preparation; C. C. Tydell, E.-S. David, L. Rowen, and E. V. R., unpublished data), in a gene discovery project based on an arrayed cDNA library from SCID thymus pro-T cells (115). It is striking that the great majority of the “new” transcription factor genes identified in this screen show less upregulation during the pro-T cell stages than GATA-3, TCF-1, and HEB, not more (E.-S. David, G. Buzi, L. Rowen, R. A. Diamond, M. K. Anderson, and E. V. R., in preparation; E.-S. David, C. C. Tydell, and E. V. R., unpublished data). At the same time, some of the most dramatic transitions in transcription factor expression from the DN1 to the DN3 stage are exemplified by the downregulation of factors like PU.1 (see Fig. 4A). Thus it seems most likely that the timing of differentiation gene activation is based not on a single “master regulator” that defines the T-lineage specification clock, but rather on the combinatorial interactions of positive regulators (Fig. 4B). Such combinatorial models could be extended to include developmentally-controlled de-repression by rapidly dropping levels of potential antagonists.
T-lineage gene expression begins in precommitment stages
Activation of the T-lineage gene expression program can be induced coordinately in vitro (116;117). However, in the adult thymus in vivo, there is evidence that individual T-lineage differentiation genes are expressed in varying combinations long before T-lineage commitment, and to some extent, in additional intrathymic cells that may have little or no T-lineage precursor activity. This noncoordinate regulation of T-lineage genes can be seen in Fig. 5, where several unconventional populations of immature lymphocytes in the thymus are examined functionally and phenotypically.
It has been evident for a long time that there are DN cells in the thymus that are not on the direct DN1-DN2-DN3 pro-T cell path as defined by markers c-kit, CD44, and CD25. Many of these obscure cells are CD44+ CD25- but not c-kit+. After ruling out contaminating TCR+ mature cells, B-lineage cells, and mature cells of other hematopoietic types, there remain several classes of thymocytes that appear to be immature lymphoid or multilineage precursors, as shown in Fig. 5 and as defined elsewhere by other combinations of markers (18;48;118)(see legend to Fig. 5 for details and additional references). The surprising fact is that substantial numbers of T-lineage genes appear to be activated even in some of these subsets that do not include canonical DN1 cells (18)(Fig. 5D, E: lanes 4, 5).
The c-kit+ DN1 cells themselves constitute a subset within the Sca-1+ Thy-1lo CD4- HSAlo population of CD44+ CD25- cells (Fig. 5A). Accordingly, the Sca-1+ Thy-1lo HSAlo subset (population 3, Fig. 5B) is also enriched for T-lineage progenitor activity (Fig. 5C). The Sca-1- HSAlo subsets (Thy-1+ and Thy-1-, populations 1 & 2) are almost uniformly positive for NK cell surface markers (119;120) and express NK-associated genes like perforin (Fig. 5D, E, lanes 1, 2). These NK-like cells, as expected, do not differentiate into DN2 or DN3 cells in fetal thymic organ culture (Fig. 5C). However, there are also two other populations (Fig. 5B: populations 4 & 5) which show substantial expression of particular T-lineage genes in vivo, including CD3ε as well as TdT and germline transcripts of Vβ8.2 (Fig. 5D, E, lanes 4 & 5) even though cells in these populations cannot develop into DN2 or DN3 cells (Fig. 5C, pop.5 = Sca+ Thy+; for pop. 4, see(121;122)). CD3ε, in spite of its dramatic upregulation within the canonical pro-T cell pathway, is clearly upregulated in non-canonical immature cells as well (Fig. 5D & E, lane 5)(18). Whether these cells are “decommissioned” T-cell precursors or cells of alternative lineages expressing T-cell genes in a “cross-lineage” fashion is not resolved, but clearly some T-cell differentiation genes are accessible to cells that are not committed to a T-cell developmental fate. In fact, sensitive transgenic and knock-in approaches have recently shown that the cis-regulatory sequences of pTα and Rag-1 become activated in multilineage lymphoid precursors even before entry into the thymus (67;112;123), even though these genes are expressed much more strongly afterwards, in the DN3 stage. These results emphasize that T-lineage differentiation gene expression is not uniquely coupled to commitment.
T-lineage gene expression is fully under way by the DN2 stage at the latest, with RAG genes, germline TCR Vβ segments, pTα, CD3ε, Lck, and TdT (in adult mice) already being transcribed (119;124;125)(Fig. 5D & E, lane 6=DN2 cells, lane 7=DN3 cells). Virtually all T-cell differentiation gene transcripts expressed by DN3 cells are already detectable in DN2 cells, although some are upregulated substantially between these two stages (e.g. pTα and Rag-1). Negative as well as positive regulation may reinforce the T-lineage identity of these cells, because the “non-T” genes expressed in other immature thymocyte subsets (Fig. 5, D, E: genes marked with asterisks) are largely or completely repressed in the DN2 as well as DN3 cells (Fig. 5D, E, lanes 6. 7).
Commitment and checkpoint imposition in the DN3 stage
Proliferation is greatest during the the DN1 and DN2 stages, and then pauses at the DN3 stage, while TCR gene rearrangement becomes maximal at the DN3 stage. Successful TCR gene rearrangement is detected by preTCR assembly, and therefore the components of the pre-TCR signaling complex must be in place by the time cells reach the DN3 stage. Accordingly, the CD3 components, signaling kinases, and the surrogate TCR chain pTα reach a peak of expression in the DN3 stage, along with expression of the Rag genes (10;112;126)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted). On the other hand, cytokine-mediated proliferation, which has continued through the DN1 and DN2 stages, is brought to a halt in the DN3 stage (127). This coincides with the downregulation of c-kit, the growth factor receptor that apparently sustains proliferation of precursors immediately before entry and in the earliest stages within the thymus (86;128;129). The main cytokine receptor driving intrathymic DN2 proliferation, the IL7R, continues to be expressed at the DN3 stage, but can no longer drive proliferation (114;130). This arrest in the DN3 stage is the checkpoint from which cells can only emerge by successful TCR rearrangement. Disabling of the cytokine-mediated proliferation apparatus appears to be important to enable the TCR-dependent phases of αβ T cell development to proceed normally (45;99;131;132).
Cell transfer experiments indicate that the DN3 cells are the first to be fully committed to the T-cell lineage (19;24;30;133;134). Commitment, therefore, is associated with proliferative arrest and with the imposition of a new requirement for pre-TCR expression before proliferation can resume. We have argued that this committed state is a result of the sharp downregulation of “non-T” regulatory genes like PU.1 in this stage (135). PU.1 expression is required for the cells to maintain DC potential and induces DC marker expression on fetal thymocytes (136). If PU.1 expression is added back to purified DN3 cells by retroviral transduction, the cells can transdifferentiate to a myeloid-like phenotype (137).
Even once committed, pro-T cells remain acutely dependent on Notch signaling for viability (138), in a role apparently distinct from the previous effects of Notch on lineage choice (24;116;139). Notch1 and Notch3 expression and activation of Notch target genes are maximal at the DN3 stage. In fact, these levels are significantly higher than in the DN1 stages when B-cell potential is presumably just being suppressed (14;140;141)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted). The shift from cytokine-dependent proliferation to Notch-dependent survival, pending pre-TCR-dependent rescue, thus imposes a distinctive gene expression signature on the DN3 cells.
Rite of passage: regulatory transitions at β-selection and exit from the pro-T cell state
The pro-T cell stages end abruptly, either with (pre)TCR-dependent selection (β-selection or γδ-selection) or with death. β-selection triggers the second major phase of proliferation in thymocyte development, normally producing an approximate hundred-fold increase in cell numbers as the cells make the transition to the CD4+ CD8+ (DP) stage. This proliferation dilutes out gene products that were expressed in the pro-T cell stages and helps clear the decks for the new physiological state, installed in the DP cells, that makes TCR-dependent positive and negative selection possible. The extent of proliferation, however, can be regulated separately from the simple triggering of β-selection or γδ-selection per se. This is a major point where effects such as those diagrammed in Fig. 3 can be manifested. Perturbations that reduce proliferation generally have a disproportionate effect on steady-state levels of DP thymocytes, whether or not they interfere with β-selection as a differentiation program. Furthermore, cells choosing the γδ T-cell fate as an alternative to β-selection can mature with much less proliferative expansion, as we have recently been able to measure directly (T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted). As a result, perturbations that block proliferation can alter the effective αβ:γδ thymocyte ratio without necessarily affecting the αβ/γδ lineage choice itself.
Much excellent work has focused on the way β-selection triggers proliferation, changes in TCR gene accessibility for recombination, and allelic exclusion (rev. by (142-147)). However, in addition to these effects, it profoundly transforms the physiology of the cells, swapping apoptosis control functions, blocking transcriptional responses to some activation pathways, enhancing responses to others, and temporarily installing a distinctive set of functions unlike those of mature T cells (rev. by (148)). Detailed comparisons of young adult thymocytes before and after β-selection show that these effects are accompanied by diverse regulatory changes, including both transient responses to the β-selection signal (149-151) and longer-term alterations that play an active role in the unique physiology of DP cells, the major population of thymocytes that is generated by β-selection (99;130;152-156). The IL7R signaling capability is further disabled, and IL7Rα mRNA expression is shut off (114;130). Certain transcription factors, including Ets-family factors SpiB and Erg, and an alternative promoter-use variant of the bHLH factor HEB, are sharply downregulated at β-selection and apparently never used in T-cell development again (114;115;157)(D. Wang, et al., ms. submitted).
Another critical change that occurs at β-selection is that thymocytes are weaned from their dependence on Notch/Delta signaling for the first time since their entry into the T-lineage pathway (45;140;158;159)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted). Among the genes that are precipitously downregulated in the process are Notch1, Notch3, HES-1, and other Notch target genes (14;45;114;140;141;160;161)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted). The end of the pro-T cell stage is therefore the point at which thymocytes exchange their dependence on Notch for a new dependence on TCR signals for subsequent developmental decisions and survival.
A DIRECT COMPARISON OF FETAL AND ADULT PRO-T CELL GENE EXPRESSION PROGRAMS
Comparison between pro-T cell gene expression patterns in fetal and postnatal thymocytes
The landmark stages in adult pro-T cell development just reviewed include many distinct regulatory functions in which differences might be found between adult and fetal programs. For the remainder of this review, we discuss evidence identifying discrete gene regulatory changes that may contribute to these differences. Adult and fetal pro-T cell development, as we have noted, differ both in the speed of differentiation relative to proliferation and in the details of the kinds of T cells produced. This suggests that a direct comparison of adult vs. fetal pro-T cell gene expression is likely to show commonality for those genes that control all the core features of the pro-T-cell developmental program, while showing differences in genes that control either the population dynamics or the generation of specific fetal γδ T cells as opposed to αβ T cells. We have therefore undertaken a systematic comparison of the expression patterns of a series of transcription factors and differentiation genes in purified pro-T cell subsets of E14.5 fetal mice and young adult mice (M. Morales, E.-S. David, M. A. Yui, and E. V. R.; Figs. 6-9 and data not shown).
Figure 6.
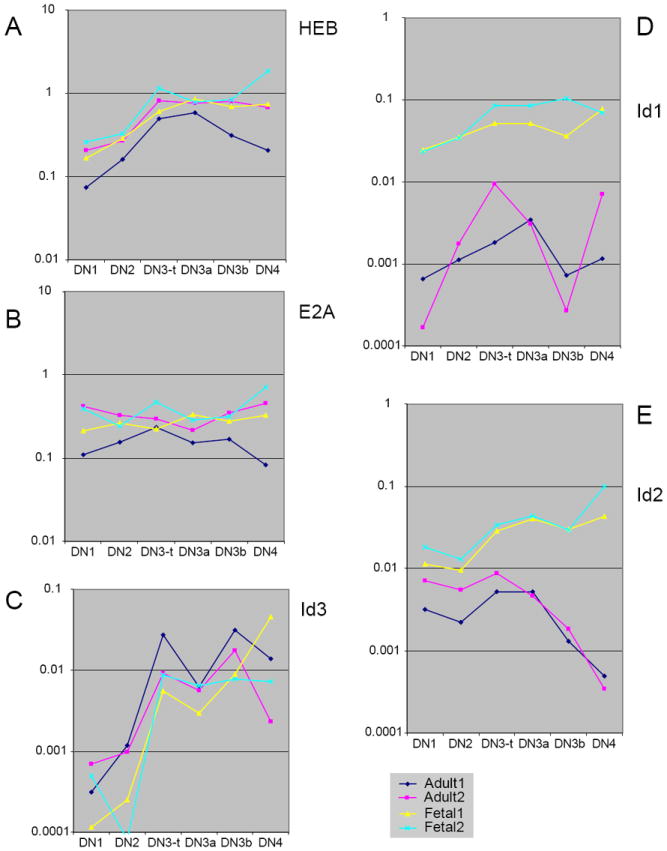
Comparison of bHLH transcription factor and antagonist gene expression in fetal and adult DN thymocytes, pro-T stages through β-selection: fetal-specific increase in Id1 and Id2. Q-PCR analyses of expression of the bHLH factors HEB (A) and E2A (B) and the antagonists Id1 (D), Id2 (E), and Id3 (C) are shown for two independent series each of E14.5 fetal and adult DN thymocyte subsets. Primers used for the realtime PCR analysis are listed in Table 1. Adult (navy and magenta) and fetal (yellow and cyan) data are presented as individual series without averaging, to distinguish systematic variation between fetal and adult series from variation between independent samples of the same type. The adult samples shown here and in Figs. 7-9 are different sets from those used to generate Fig. 4. However, all the samples used in Figs. 6-9 are the same and the results shown are directly comparable. Adult and fetal thymocytes were both prepared by first depleting CD8+, CD3+, TCRγδ+, and Ter-119+ cells, and then performing two sorts on the FACSAria. DN1, DN2, DN3a, and DN3b cells were sorted based on c-kit, CD27, CD44, and CD25 expression from one aliquot, while total DN3 (DN3-t) and DN4 cells were sorted based on c-kit, CD44, CD25, and HSA expression from another aliquot or another cell preparation. In the graphs, DN3-t samples are ordered between the DN2 and DN3a samples to reproduce the DN1-DN2-DN3 (total) succession shown in Fig. 4, to permit the most direct comparison between DN3a and DN3b, and to juxtapose DN3b samples with the DN4 samples that represent their immediate descendents. Note that for genes that are strongly upregulated at β-selection like Id3 (C), the inclusion of DN3b-type cells in the DN3-t sample can make the trend lines appear jagged. As we report elsewhere in detail (T. Taghon et al., submitted), the effects of β-selection in adult thymocytes are seen most clearly in the comparison between DN3a (preselection) and DN3b (newly selected) cells.
In these panels, note that the variability between series in panels (A) and (B) (especially adult samples after β-selection, magenta and navy) fully encompasses the differences between adult and fetal samples. For calibration of these differences, compare the similar patterns and levels of expression of HEB (A) with those from independent samples in Fig. 4A. In contrast, the fetal expression patterns (yellow and cyan) lie distinctly outside the adult patterns (magenta and navy) in panels (D) and (E).
Figure 9.
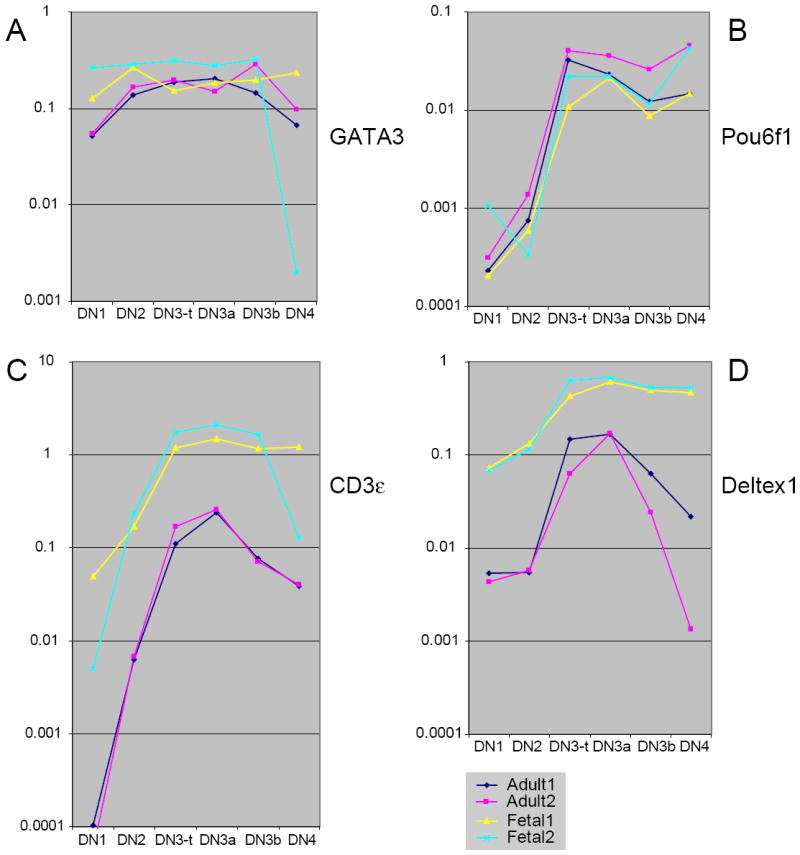
Comparison between adult and fetal pro-T cell expression of distinctly regulated components of the T-cell specification gene set.
(A) GATA-3 expression. (B) Pou6f1 expression. (C) CD3ε expression. (D) Deltex1 expression. See legend to Fig. 6 for explanation of samples. For adult samples, compare the similarity between the GATA-3 and CD3ε levels and patterns of expression in (A) and (C) with those shown in independent samples in Fig. 4A. Although both are tightly associated with the T-cell specification mechanism, both CD3ε and Deltex1 are expressed at remarkably different levels in fetal and adult thymocytes from the earliest DN1 stage throughout the pro-T cell stages.
DN1, DN2, DN3 and DN4 subsets were therefore purified from fetal thymocytes or from Lin-depleted adult thymocytes (114) by a modification of the method of Ceredig and Rolink (29)(E.-S. David, G. Buzi, L. Rowen, R. A. Diamond, M. K. Anderson, and E. V. R., in preparation). Details are described in the legend to Fig. 6. This method includes rigorous selection for c-kit+ progenitor-type cells to define the DN1 and DN2 populations. DN4 cells, produced by β-selection or γδ-selection, were purified as the HSA (CD24)+ CD25- CD44- c-kit- fraction while DN3 cells in total were purified as the HSA+ CD25+ CD44- c-kit- fraction. To obtain further insight into immediate consequences of β-selection signaling, some preparations of DN3 cells were further subdivided into DN3a and DN3b populations, using a new method we have described elsewhere (T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted for publication) to separate DN3 cells that have not yet received β-selection or γδ-selection signals (DN3a) from those that have (DN3b). cDNA samples from these fractions were then probed for expression of regulatory genes by quantitative real-time PCR, using the primer pairs listed in Table 1. All fetal/adult comparisons were made for each primer pair by running two independent series each of fetal and adult samples together in the same analysis. β-actin measurements from the same sample dilutions were used to normalize the results, as previously described (114). All the data shown are measurements of gene expression in the same four series of cDNA preparations, and they therefore serve as controls for the specificity of changes in each other’s expression. Moreover, the gene expression patterns illustrated here are all supported by additional independent analyses of separate adult and/or fetal samples (114)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted for publication)(E.-S. David, G. Buzi, L. Rowen, R. A. Diamond, M. K. Anderson, and E. V. R., in preparation). Detailed methods for the gene expression measurements are to be presented elsewhere (E.-S. David, G. Buzi, L. Rowen, R. A. Diamond, M. K. Anderson, and E. V. R., in preparation).
TABLE 1.
PRIMERS FOR Q-PCR ANALYSIS
| GENE NAME | Forward Primer | Reverse Primer | |
|---|---|---|---|
| 1 | Aiolos | CCGAGATGGGAAGTGAGAGA | CGCTTCTCACCGATGAATTT |
| 2 | β-actin | ACACCCGCCACCAGTTC | TACAGCCCGGGGAGCAT |
| 3 | CD3ε | CGTCCGCCATCTTGGTAGAG | ATTCAATGTTCTCGGCATCGT |
| 4 | C/ebpα | CGGTCATTGTCACTGGTCAACT | GGACAAGAACAGCAACGAGTACC |
| 5 | C/ebpβ | GTTTCGGGACTTGATGCAATC | CGCAGGAACATCTTTAAGGTGAT |
| 6 | C/ebpδ | TCCACGACTCCTGCCATGTA | TGAAGAGGTCGGCGAAGAGTT |
| 7 | C/ebpγ | GCGCAGGTACATGTGAAGATT | CTGCGACAGCTTGCTCATT |
| 8 | Deltex1 | GAGGATGTGGTTCGGAGGTA | CCCTCATAGCCAGATGCTGT |
| 9 | E2A | CAGATGGTGGCCTGGATACT | CATCCCTGCTGTAGCTGTCA |
| 10 | Ets1 | AAAAGTGGATCTCGAGCTTTTCC | CTTTCAAGGCTTGGGACATCA |
| 11 | Ets2 | GCAAGGCAAACCAGTTATTCCT | ACTTGTCAGAGAGTAGCTCCAGAAGAA |
| 12 | Gapdh | ACTCCACTCACGGCAAATTCA | GCCTCACCCCATTTGATGTT |
| 13 | Gata2 | ACCACAAGATGAATGGACAGAA | GTCGTCTGACAATTTGCACAAC |
| 14 | Gata3 | GAGGTGGTTGTCTGC | TTTCACAGCACTAGAGACCCTGTTA |
| 15 | Gfi1 | CGAGTTCGAGGACTTTTGGA | CATGCATAGGGCTTGAAAGG |
| 16 | Gfi1b | CCTTTGCCTGTGATGTCTGT | ATGAACGCTTGAAGGCTTTG |
| 17 | HEB | GAGAAGAAGACCGCTCCATGAT | TGGCTTGGGAGATGGGTAAC |
| 18 | Helios | CACCTCAGGACCCATTCTGT | TGACAGCGTTCCTTGTGTTC |
| 19 | Id1 | GGCGAGATCAGTGCCTTG | AAGGGCTGGAGTCCATCTG |
| 20 | Id2 | GTCCTTGCAGGCATCTGAAT | CTCCTGGTGAAATGGCTGAT |
| 21 | Id3 | AGAGGAGCTTTTGCCACTGA | TGGAGAGAGGGTCCCAGAGT |
| 22 | Ikaros | TCCCAAGTTTCAGGAAAGGA | TCTGCTGTGCTCCAGAGGTA |
| 23 | IL2-UP | ACCTTGGGAGCTGAAATCCT | TTTTGAGGGATCGCTAATGG |
| 24 | Lef1 | ACCTACAGCGACGAGCACTT | GGGTAGAAGGTGGGGATTTC |
| 25 | MEF | CAGGCTCACCAAAACTGTGA | TTGGTCAGCACCGTAGTCAG |
| 26 | Pou6f1 | CTGCAACTCCCATCCCAATC | CGCAAACTCCCGGATCTCTTCT |
| 27 | PU.1 | CCCGGATGTGCTTCCCTTAT | TCCAAGCCATCAGCTTCTCC |
| 28 | Rag1 | GTTAACAACCAAGCTGCAGACATT | TCATCGGGTGCAGAACTGAAG |
| 29 | Runx-1 | CTCGGCAGAACTGAGAAATG | GGTGATGGTCAGAGTGA |
| 30 | Runx-3 | GGTTCAACGACCTTCGATTC | GGTCCATCCACAGTGACCTT |
| 31 | SpiB | CTTGCTCTGGAGGCTGCAC | CCCCCATCTGAATCTGGGTA |
| 32 | Tbet | AGGTGTCTGGGAAGCTGAGA | ATTCGCCGTCCTTGCTTAGT |
| 33 | Tcf7 | CAAGGCAGAGAAGGAGGCTAAG | GGCAGCGCTCTCCTTGAG |
The results (Figs. 6-9) show overall consistency between the fetal and adult pro-T cell gene expression patterns, but with a few remarkable differences. The genes analyzed fall into three general classes in terms of the fetal/adult comparison. “Class 1” genes are expressed indistinguishably between corresponding adult and fetal samples; any variation falls within the range of error of separate preparations of pro-T cells from the same stage of life. “Class 2” genes are expressed in parallel ways in fetal and adult thymocytes throughout the DN1 and DN2 stages, but then show widening differences between fetal and adult samples at the DN3 and DN4 stages. Many of these are genes that are downregulated in adult thymocytes during β-selection: these tend to continue to be expressed at pre-β-selection levels in all fetal DN3 and DN4 cells. “Class 3” genes are genes that are expressed at greatly different levels in fetal and adult pro-T cells throughout all the pro-T cell stages. These “class 3” genes are generally expressed at least tenfold more strongly in fetal thymocytes than in adult thymocytes, as compared to β-actin. Many of these “class 3” genes also show the more persistent pattern of expression in fetal DN3 and DN4 thymocytes that is characteristic of the “class 2” genes. To interpret these expression patterns in terms of transcription factor function, the results are discussed in groups according to gene family.
bHLH family transcription factors: selective modulation of Id1 and Id2
E2A and HEB bHLH transcription factors are important positive drivers of T-cell gene expression, both as homodimers and as heterodimers (rev. by (162;163)). They are expressed at the RNA level throughout the pro-T cell stages at high, comparable, but relatively unchanging levels. There is a gentle rise in HEB from the DN1 to the DN3 stage and a flatter pattern of E2A RNA expression in these stages as shown in Fig. 6A and B (also see Fig. 4A). As is often the case in these analyses, quantitative PCR of the independently prepared sample series from adult (pink and navy) and fetal (yellow and cyan) pro-T cells gives some inter-series scatter. However, the results for the fetal samples lie within the range of variation between the adult samples. These genes are therefore “class 1” in terms of fetal/adult similarity. The activity of bHLH factors in vivo, however, depends on the levels of their antagonists, including the HLH Inhibitors of DNA Binding Id1, Id2, and Id3. Id3 plays an important role in proliferative expansion at β-selection as well as in activating the later maturation events of positive selection, whereas Id2 promotes proliferation in a number of contexts and is a critical regulator of NK cell development (163-165). For these reasons, and in view of the high level of NK potential of fetal pro-T cells, the Id factor expression patterns are of great interest. In the adult thymus, only Id2 and Id3 are significantly expressed, but the Id factors show a different pattern in the fetal thymus. Like E2A and HEB, Id3 is expressed very similarly in the adult and fetal thymocytes. In both adult and fetal samples, Id3 is insignificantly expressed in the DN1 and DN2 stages but upregulated in the DN3 stage, especially the post-selection DN3b cells, and sustained in DN4 cells (Fig. 6C). However, Id1 and Id2 are expressed in dramatically different ways, defining “class 3” relationships between fetal and adult. Id1, which is expressed at extremely low and irreproducible levels in adult pro-T cells, is expressed throughout the fetal pro-T cell stages at 10-100 fold higher levels (Fig. 6D). Id2, on the other hand, is expressed in the DN1-3 stages but downregulated at β-selection in adult cells, yet it begins expression at a higher level in fetal DN1 and DN2 cells and then sustains equal or increasing expression through the DN3 and DN4 stages (Fig. 6E). By the DN3b and DN4 stages, normalized Id2 levels are 20-50 fold higher in the fetal pro-T cells than in the adult pro-T cells.
Thus in spite of consistent bHLH expression, there is a substantially higher level of potential bHLH antagonism in fetal pro-T cells, starting with the high level of Id1 in all fetal pro-T subsets and including an elevated and sustained expression of Id2. The difference in overall bHLH activity predicted from these expression patterns could account both for the high efficiency of NK cell development from fetal pro-T cells (69) and for the highly specific production of TCRγ rearrangements in fetal thymocytes that can be inhibited by E2A activity (61).
Calibrating the significance of quantitative differences: the TCF/LEF family transcription factors
The postnatal-specific defects in TCF-1 deficient mutants, discussed above, raise the question of why fetal pro-T cell development appears to be able to use LEF-1 as a substitute for TCF-1, while adult pro-T cell development cannot. Direct quantitative comparison suggests a simple explanation. TCF-1 levels rise gently (about 3-5 fold) in adult pro-T cells from the DN1 to the DN3 stage, as shown in Fig. 7A and Fig. 4A, and the levels for fetal thymocytes are completely within the range of variation between adult sample series, a perfect example of a “class 1” pattern. However, LEF-1 regulation is modestly but informatively different. It is upregulated sharply between the DN2 and DN3 stages in both adult and fetal pro-T cells, but in all cases its levels in the fetal cells are higher, by 3-10 fold (Fig. 7B). The difference is not as dramatic as for the Id factors (cf. Fig. 6), but it puts LEF-1 into the same expression level range as TCF-1 in the fetal case while it is always the minority factor in the adult case, at least based on RNA levels. The fact that this difference is correlated with a biologically significant difference in impairment in fetal and adult TCF-1 knockout animals (106-108) implies that the greater quantitative differences seen in more definitive “class 3” cases may also have a functional impact.
Figure 7.
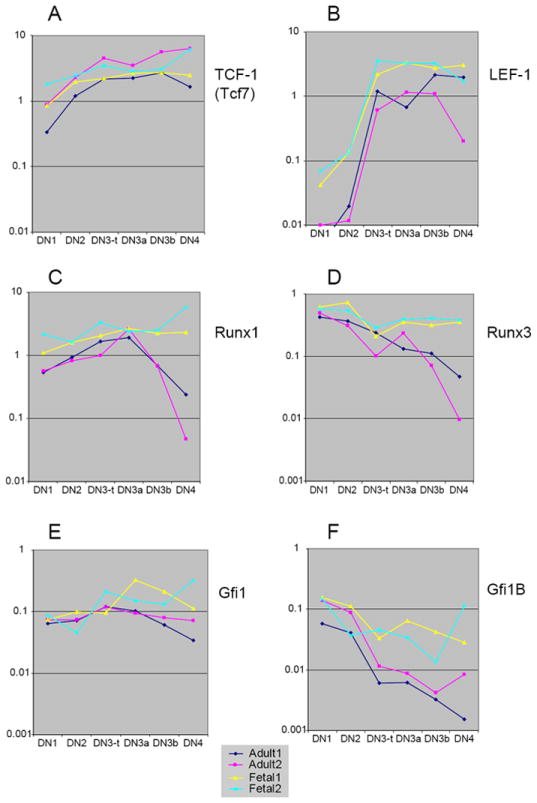
Comparison between adult and fetal pro-T cell expression of TCF/LEF, Runx, and Gfi1 family transcription factors: differences in magnitude and regulation post β-selection. (A) TCF-1 (Tcf7) expression. (B) LEF-1 expression. (C) Runx1 expression. (D) Runx3 expression. (E) Gfi1 expression. (F) Gfi1B expression. See legend to Fig. 6 for explanation of samples. Primers used in Q-PCR analyses are listed in Table 1. Compare the similar levels and patterns of TCF-1 expression (A) with those in independent samples in Fig. 4A.
Similarities in the DN1 and DN2 stages defined by Runx and Gfi1 family transcription factors
Important roles in proliferation and differentiation of pro-T cells are served by Runx and Gfi1 family transcription factors (rev. by (1)). While Runx1 and Runx3 have been studied most in the context of CD4 gene regulation later in T-cell development, Runx1 is crucial for the differentiation of pro-T cells (44;166-168). Gfi1 family factors are likely to act as key regulators of population expansion and quiescence in pro-T cells from the stem-cell stages onward (169-172). Two related genes from each of these families are expressed strongly in the pro-T cell stages: Runx1 and Runx3 (Fig. 7C, D), and Gfi1 and Gfi1B (Fig. 7E, F).
Three of these four genes exemplify “class 2” comparisons. All four transcription factors are expressed in both adult and fetal pro-T cells, and their mRNA levels are very similar in both kinds of samples in the DN1 and DN2 stages. Runx1 levels gently rise from the DN1 through the DN3a stage, then fall steeply after β-selection in the adult samples (DN3b, DN4; Fig. 7C). However, in the fetus, the high levels of Runx1 expression seen in DN2 and DN3 cells are also sustained through the DN4 stage. Runx3 is expressed in a slightly different pattern, first at a high level in DN1 and DN2 cells and then, in the adult, showing a decline that begins in the DN3 stage before β-selection (Fig. 7D, DN3a vs. DN2). In the fetus, however, Runx3 is sustained at high levels all the way through the DN4 stage, already diverging from the adult pattern by the DN3a stage (Fig. 7D). A very similar pattern to that of Runx3 is shown by Gfi1B (Fig. 7F), which starts at a high level in DN1 and DN2 and declines only minimally after that in the fetal pro-T cells. In adult samples, on the other hand, Gfi1B expression starts at similar levels to those in fetal DN1 and DN2 cells, but then drops precipitously beginning at the DN3 (DN3a) stage. In contrast to all these “class 2” examples, Gfi1 represents a “class 1” gene, with only small and irreproducible changes between the adult and fetal series (Fig. 7E).
The “class 2” expression pattern of Gfi1B, Runx1, Runx3, and genes to be discussed below provide an initial clue that regulatory differences between fetal and adult early T cells are concentrated at the stages of β-selection checkpoint arrest and β-selection itself, rather than in the earlier specification stages.
ETS family factors and the non-redundancy of PU.1 and SpiB
The Ets transcription factor family is particularly interesting because it includes many factors with potentially redundant function (173) and overlapping expression patterns in pro-T cells (115), yet there are specific genetic requirements for individual Ets-family factors in T-cell development (174-180). The positive requirement for the Ets factor PU.1 is one of the features that appears to shift between fetal and postnatal pro-T cells (75), even though excess expression of this factor interferes with pro-T cell development in adult and fetal cells alike (135-137). The family includes striking examples of “class 1”, “class 2”, and “class 3” relationships between adult and fetal expression patterns.
Both Ets1 and Ets2 are expressed with a 5-10 fold upsurge between the DN2 and DN3 stages, followed by a relative plateau (Fig. 8A, B). Ets1 is expressed almost indistinguishably between adult and fetal pro-T cell samples of corresponding stages, a perfect example of a “class 1” gene (Fig. 8A). Ets2 is also expressed similarly, though at a slightly higher level throughout in the fetal cells (Fig. 8B), reminiscent of the regulation of LEF-1 (cf. Fig. 7B). At most stages, however, the difference between average fetal and adult levels of Ets2 is less than three fold.
Figure 8.
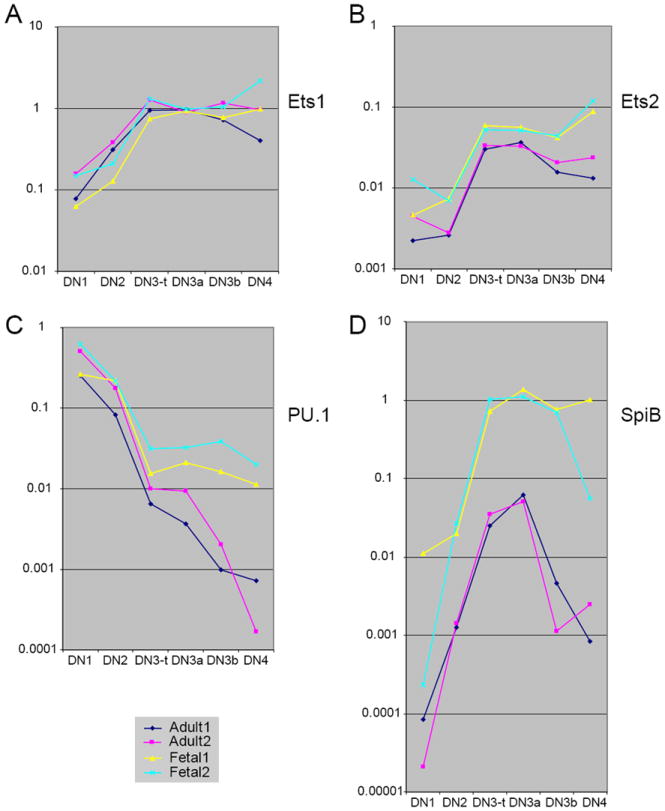
Comparison between adult and fetal pro-T cell expression of Ets family transcription factors: fetal-specific upregulation of SpiB.
(A) Ets1 expression. (B) Ets2 expression. (C) PU.1 expression. (D) SpiB expression. See legend to Fig. 6 for explanation of samples. Compare the similar levels and patterns of PU.1 expression in the adult samples in (C) with those shown in independent samples in Fig. 4A.
Another Ets-family transcription factor, MEF, was examined because of its importance for NK cell maturation (181), but showed relatively constant expression and similar levels in adult and fetal pro-T cells (data not shown).
PU.1 and SpiB are both expressed in adult pro-T cells, but in different patterns, with SpiB maximal in the newly committed cells at the DN3 (DN3a) stage as PU.1 is being sharply repressed (114;115;135;136). They have extremely similar DNA binding specificities but differences in protein-interaction domains outside of the DNA binding domain (rev. by (173)). There is some evidence that these factors can act redundantly (182), and at artificially high levels both of them can interfere with T-cell development similarly (135;136;183), but this redundancy is not seen in all contexts (136;184). In particular, it is not known whether SpiB is structurally competent to fulfil the early positive role for PU.1 that is needed for pro-T cell development. If the factors could act redundantly in the pro-T cell context, then one way that postnatal T-cell development could become less dependent on PU.1 might be by expressing higher levels of SpiB during the DN1 and DN2 stages than in fetal pro-T cells.
The results shown in Fig. 8C and D completely refute this prediction. The surprise is not with PU.1, which shows a close match between its expression levels in fetal and adult pro-T cells throughout the DN1 and DN2 stages, then declines sharply (Fig. 8C; cf. Fig. 4A). There is some difference between adult and fetal patterns for PU.1, as it continues to drop from the DN3a stage onward in the adult (>200 fold overall) but stabilizes expression at the DN3a level in the fetus (Fig. 8C), a “class 2” like relationship. Nevertheless, the 30-fold overall drop in PU.1 levels in the fetus from DN1 to DN3 are presumably sufficient to allow T-lineage commitment. In contrast, the SpiB expression is unexpectedly different in fetal and adult cells, in the opposite direction of that predicted. A striking example of a “class 3” gene, SpiB is expressed at a much higher level in the fetal pro-T cells than in the adult pro-T cells (at least tenfold higher; Fig. 8D). Because of the extensive alternative splicing of SpiB in pro-T cells (115), its expression was tested with two different sets of primers designed to cross a different intron, and the results were the same (data not shown). While SpiB RNA levels in the adult pro-T cells are always relatively low, the level of SpiB RNA expression in the fetal DN3 cells appears to be high in relation to the highest levels of PU.1 or other Ets factors at any stage of pro-T cell development. Thus, if SpiB levels have any relationship to the change in PU.1 dependence between fetal and postnatal pro-T cell development, then it must be by acting as an antagonist of PU.1 rather than as a redundant agonist.
The PU.1/SpiB relationship marks a specific alteration in pro-T cell developmental regulation between fetal and adult. As noted above, Ikaros also appears to be more essential for thymocyte development in fetal life than after birth. However, in contrast to PU.1 and SpiB, expression patterns of the Ikaros family genes Ikaros, Aiolos, and Helios did not show substantial differences in pattern or level between corresponding adult and fetal samples beyond the scatter of the data (data not shown).
T-lineage signature genes: splitting the specification process
The OP9-DL1 system for inducing T-cell development in monolayer culture has been extremely useful to identify genes that are turned on in lymphoid precursors very specifically in response to Notch/Delta signaling (116;117;139): these define a “specification group” of T-cell genes. Among these genes is TCF-1, which we have already examined (Figs. 4A, 7A). Another of these genes is the essential T-lineage transcription factor GATA-3, previously encountered in Fig. 4A. As shown in Fig. 9A, like TCF-1, GATA-3 is also expressed with a very gentle increase from DN1 to DN3, and with little difference between fetal and adult pro-T cells (disregarding a stray point in one fetal sample). Thus GATA-3 falls into the “class 1” category as well. The constant regulatory features between fetal and adult programs are not confined to these two genes induced at the earliest stages of the T-cell specification process. We have seen that Ets1, which is upregulated only at the DN3 stage, is also a “class 1” gene, and Fig. 9B shows that another gene upregulated at the same time, encoding the POU domain factor Pou6f1 (Brn-5), is also upregulated indistinguishably between adult and fetal pro-T cells. Thus there is a regulatory core of the T-cell specification process induced by Notch which appears to be qualitatively and quantitatively invariant between fetal and adult pro-T cell development.
Thus it is particularly surprising that two other major Notch-activated genes, CD3ε and Deltex1, are not regulated similarly in adult and fetal pro-T cells, but instead provide dramatic examples of “class 3” genes (Fig. 9C, D). In both adult and fetal pro-T cells, expression of these genes rises to a peak in the DN3/DN3a stage, as expected for the known importance of Notch signaling at this point (138) and in agreement with the expression of known Notch target genes (T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. R., ms. submitted for publication). However, there is a substantial quantitative difference between the magnitudes of CD3ε and Deltex1 expression in fetal and adult thymocytes, with about tenfold higher mRNA levels in each case in the fetal cells. While CD3ε expression follows the same relative pattern of expression in fetal and adult cells, the higher overall levels shown in Fig. 9C could predict a more efficient assembly or a higher signaling capability for TCRγδ or pre-TCR complexes in the fetal DN3 cells than in the adult. In the case of Deltex1, not only is the level altered but also the pattern of expression, with what appears to be a persistence of Deltex1 expression after β-selection in the fetal case that contrasts sharply with the adult case (Fig. 9D). The role of Deltex1 in T-lineage differentiation is not clear, but as an indicator of Notch signaling (161) it implies that Notch signals themselves may be sustained past the β-selection checkpoint in the fetus in a way that they are not in the adult.
Summary of comparisons
These results have several implications. First, they indicate that there is an invariant core of events in T-cell specification, involving relatively fixed benchmarks for expression of GATA-3, TCF-1, HEB, E2A, and other genes, and that key transitions such as that from DN2 to DN3 are accompanied by an almost-invariant set of regulatory changes, with upregulation of genes like Ets1, Ets2, LEF-1, and Pou6f1. Second, however, other regulatory molecules in the T-cell specification process are not used in quantitatively similar ways even between adult and fetal T-cell development in the same species, and these include both the critical TCR complex signaling component, CD3ε and a direct indicator of Notch signaling, Deltex1. Third, the results indicate that in some ways the fetal DN1 and DN2 stages are more similar to those of the adult than the DN3 and DN4 stages. It is at the DN3 stage and at β-selection that the most consistent differences between fetal and adult appear. Fourth, while part of the fetal-adult difference may reflect the greater presence of γδ cells in the fetal DN4 subset, it is notable that several of the gene regulatory differences between adult and fetal pro-T cells begin before β-selection, in the DN3/DN3a cells. Most of them have the character of blurring the discontinuities that occur in the adult at β-selection: especially, deferring the repression of genes like Runx3, Gfi1B, Deltex1, and even PU.1. Fetal pro-T cell development appears to preserve more “pre-commitment” regulatory character through commitment and into the DN4 stage than in the adult.
Conclusions
The developmental changes that occur in the pro-T cell stages include both a central differentiative core, that defines these cells as T-lineage precursors, and variable features that can be modulated in different stages of ontogeny or in different pathological situations. Among the features that are known to differ, there is the variable extent of cytokine-driven proliferation in the DN1 and DN2 stages, the ragged onset of T-cell differentiation gene expression with respect to the specification process, the varying predisposition to give rise to different αβ or γδ T cell progeny, and the variable extent of proliferation after pre-TCR or TCRγδ-dependent selection. Here a comparison between patterns of regulatory gene expression in the adult and fetal cases provides evidence to begin to categorize those functions that are truly constant in the T-cell specification process and distinguish them from those which may control these variations.
A strikingly constant regulatory ensemble appears to be provided by the expression patterns of TCF-1 and GATA-3, which are induced early in response to Notch signaling and show a gradual increase through the pro-T cell stages; by E2A and HEB, which are expressed at high levels throughout pro-T cell development; by Ets1 and Pou6f1, which are crisply upregulated between the DN2 and DN3 stages; and by the declining level of PU.1, which is repressed with increasing severity from the DN1 to the DN3 stage. Ikaros and Helios, expressed throughout the pro-T cell stages, and Aiolos, induced predominantly at β-selection, are also similar in level in fetal and adult pro-T cells (M. M. and E.-S. D., data not shown)(T. Taghon, M. A. Yui, R. Pant, R. A. Diamond, and E. V. Rothenberg, ms. submitted). Also similar are the absences of expression of C/EBPα and C/EBPβ transcription factors throughout these stages and the initially low, rapidly extinguished expression of GATA-2 in fetal and adult pro-T cells (M. M. and E.-S. D., data not shown). The constant significance of the transition from the DN2 to the DN3 stage is also underlined by the consistent upregulation of LEF-1 and Ets2 at this point, albeit to somewhat different levels in fetus and adult.
The results present some surprises about the stages that are most divergent. In spite of the differences in transit rate from DN1 to DN3 stages, the majority of regulatory genes we examined were expressed at virtually indistinguishable levels between fetus and adult through the DN1 and DN2 stages, often into the DN3a (preselection) stage. The differences were most common and most pronounced in the stages immediately attending β-selection or γδ-selection. This indicates that a constant regulatory core is likely to be critical for successful T-lineage specification itself, i.e. the accurate funneling of cells into the T-lineage program from what may be diverse progenitor-cell states. On the other hand, the regulatory features of β-selection checkpoint enforcement may be profoundly variable between E14.5 fetus and adult. This was not anticipated and is rarely addressed in the literature. Part of the difference may reflect the differentiation of first-wave γδ cells, unique to the fetus, which would make the composition of the DN4 and DN3b populations more γδ-dominated than in the adult. However, so many of the changes effectively perpetuate DN2/DN3a gene expression patterns into the DN4 stage, it appears that the fetal thymocytes lack the entire regulatory discontinuity that β-selection entails in the adult. Even the “class 1” genes, which are expressed similarly between fetal and adult thymocytes, do not reveal any clear discontinuity in fetal thymocytes at β-selection, because all the genes in this category are expressed at fairly constant levels from the DN3a to DN4 transition in adult cells as well. It is possible that β-selection, in its sense of conferring a distinctive regulatory profile on DP thymocytes, may not be the same process in adult and fetus at all.
Finally, overall, the fetal pro-T cell specification process is shown to deploy a highly distinctive balance of potent regulators that should make it work differently from the adult process in several respects. One can only speculate at this point about the consequences of these differences. The genes that are expressed differentially between fetus and adult include factors implicated in proliferation, such as Runx1, Runx3, Gfi1B, LEF-1, Ets2, and the Id factors. This set of genes may indeed underlie fetal-specific regulation of population dynamics. It is important to emphasize also that these genes are likely to control other functions besides proliferation. Runx factors control the timing of CD4 expression (168;185;186). The abundance of Id1 and Id2 expression could contribute directly to the use of fetal-type TCRγ and TCRδ segments in early gene rearrangements, facilitating the predominance of cells with these receptors in the first wave of fetal thymocyte development (61;187).The massively higher expression of Id1 and Id2 could possibly relax some checkpoint controls (163), even while the massively higher expression of SpiB might tend to restrain some of that progression (136). At the same time, the augmented expression of CD3ε in fetal pro-T cells could facilitate strong signals that might assist in the selection of γδ cells in the first wave of T-cell development, according to an emerging strength-of-signal model for γδ-T cell lineage choice (188-190). The functional impact of the strikingly elevated Deltex1 expression is harder to guess, but it suggests at a minimum that Notch signaling is even more powerful and sustained in the fetal thymus than in the adult. The suggestion that Notch signaling continues through β-selection would mark a sharp qualitative difference between fetal and adult thymocyte development, one that could dwarf the differences seen in the pro-T cell stages.
The distinctive features of pro-T cell development in the fetal thymus provide concrete evidence to explain why the behavior of fetal cells may fail to match their adult counterparts experimentally. They also provide an invitation to explore the potential for regulatory flexibility that is inherent in the T cell program. The results described here may begin to indicate points of separation between pure differentiation functions and population expansion regulators, and shed some light on the ways T-cell development reconciles the need to maintain a robust and sophisticated differentiation program with contexts of highly varying population dynamics.
Acknowledgments
The authors wish to thank Michele Anderson, Christopher Franco, and Rashmi Pant for valuable contributions to the foundations of this review when they were in the Rothenberg Lab, Satoko Adachi for genomic DNA-specific primers and advice, Stephanie Adams for outstanding multiparameter fluorescence-activated cell sorting, and Ruben Bayon and the staff of the Office for Laboratory Animal Research for expert care of numerous mutant mouse strains used in the research. We also thank Lee Rowen (Institute for Systems Biology, Seattle) for sequencing multiple pro-T cell cDNA clones. M. M. gratefully acknowledges funding from the Minority Undergraduate Research Fellowships Program at the California Institute of Technology. The work discussed in this review was supported by grants from the USPHS, CA90233 and CA98925, by NASA grant NAG2-1588, and by the DNA Sequencer Royalty Fund at Caltech.
References
- 1.Rothenberg EV, Taghon T. Molecular Genetics of T Cell Development. Annu Rev Immunol. 2005;23:601–49. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg EV, Anderson MK. Elements of transcription factor network design for T-lineage specification. Devel Biol. 2002;246:29–44. doi: 10.1006/dbio.2002.0667. [DOI] [PubMed] [Google Scholar]
- 3.Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–45. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 4.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–22. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 5.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 6.Warren LA, Rothenberg EV. Regulatory coding of lymphoid lineage choice by hematopoietic transcription factors. Curr Opin Immunol. 2003;15:166–75. doi: 10.1016/s0952-7915(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 7.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 8.Guidos CJ. Notch signaling in lymphocyte development. Semin Immunol. 2002;14:395–404. doi: 10.1016/s104453230200074x. [DOI] [PubMed] [Google Scholar]
- 9.Radtke F, Wilson A, Ernst B, MacDonald HR. The role of Notch signaling during hematopoietic lineage commitment. Immunol Rev. 2002;187:65–74. doi: 10.1034/j.1600-065x.2002.18706.x. [DOI] [PubMed] [Google Scholar]
- 10.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–6. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 11.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–74. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 12.Pelayo R, Welner R, Perry SS, Huang J, Baba Y, Yokota T, Kincade PW. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr Opin Immunol. 2005;17:100–7. doi: 10.1016/j.coi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–70. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 14.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–9. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 15.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signalling. Blood. 2005;106:886–92. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 17.Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J Immunol. 2005;174:2525–32. doi: 10.4049/jimmunol.174.5.2525. [DOI] [PubMed] [Google Scholar]
- 18.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–45. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, Katsura Y, Kawamoto H. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol. 2003;171:3401–6. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 20.Bhandoola A, Sambandam A, Allman D, Meraz A, Schwarz B. Early T lineage progenitors: new insights, but old questions remain. J Immunol. 2003;171:5653–8. doi: 10.4049/jimmunol.171.11.5653. [DOI] [PubMed] [Google Scholar]
- 21.Katsura Y. Redefinition of lymphoid progenitors. Nat Rev Immunol. 2002;2:127–32. doi: 10.1038/nri721. [DOI] [PubMed] [Google Scholar]
- 22.Lu M, Kawamoto H, Katsube Y, Ikawa T, Katsura Y. The common myelolymphoid progenitor: a key intermediate stage in hemopoiesis generating T and B cells. J Immunol. 2002;169:3519–25. doi: 10.4049/jimmunol.169.7.3519. [DOI] [PubMed] [Google Scholar]
- 23.Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zúñiga-Pflücker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004;172:5230–9. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004;200:469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenberg EV, Dionne CJ. Lineage plasticity and commitment in T-cell development. Immunol Rev. 2002;187:96–115. doi: 10.1034/j.1600-065x.2002.18709.x. [DOI] [PubMed] [Google Scholar]
- 26.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ lympho-myeloid stem cells lacking erythromegakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Mebius RE, Miyamoto T, Christensen J, Domen J, Cupedo T, Weissman IL, Akashi K. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3- cells, as well as macrophages. J Immunol. 2001;166:6593–601. doi: 10.4049/jimmunol.166.11.6593. [DOI] [PubMed] [Google Scholar]
- 28.Singh H. Gene targeting reveals a hierarchy of transcription factors regulating specification of lymphoid cell fates. Curr Opin Immunol. 1996;8:160–5. doi: 10.1016/s0952-7915(96)80053-7. [DOI] [PubMed] [Google Scholar]
- 29.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol. 2002;2:888–97. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- 30.Lee C-K, Kim JK, Kim Y, Lee MK, Kim K, Kang JK, Hofmeister R, Durum SK, Han SS. Generation of macrophages from early T progenitors in vitro. J Immunol. 2001;166:5964–9. doi: 10.4049/jimmunol.166.10.5964. [DOI] [PubMed] [Google Scholar]
- 31.Michie AM, Carlyle JR, Schmitt TM, Ljutic B, Cho SK, Fong Q, Zúñiga-Pflücker JC. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J Immunol. 2000;164:1730–3. doi: 10.4049/jimmunol.164.4.1730. [DOI] [PubMed] [Google Scholar]
- 32.Ikawa T, Kawamoto H, Fujimoto S, Katsura Y. Commitment of common T/natural killer (NK) progenitors to unipotent T and NK progenitors in the murine fetal thymus revealed by a single progenitor assay. J Exp Med. 1999;190:1617–25. doi: 10.1084/jem.190.11.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas K, Vremec D, Wu L, Shortman K. A linkage between dendritic cell and T-cell development in the mouse thymus: the capacity of sequential T-cell precursors to form dendritic cells in culture. Dev Comp Immunol. 1998;22:339–49. doi: 10.1016/s0145-305x(98)00012-3. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto H, Ohmura K, Fujimoto S, Lu M, Ikawa T, Katsura Y. Extensive proliferation of T cell lineage-restricted progenitors in the thymus: an essential process for clonal expression of diverse T cell receptor beta chains. Eur J Immunol. 2003;33:606–15. doi: 10.1002/eji.200323461. [DOI] [PubMed] [Google Scholar]
- 35.Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–62. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrie HT. Cell migration and the control of post-natal T-cell lymphopoiesis in the thymus. Nat Rev Immunol. 2003;3:859–66. doi: 10.1038/nri1223. [DOI] [PubMed] [Google Scholar]
- 37.Huesmann M, Scott B, Kisielow P, von Boehmer H. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 1991;66:533–40. doi: 10.1016/0092-8674(81)90016-7. [DOI] [PubMed] [Google Scholar]
- 38.Shortman K, Vremec D, Egerton M. The kinetics of T cell antigen receptor expression by subgroups of CD4+8+ thymocytes: delineation of CD4+8+32+ thymocytes as post-selection intermediates leading to mature T cells. J Exp Med. 1991;173:323–32. doi: 10.1084/jem.173.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diamond RA, Ward SB, Owada-Makabe K, Wang H, Rothenberg EV. Different developmental arrest points in RAG2-/- and scid thymocytes on two genetic backgrounds: developmental choices and cell death mechanisms before TCR gene rearrangement. J Immunol. 1997;158:4052–64. [PubMed] [Google Scholar]
- 40.Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J Immunol. 2004;173:1604–11. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- 41.Laurent J, Bosco N, Marche PN, Ceredig R. New insights into the proliferation and differentiation of early mouse thymocytes. Int Immunol. 2004;16:1069–80. doi: 10.1093/intimm/dxh108. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 2002;17:749–56. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 43.Zúñiga-Pflücker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 44.Kawazu M, Asai T, Ichikawa M, Yamamoto G, Saito T, Goyama S, Mitani K, Miyazono K, Chiba S, Ogawa S, Kurokawa M, Hirai H. Functional domains of Runx1 are differentially required for CD4 repression, TCRbeta expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J Immunol. 2005;174:3526–33. doi: 10.4049/jimmunol.174.6.3526. [DOI] [PubMed] [Google Scholar]
- 45.Balciunaite G, Ceredig R, Fehling HJ, Zúñiga-Pflücker JC, Rolink AG. The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur J Immunol. 2005;35:1292–300. doi: 10.1002/eji.200425822. [DOI] [PubMed] [Google Scholar]
- 46.Zamisch M, Moore-Scott B, Su DM, Lucas PJ, Manley N, Richie ER. Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J Immunol. 2005;174:60–7. doi: 10.4049/jimmunol.174.1.60. [DOI] [PubMed] [Google Scholar]
- 47.Su D-M, Navarre S, Oh W, Condie BG, Manley NR. A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat Immunol. 2003;4:1128–35. doi: 10.1038/ni983. [DOI] [PubMed] [Google Scholar]
- 48.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–74. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 49.Taghon T, De Smedt M, Stolz F, Cnockaert M, Plum J, Leclercq G. Enforced expression of GATA-3 severely reduces human thymic cellularity. J Immunol. 2001;167:4468–75. doi: 10.4049/jimmunol.167.8.4468. [DOI] [PubMed] [Google Scholar]
- 50.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias A, Chen D, Rothenberg EV. Definition of regulatory network elements for T-cell development by perturbation analysis with PU.1 and GATA-3. Devel Biol. 2002;246:103–21. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 51.Kincade PW, Owen JJ, Igarashi H, Kouro T, Yokota T, Rossi MI. Nature or nurture? Steady-state lymphocyte formation in adults does not recapitulate ontogeny. Immunol Rev. 2002;187:116–25. doi: 10.1034/j.1600-065x.2002.18710.x. [DOI] [PubMed] [Google Scholar]
- 52.Ling KW, Dzierzak E. Ontogeny and genetics of the hemato/lymphopoietic system. Curr Opin Immunol. 2002;14:186–91. doi: 10.1016/s0952-7915(02)00320-5. [DOI] [PubMed] [Google Scholar]
- 53.Havran WL, Allison JP. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 1990;344:68–70. doi: 10.1038/344068a0. [DOI] [PubMed] [Google Scholar]
- 54.Ikuta K, Kina T, NacNeil I, Uchida N, Péault B, Chien Y, Weissman IL. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–74. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 55.Shortman K, Wu L, Kelly KA, Scollay R. The beginning and the end of the development of TCRγδ cells in the thymus. Curr Top Microbiol Immunol. 1991;173:71–80. [PubMed] [Google Scholar]
- 56.Rothenberg EV. The development of functionally responsive T cells. Adv Immunol. 1992;51:85–214. doi: 10.1016/s0065-2776(08)60487-3. [DOI] [PubMed] [Google Scholar]
- 57.Haas W, Pereira P, Tonegawa S. Gamma/delta cells. Annu Rev Immunol. 1993;11:637–85. doi: 10.1146/annurev.iy.11.040193.003225. [DOI] [PubMed] [Google Scholar]
- 58.De Creus A, Van Beneden K, Stevenaert F, Debacker V, Plum J, Leclercq G. Developmental and functional defects of thymic and epidermal Vγ3 cells in IL-15-deficient and IFN regulatory factor-1-deficient mice. J Immunol. 2002;168:6486–93. doi: 10.4049/jimmunol.168.12.6486. [DOI] [PubMed] [Google Scholar]
- 59.Ye S-K, Maki K, Lee H-C, Ito A, Kawai K, Suzuki H, Mak TW, Chien Y, Honjo T, Ikuta K. Differential roles of cytokine receptors in the development of epidermal γδ T cells. J Immunol. 2001;167:1929–34. doi: 10.4049/jimmunol.167.4.1929. [DOI] [PubMed] [Google Scholar]
- 60.Leclercq G, Debacker V, De Smedt M, Plum J. Differential effects of interleukin-15 and interleukin-2 on differentiation of bipotential T/natural killer progenitor cells. J Exp Med. 1996;184:325–36. doi: 10.1084/jem.184.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–8. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 63.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–5. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 64.Kawamoto H, Ohmura K, Katsura Y. Direct evidence for the commitment of hematopoietic stem cells to T, B, and myeloid lineages in murine fetal liver. Int Immunol. 1997;9:1011–9. doi: 10.1093/intimm/9.7.1011. [DOI] [PubMed] [Google Scholar]
- 65.Katsura Y, Kawamoto H. Stepwise lineage restriction of progenitors in lymphomyelopoiesis. Int Rev Immunol. 2001;20:1–20. doi: 10.3109/08830180109056720. [DOI] [PubMed] [Google Scholar]
- 66.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–72. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 67.Martin CH, Aifantis I, Scimone ML, von Andrian UH, Reizis B, von Boehmer H, Gounari F. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat Immunol. 2003;4:866–73. doi: 10.1038/ni965. [DOI] [PubMed] [Google Scholar]
- 68.Rodewald H-R, Moingeon P, Lucich JL, Dosiou C, Lopez P, Reinherz EL. A population of early fetal thymocytes expressing FcgammaRII/III contains precursors of T lymphocytes and natural killer cells. Cell. 1992;69:139–50. doi: 10.1016/0092-8674(92)90125-v. [DOI] [PubMed] [Google Scholar]
- 69.Carlyle JR, Michie AM, Furlonger C, Nakano T, Lenardo MJ, Paige CJ, Zúñiga-Pflücker JC. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J Exp Med. 1997;186:173–82. doi: 10.1084/jem.186.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michie AM, Carlyle JR, Schmitt TM, Ljutic B, Cho SK, Fong Q, Zúñiga-Pflücker JC. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J Immunol. 2000;164:1730–3. doi: 10.4049/jimmunol.164.4.1730. [DOI] [PubMed] [Google Scholar]
- 71.Wang J-H, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 72.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–7. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 73.Back J, Dierich A, Bronn C, Kastner P, Chan S. PU.1 determines the self-renewal capacity of erythroid progenitor cells. Blood. 2004;103:3615–23. doi: 10.1182/blood-2003-11-4089. [DOI] [PubMed] [Google Scholar]
- 74.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–58. [PMC free article] [PubMed] [Google Scholar]
- 76.Georgopoulos K, Bigby M, Wang J-H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–56. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 77.Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno SI, Arinobu Y, Geary K, Zhang P, Dayaram T, Fenyus ML, Elf S, Chan S, Kastner P, Huettner CS, Murray R, Tenen DG, Akashi K. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005 doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pallard C, Stegmann AP, van Kleffens T, Smart F, Venkitaraman A, Spits H. Distinct roles of the phosphatidylinositol 3-kinase and STAT5 pathways in IL-7-mediated development of human thymocyte precursors. Immunity. 1999;10:525–35. doi: 10.1016/s1074-7613(00)80052-7. [DOI] [PubMed] [Google Scholar]
- 79.Hozumi K, Kondo M, Nozaki H, Kobori A, Nishimura T, Nishikawa S, Sugamura K, Habu S. Implication of the common γ chain of the IL-7 receptor in intrathymic development of pro-T cells. Int Immunol. 1994;6:1451–4. doi: 10.1093/intimm/6.9.1451. [DOI] [PubMed] [Google Scholar]
- 80.Bhatia SK, Tygrett LT, Grabstein KH, Waldschmidt TJ. The effect of in vivo IL-7 deprivation on T cell maturation. J Exp Med. 1995;181:1399–409. doi: 10.1084/jem.181.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore TA, von Freeden-Jeffry U, Murray R, Zlotnik A. Inhibition of γδ T cell development and early thymocyte maturation in IL-7 -/- mice. J Immunol. 1996;157:2366–73. [PubMed] [Google Scholar]
- 82.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor- deficient mice. Cell. 1997;89:1033–41. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 83.He Y-W, Nakajima H, Leonard WJ, Adkins B, Malek TR. The common γ-chain of cytokine receptors regulates intrathymic T cell development at multiple stages. J Immunol. 1997;158:2592–9. [PubMed] [Google Scholar]
- 84.Malissen M, Pereira P, Gerber DJ, Malissen B, DiSanto JP. The common cytokine receptor γ chain controls survival of γ/δ T cells. J Exp Med. 1997;186:1277–85. doi: 10.1084/jem.186.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1-/- mice. Cell. 1997;89:1011–9. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 86.Rodewald H-R, Ogawa M, Haller C, Waskow C, DiSanto JP. Pro-thymocyte expansion by c-kit and the common cytokine receptor γ chain is essential for repertoire formation. Immunity. 1997;6:265–72. doi: 10.1016/s1074-7613(00)80329-5. [DOI] [PubMed] [Google Scholar]
- 87.von Freeden-Jeffry U, Solvason N, Howard M, Murray R. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity. 1997;7:147–54. doi: 10.1016/s1074-7613(00)80517-8. [DOI] [PubMed] [Google Scholar]
- 88.Di Santo JP, Aifantis I, Rosmaraki E, Garcia C, Fainberg J, Fehling HJ, Fischer A, von Boehmer H, Rocha B. The common cytokine receptor γ chain and the pre-T cell receptor provide independent but critically overlapping signals in early α/β T cell development. J Exp Med. 1999;189:563–73. doi: 10.1084/jem.189.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Porter BO, Scibelli P, Malek TR. Control of T cell development in vivo by subdomains within the IL-7 receptor α-chain cytoplasmic tail. J Immunol. 2001;166:262–9. doi: 10.4049/jimmunol.166.1.262. [DOI] [PubMed] [Google Scholar]
- 90.Rodewald H-R, Waskow C, Haller C. Essential requirement for c-kit and common γ chain in thymocyte development cannot be overruled by enforced expression of Bcl-2. J Exp Med. 2001;193:1431–7. doi: 10.1084/jem.193.12.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Durum SK, Candéias S, Nakajima H, Leonard WJ, Baird AM, Berg LJ, Muegge K. Interleukin 7 receptor control of T cell receptor γ gene rearrangement: role of receptor-associated chains and locus accessibility. J Exp Med. 1998;188:2233–41. doi: 10.1084/jem.188.12.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ye S-K, Maki K, Kitamura T, Sunaga S, Akashi K, Domen J, Weissman IL, Honjo T, Ikuta K. Induction of germline transcription in the TCRγ locus by Stat5: implications for accessibility control by the IL-7 receptor. Immunity. 1999;11:213–23. doi: 10.1016/s1074-7613(00)80096-5. [DOI] [PubMed] [Google Scholar]
- 93.Kang J, Coles M, Raulet DH. Defective development of γ/δ T cells in interleukin 7 receptor- deficient mice is due to impaired expression of T cell receptor γ genes. J Exp Med. 1999;190:973–82. doi: 10.1084/jem.190.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlissel MS, Durum SD, Muegge K. The interleukin 7 receptor is required for T cell receptor gamma locus accessibility to the V(D)J recombinase. J Exp Med. 2000;191:1045–50. doi: 10.1084/jem.191.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang J, Durum SK, Muegge K. Cutting edge: histone acetylation and recombination at the TCRγ γlocus follows IL-7 induction. J Immunol. 2001;167:6073–7. doi: 10.4049/jimmunol.167.11.6073. [DOI] [PubMed] [Google Scholar]
- 96.Lee H-C, Ye S-K, Honjo T, Ikuta K. Induction of germline transcription in the human TCRγ locus by STAT5. J Immunol. 2001;167:320–6. doi: 10.4049/jimmunol.167.1.320. [DOI] [PubMed] [Google Scholar]
- 97.Ye S-K, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, Honjo T, Ikuta K. The IL-7 Receptor controls the accessibility of the TCRγ Locus by Stat5 and histone acetylation. Immunity. 2001;15:813–23. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- 98.Kang J, DiBenedetto B, Narayan K, Zhao H, Der SD, Chambers CA. STAT5 Is required for thymopoiesis in a development stage-specific manner. J Immunol. 2004;173:2307–14. doi: 10.4049/jimmunol.173.4.2307. [DOI] [PubMed] [Google Scholar]
- 99.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORγt: impact on thymocyte development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crompton T, Outram SV, Buckland J, Owen MJ. Distinct roles of the interleukin-7 receptor α chain in fetal and adult thymocyte development revealed by analysis of interleukin-7 receptor α-deficient mice. Eur J Immunol. 1998;28:1859–66. doi: 10.1002/(SICI)1521-4141(199806)28:06<1859::AID-IMMU1859>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 101.Porter BO, Malek TR. Thymic and intestinal intraepithelial T lymphocyte development are each regulated by the gammac-dependent cytokines IL-2, IL-7, and IL-15. Semin Immunol. 2000;12:465–74. doi: 10.1006/smim.2000.0264. [DOI] [PubMed] [Google Scholar]
- 102.Laky K, Lewis JM, Tigelaar RE, Puddington L. Distinct requirements for IL-7 in development of TCRγδ cells during fetal and adult life. J Immunol. 2003;170:4087–94. doi: 10.4049/jimmunol.170.8.4087. [DOI] [PubMed] [Google Scholar]
- 103.Gadina M, Sudarshan C, Visconti R, Zhou YJ, Gu H, Neel BG, O’Shea JJ. The docking molecule gab2 is induced by lymphocyte activation and is involved in signaling by interleukin-2 and interleukin-15 but not other common γ chain-using cytokines. J Biol Chem. 2000;275:26959–66. doi: 10.1074/jbc.M004021200. [DOI] [PubMed] [Google Scholar]
- 104.Ozaki K, Leonard WJ. Cytokine and cytokine receptor pleiotropy and redundancy. J Biol Chem. 2002;277:29355–8. doi: 10.1074/jbc.R200003200. [DOI] [PubMed] [Google Scholar]
- 105.Kovanen PE, Rosenwald A, Fu J, Hurt EM, Lam LT, Giltnane JM, Wright G, Staudt M, Leonard WJ. Analysis of γc-family cytokine target genes. Identification of dual-specificity phosphatase 5 (DUSP5) as a regulator of mitogen-activated protein kinase activity in interleukin-2 signaling. J Biol Chem. 2003;278:5205–13. doi: 10.1074/jbc.M209015200. [DOI] [PubMed] [Google Scholar]
- 106.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–4. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 107.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PAJ, Van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–4. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 108.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 109.Gounari F, Aifantis I, Khazaie K, Hoeflinger S, Harada N, Taketo MM, von Boehmer H. Somatic activation of β-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat Immunol. 2001;2:863–9. doi: 10.1038/ni0901-863. [DOI] [PubMed] [Google Scholar]
- 110.Ioannidis V, Beermann F, Clevers H, Held W. The β-catenin--TCF-1 pathway ensures CD4+CD8+ thymocyte survival. Nat Immunol. 2001;2:691–7. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 111.Staal FJT, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, van Dongen JJ. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172:1099–108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- 112.Gounari F, Aifantis I, Marti C, Fehling HJ, Hoeflinger S, Leder P, von Boehmer H, Reizis B. Tracing lymphopoiesis with the aid of a pTα-controlled reporter gene. Nat Immunol. 2002;3:489–96. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- 113.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–91. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- 115.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–48. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 116.Taghon TN, David E-S, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–78. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmitt TM, De Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–7. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 118.Wu L, Scollay R, Egerton M, Pearse M, Spangrude GJ, Shortman K. CD4 expressed on earliest T-lineage precursor cells in the adult murine thymus. Nature. 1991;349:71–4. doi: 10.1038/349071a0. [DOI] [PubMed] [Google Scholar]
- 119.Wang H, Diamond RA, Yang-Snyder JA, Rothenberg EV. Precocious expression of T-cell functional response genes in vivo in primitive thymocytes before T-lineage commitment. Int Immunol. 1998;10:1623–35. doi: 10.1093/intimm/10.11.1623. [DOI] [PubMed] [Google Scholar]
- 120.Telfer JC, Rothenberg FV. Expression and function of a stem-cell promoter for the murine CBFα2 gene: distinct roles and regulation in natural killer and T cell development. Devel Biol. 2001;229:363–82. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 121.Antica M, Wu L, Shortman K, Scollay R. Intrathymic lymphoid precursor cells during fetal thymus development. J Immunol. 1993;151:5887–95. [PubMed] [Google Scholar]
- 122.Michie AM, Carlyle JR, Zúñiga-Pflücker JC. Early intrathymic precursor cells acquire a CD4low phenotype. J Immunol. 1998;160:1735–41. [PubMed] [Google Scholar]
- 123.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–30. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 124.Chen F, Rowen L, Hood L, Rothenberg EV. Differential transcriptional regulation of individual TCR Vβ segments before gene rearrangement. J Immunol. 2001;166:1771–80. doi: 10.4049/jimmunol.166.3.1771. [DOI] [PubMed] [Google Scholar]
- 125.Buckland J, Pennington DJ, Bruno L, Owen MJ. Co-ordination of the expression of the protein tyrosine kinase p56lck with the pre-T cell receptor during thymocyte development. Eur J Immunol. 2000;30:8–18. doi: 10.1002/1521-4141(200001)30:1<8::AID-IMMU8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 126.Wilson A, Capone M, MacDonald HR. Unexpectedly late expression of intracellular CD3ε and TCR γδ proteins during adult thymus development. Int Immunol. 1999;11:1641–50. doi: 10.1093/intimm/11.10.1641. [DOI] [PubMed] [Google Scholar]
- 127.Kang J, Der SD. Cytokine functions in the formative stages of a lymphocyte’s life. Curr Opin Immunol. 2004;16:180–90. doi: 10.1016/j.coi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 128.Perry SS, Pierce LJ, Slayton WB, Spangrude GJ. Characterization of thymic progenitors in adult mouse bone marrow. J Immunol. 2003;170:1877–86. doi: 10.4049/jimmunol.170.4.1877. [DOI] [PubMed] [Google Scholar]
- 129.Waskow C, Paul S, Haller C, Gassmann M, Rodewald H. Viable c-KitW/W mutants reveal pivotal role for c-Kit in the maintenance of lymphopoiesis. Immunity. 2002;17:277–88. doi: 10.1016/s1074-7613(02)00386-2. [DOI] [PubMed] [Google Scholar]
- 130.Van De Wiele CJ, Marino JH, Murray BW, Vo SS, Whetsell ME, Teague TK. Thymocytes between the β-selection and positive selection checkpoints are nonresponsive to IL-7 as assessed by STAT-5 phosphorylation. J Immunol. 2004;172:4235–44. doi: 10.4049/jimmunol.172.7.4235. [DOI] [PubMed] [Google Scholar]
- 131.El Kassar N, Lucas PJ, Klug DB, Zamisch M, Merchant M, Bare CV, Choudhury B, Sharrow SO, Richie E, Mackall CL, Gress RE. A dose effect of IL-7 on thymocyte development. Blood. 2004;104:1419–27. doi: 10.1182/blood-2004-01-0201. [DOI] [PubMed] [Google Scholar]
- 132.Yasuda Y, Kaneko A, Nishijima I, Miyatake S, Arai K. Interleukin-7 inhibits pre-T-cell differentiation induced by the pre-T-cell receptor signal and the effect is mimicked by hGM-CSF in hGM-CSF receptor transgenic mice. Immunology. 2002;106:212–21. doi: 10.1046/j.1365-2567.2002.01402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A. 2002;99:4508–13. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shortman K, Vremec D, Corcoran LM, Georgopoulos K, Lucas K, Wu L. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol Rev. 1998;165:39–46. doi: 10.1111/j.1600-065x.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 135.Anderson MK, Weiss A, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T-cell development at the pro-T stage. Immunity. 2002;16:285–96. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 136.Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–94. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- 137.Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–66. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 138.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005 doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 139.Höflinger S, Kesavan K, Fuxa M, Hutter C, Heavey B, Radtke F, Busslinger M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J Immunol. 2004;173:3935–44. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- 140.Huang EY, Gallegos AM, Richards SM, Lehar SM, Bevan MJ. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J Immunol. 2003;171:2296–304. doi: 10.4049/jimmunol.171.5.2296. [DOI] [PubMed] [Google Scholar]
- 141.Choi JW, Pampeno C, Vukmanovic S, Meruelo D. Characterization of the transcriptional expression of Notch-1 signaling pathway members, Deltex and HES-1, in developing mouse thymocytes. Dev Comp Immunol. 2002;26:575–88. doi: 10.1016/s0145-305x(01)00095-7. [DOI] [PubMed] [Google Scholar]
- 142.Borowski C, Martin C, Gounari F, Haughn L, Aifantis I, Grassi F, von Boehmer H. On the brink of becoming a T cell. Curr Opin Immunol. 2002;14:200–6. doi: 10.1016/s0952-7915(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 143.Michie AM, Zuniga-Pflucker JC. Regulation of thymocyte differentiation: pre-TCR signals and β-selection. Semin Immunol. 2002;14:311–23. doi: 10.1016/s1044-5323(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 144.Kruisbeek AM, Haks MC, Carleton M, Michie AM, Zúñiga-Pflücker JC, Wiest DL. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol Today. 2000;21:637–44. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- 145.Iritani BM, Alberola-Ila J, Forbush KA, Perimutter RM. Distinct signals mediate maturation and allelic exclusion in lymphocyte progenitors. Immunity. 1999;10:713–22. doi: 10.1016/s1074-7613(00)80070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Michie AM, Trop S, Wiest DL, Zúñiga-Pflücker JC. Extracellular signal-regulated kinase (ERK) activation by the pre-T cell receptor in developing thymocytes in vivo. J Exp Med. 1999;190:1647–56. doi: 10.1084/jem.190.11.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.von Boehmer H, Aifantis I, Feinberg J, Lechner O, Saint-Ruf C, Walter U, Buer J, Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol. 1999;11:135–42. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- 148.Rothenberg EV, Yui MA, Telfer JC. T-Cell Developmental Biology. In: Paul WE, editor. Fundamental Immunology. 5. Philadelphia: Lippincott, Williams & Wilkins; 2003. pp. 259–301. [Google Scholar]
- 149.Xi H, Kersh GJ. Sustained early growth response gene 3 expression inhibits the survival of CD4/CD8 double-positive thymocytes. J Immunol. 2004;173:340–8. doi: 10.4049/jimmunol.173.1.340. [DOI] [PubMed] [Google Scholar]
- 150.Carleton M, Haks MC, Smeele SA, Jones A, Belkowski SM, Berger MA, Linsley P, Kruisbeek AM, Wiest DL. Early growth response transcription factors are required for development of CD4-CD8- thymocytes to the CD4+CD8+ stage. J Immunol. 2002;168:1649–58. doi: 10.4049/jimmunol.168.4.1649. [DOI] [PubMed] [Google Scholar]
- 151.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–46. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen D, Rothenberg EV. Molecular basis for developmental changes in interleukin-2 gene inducibility. Mol Cell Biol. 1993;13:228–37. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rincon M, Flavell RA. Regulation of AP-1 and NFAT transcription factors during thymic selection of T cells. Mol Cell Biol. 1996;16:1074–84. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen F, Chen D, Rothenberg EV. Specific regulation of Fos family transcription factors in thymocytes at two developmental checkpoints. Int Immunol. 1999;11:677–88. doi: 10.1093/intimm/11.5.677. [DOI] [PubMed] [Google Scholar]
- 155.He Y-W, Beers C, Deftos ML, Ojala EW, Forbush KA, Bevan MJ. Down-regulation of the orphan nuclear receptor RORγt is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–74. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- 156.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of γδ cell differentiation by αβ T cell progenitors. Science. 2005;307:925–8. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 157.Rothenberg EV. T-lineage specification and commitment: a gene regulation perspective. Semin Immunol. 2002;14:431–40. doi: 10.1016/s1044532302000787. [DOI] [PubMed] [Google Scholar]
- 158.Schmitt TM, Zúñiga-Pflücker JC. Thymus-derived signals regulate early T-cell development. Crit Rev Immunol. 2005;25:141–60. doi: 10.1615/critrevimmunol.v25.i2.40. [DOI] [PubMed] [Google Scholar]
- 159.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 2002;16:869–79. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 160.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Deftos ML, He Y-W, Ojala EW, Bevan MJ. Correlating Notch signaling with thymocyte maturation. Immunity. 1998;9:777–86. doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Greenbaum S, Zhuang Y. Regulation of early lymphocyte development by E2A family proteins. Semin Immunol. 2002;14:405–14. doi: 10.1016/s1044532302000751. [DOI] [PubMed] [Google Scholar]
- 163.Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nature Rev Immunol. 2002;1:193–9. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- 164.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–71. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 165.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci U S A. 2001;98:5164–9. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nishimura M, Fukushima-Nakase Y, Fujita Y, Nakao M, Toda S, Kitamura N, Abe T, Okuda T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–70. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- 167.Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Hirai H, Ogawa S, Kurokawa M. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 168.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae S-C, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 169.Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–44. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–19. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zeng H, Yücel R, Kosan C, Klein-Hitpass L, Möröy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–25. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 173.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 174.Spain LM, Guerriero A, Kunjibettu S, Scott EW. T cell development in PU.1-deficient mice. J Immunol. 1999;163:2681–7. [PubMed] [Google Scholar]
- 175.Zaldumbide A, Carlotti F, Pognonec P, Boulukos KE. The role of the Ets2 transcription factor in the proliferation, maturation, and survival of mouse thymocytes. J Immunol. 2002;169:4873–81. doi: 10.4049/jimmunol.169.9.4873. [DOI] [PubMed] [Google Scholar]
- 176.Xue H-H, Bollenbacher J, Rovella V, Tripuraneni R, Du Y-B, Liu C-Y, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nat Immunol. 2004;5:1036–44. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- 177.Bories J-C, Willerford DM, Grévin D, Davidson L, Camus A, Martin P, Stéhelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–8. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 178.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 179.Barton K, Muthusamy N, Fischer C, Ting C-N, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 180.Eyquem S, Chemin K, Fasseu M, Bories JC. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor β locus. Proc Natl Acad Sci U S A. 2004;101:15712–7. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS protein MEF plays a critical role in perforin gene expression and the development of Natural Killer and NK-T cells. Immunity. 2002;17:437–49. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 182.Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood. 2001;97:2908–12. doi: 10.1182/blood.v97.9.2908. [DOI] [PubMed] [Google Scholar]
- 183.Schotte R, Rissoan MC, Bendriss-Vermare N, Bridon JM, Duhen T, Weijer K, Briere F, Spits H. The transcription factor Spi-B is expressed in plasmacytoid DC precursors and inhibits T-, B-, and NK-cell development. Blood. 2003;101:1015–23. doi: 10.1182/blood-2002-02-0438. [DOI] [PubMed] [Google Scholar]
- 184.Dahl R, Ramirez-Bergeron DL, Rao S, Simon MC. Spi-B can functionally replace PU.1 in myeloid but not lymphoid development. EMBO J. 2002;21:2220–30. doi: 10.1093/emboj/21.9.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–70. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- 186.Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, Moroy T, Taniuchi I, Steimle V, Littman DR, Ehlers M. Runx3 regulates Integrin αE/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J Immunol. 2005;175:1694–705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 187.Blom B, Heemskerk MHM, Verschuren MCM, van Dongen JJM, Stegmann APA, Bakker AQ, Couwenberg F, Res PCM, Spits H. Disruption of αβ but not of γδ T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO J. 1999;18:2793–802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of γδ TCR signaling efficiently diverts thymocytes to the αβ lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 189.Hayes SM, Shores EW, Love PE. An architectural perspective on signaling by the pre-, αβ and γδ T cell receptors. Immunol Rev. 2003;191:28–37. doi: 10.1034/j.1600-065x.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 190.Hayes SM, Li L, Love PE. TCR signal strength influences αβ/γδ lineage fate. Immunity. 2005;22:583–93. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 191.Adachi S, Rothenberg EV. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in competent, nontranscribing T-cells. Nucleic Acids Res. 2005;33:3200–10. doi: 10.1093/nar/gki637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang H, Diamond RA, Rothenberg EV. Cross-lineage expression of Ig-β (B29) in thymocytes: positive and negative gene regulation to establish T-cell identity. Proc Natl Acad Sci USA. 1998;95:6831–6. doi: 10.1073/pnas.95.12.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]


