Abstract
Although the placebo effect is known to have a strong impact on the outcomes of clinical trials, methods for measuring it are limited to physiological observations. We propose a method of localizing, identifying and measuring placebo and treatment-induced networks in the brain using functional neuroimaging. Measuring the relative activation of these “placebo” brain networks serves as a proxy for the placebo effect contained within the variable of interest (depression rating, blood pressure, etc). Analogous to the difference between a paired and unpaired t-test, this allows for a sharp gain in power and reduction in the sample sizes needed in clinical trials, potentially leading to a drastically smaller sample sizes for establishing efficacy, a shorter time-to-market for a drug, and a drastic reduction in the cost of bringing new drugs into the market.
Keywords: placebo effect, clinical trials, spectral clustering, fMRI, ICA
1. Introduction
The “placebo effect” refers to the remarkable finding that many inert medical interventions, especially simulated drugs, are nevertheless powerful in reducing disease symptoms and occasionally curing disease. It has been described in medical texts as early as 1785, but the physical basis of the placebo effect is not known. The negative impacts of placebo affect all of healthcare, as the effects of otherwise useful drugs may be only small compared to placebo, thereby requiring extremely large clinical trials in order to achieve a statistically significant effect. The consequences include costs in the hundreds of millions of dollars, per new drug, as well as the inability to release chemically effective compounds that might be useful for patients. The problems of placebo are especially relevant to neuropsychiatric disorders such as depression (itself a major cause of death and disability), where placebo effects are particularly large, making it exceedingly difficult to study the relative efficacy of new compounds.
In an fMRI study on nicotine cravings induced in smokers, three sets of patients were divided into three treatment groups. The first group was medicinal (bupropion), the second group was an inert treatment (placebo pill) group, and the third group received counseling for smoking cessation. These 51 subjects were scanned up to three times, both pre- and post-treatment for a total of 304 fMRI scans. These patients were scanned while viewing video-cues meant to induce craving. Using the counseling group as a control for both the inert and active treatment groups, we present a method for isolating brain networks which tend to show up in patients post-treatment after receiving either an inert or active pill, yet occur infrequently pre-treatment and post-treatment for the counseling group. It is this network which we posit as being a possible “placebo” network, and propose a methodology for using the relative weighting of this network during scan-time to model the placebo effects within all patients.
We describe a powerful computational method which, when applied to images of the brain, is able to detect and to quantify the placebo effect using changes in brain activity, making it possible to measure on single individuals the added effect of drugs over the placebo. The result is that the pure treatment effect in the brain can be captured, when biologically justified. By measuring the placebo effect within the brain, the number of patients needed in both the placebo and control groups is diminished drastically by capturing the placebo effect within both the treatment and control group. Our method has been validated already within a set of typical functional magnetic resonance imaging (fMRI) data. A provisional patent for this work has been filed with the USPTO.
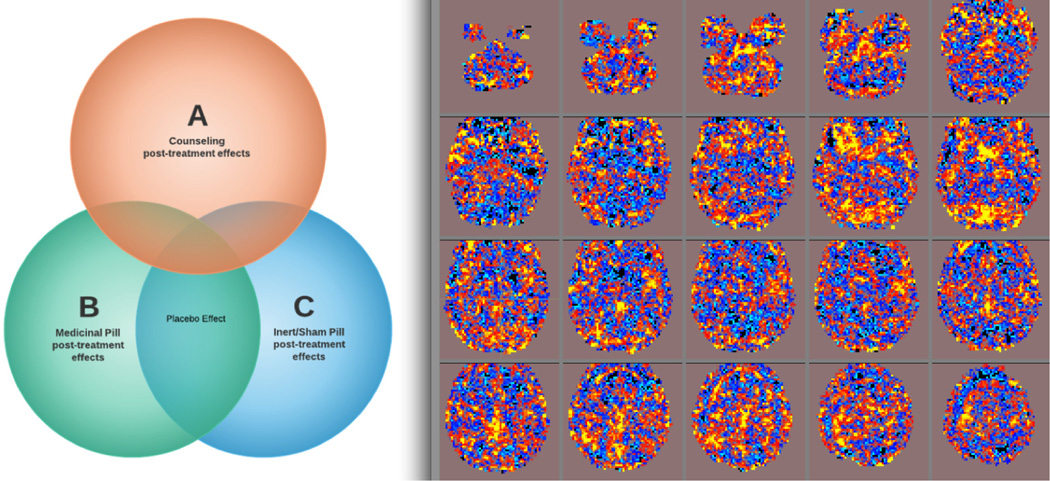
2. Nominating Placebo Networks in the Brain
Let a patient’s brain scan be modeled as a set of networks, operating longitudinally and additively over time. We can describe the placebo effect(s) as an event (network) occurring in the post-treatment groups that is only minimally present during pre-treatment. The cumulative placebo effect can be divided into sub-effects: (1) the effect of the actual pill which is induced by the patient knowing that he or she is receiving a possible powerful medication, and (2) the effect of a patient’s mental status (which we call the optimism effect). If A is the set of counseling post-treatment effects, B is the set of medicinal pill post-treatment effects, and C is the inert/sham pill post-treatment effects, we can formally define the pill effect as an event occurring in A′ ∩ (B∩C) where A′ denotes the complement of A (Figure 1). The events contained in A ∩ B ∩ C are post-treatment effects common to receiving any interventions, possibly identifiable as either the optimism effect or the natural history effect.
Figure 1.
(L) Conceptual localization of the placebo effect: a post-treatment network occurring in inert and active medication groups, yet not in the counseling group. (R) Component map as placebo nominee, identified by spectral clustering and post-hoc identification of networks typically present only after subjects have been treated with a pill (active or sham).
The blind signal separation method of independent component analysis (ICA) is a machine-learning technique that decomposes a four-dimensional fMRI scan into a set of three-dimensional volumes [1]. We use this method to nominate events (brain networks) that are present within a brain scan using the FAST-ICA routine of MELODIC in FSL [2], where each component is temporally modulated by a unique time-course. This procedure is done within-scan on the fMRI scans both pre and post-treatment, to yield 21,256 spatial maps pooled across subjects. These elements form an event space consisting of functional networks present over all scans, for all treatment groups, corresponding to the stage (pre, post-treatment) and subgroup (medicinal, inert or counseling) from which they have originated.
The normalized component maps are realigned to the standard MNI atlas space. We then reduce the spatial dimensionality of the ICA components as follows: the ICA maps from five subjects within each treatment group are concatenated together, providing a single fMRI image containing the IC maps from 15 subjects total. PCA is run on these scans, extracting 50 prototype maps across the 15 subjects. All components then are regressed onto this set of PCA maps, thereby compressing each component’s spatial dimension into 50 dimensions. This results in 21,256 total ICs which are each 50-dimensional observations. Conceptually, the PCA maps are each considered a factor, and the 50 loadings of each of the ICA maps on the PCA factors are used to reduce the total spatial dimensionality of the ICA images.
These maps are then clustered using spectral clustering [3]. In the spectral clustering framework, the eigenvalues of a distance matrix are used to partition the observations into clusters. Often non-linear distance metrics are used, which have proven more effective in identifying underlying cluster since they do not require harsh, unrealistic assumptions such as Gaussianity of cluster density. Since it is not computationally possible to run spectral clustering on all 21,256 dimension-reduced components simultaneously, we perform the clustering in partitions. We run the spectral clustering algorithm with a radial basis kernel Gaussian function. We first create 100 clusters on subsets of roughly 5000 components, next pool the centroids (center of each cluster), and then recluster these to map the centroids’ identities across the four partitions. This mapping is used to establish 100 clusters that are common and systematic across all subjects and scans.
Each scan can then be described categorically by the cluster membership of its components. These clusters correspond to spatial maps denoting frequent patterns of brain activity across subjects and scans. We wish to map which cluster(s) happen most frequently in the event of interest: post-treatment for subjects who have had either an active or inert treatment, but not in any other subgroup. We do this by identifying which cluster(s) correspond to the maximal purity and coverage. Purity is defined by the percentage of components within the cluster that are from the desired group, while coverage represents what fraction of subjects are represented within that cluster. An “ideal” placebo cluster would contain components that are mainly from patients who have received a pill, post-treatment (purity). Most post-treatment pill scans would be contained within that cluster (coverage). We then isolate the centroid which maximizes the purity and the coverage, and reconstruct this centroid as a full image map using the original principal components.
Using this technique with 100 clusters, we dimension reduce the 21,256 components using PCA, cluster using spectral clustering, and then compute the purity and coverage for each observed cluster. The cluster with the maximal purity and coverage we hypothesize as being the most likely placebo network. This map is shown in Figure 1. In practice, this map would be used to measure the placebo network before and after the intervention, by using it as a regressor. The differential effect weighting would be an indirect placebo effect measure. We simulate this model in the following section, showing the increased power, and necessarily decreased sample sizes, needed to obtain it, using the indirect placebo effect obtained from imaging.
3. Simulating the Increase in Detection Power from Adding Placebo Imaging
Let A by a subset of patients assigned to an active treatment, and B the patient group assigned to the inert (placebo) treatment. Let aij represent the measurement of patient i during phase j = pre, post of the trial. We wish to infer whether a treatment is effective, given the difference between the within-subject measurement before and after the treatment is applied. The placebo effect ρ is assumed to be present in both groups, while the treatment effect τ is assumed to exist only within the group receiving active medication. We assume the effects are additive, independent, and linear. Then the expected intervention effect for the inert group is E(bpre − bpost) = ρ + ε, while the expected intervention effect for the active medication group is E(bpre − bpost) = ρ + τ + ε.
We simulate the efficiency gain in a clinical trial using the proposed method. The “active” treatment group A is simulated pretreatment by apre ~ N(0,1), and the “inert” treatment group is modeled by bpre ~ N(0,1). The placebo effects ρ = (ρ1,ρ2) are simulated using Gibbs sampling from a bivariate Normal distribution such that
The first dimension ρ1 represents the direct placebo effect, which is measured within the value apost and bpost, while ρ2 is the indirect placebo effect measured using brain imaging. The treatment effect for patient i is simulated by ti ~ N(τ,1). The post-treatment measurement for patient i within the active group is , while the post-treatment measurement for patient i of the inert group is . We wish to test the efficiency gained by introducing the indirect measure of the placebo effect, p2, into the model. Therefore, we test two models under different conditions, to assess how the treatment effect size τ, within-group sample size n, and covariance σ change the detection rate of non-zero treatment effects. These models are:
Model 1: Traditional t-test, unequal variances of groups A and B
Model 2: xpost − xpre = GLM(𝟙Active + p2), where regressor 𝟙 denotes the indicator function of whether the patient x is a member of the active or inert treatment groups, regressor p2 is the indirect placebo effect measured using brain imaging, and GLM is the general linear model.
We simulate data and test these models under three conditions to assess the change in power, varying separately the effect size, covariance of the indirect and direct placebos, and the sample sizes. These conditions are:
Condition 1: n1 = n2 = 1000, σ = .8, ρ1 = ρ2 = .1. The treatment effect τ is varied from (0, .01, .02, …, 1). For each value of τ, samples of 2000 total patients are simulated using the specified parameters, and Model 1 and Model 2 are used to test the hypothesis that τ = 0. 1000 realizations and tests are performed at each such τ.
Condition 2: n1 = n2 = 1000, τ = .1, ρ1 = ρ2 = .1. The covariance between the direct and indirect placebo effects, σ, is varied from (0, .01, .02, …, 1) For each value of σ, 2000 patients are generated 1000 times. For each generation, Model 1 and Model 2 are used to test the hypothesis that τ = 0. This trivial effect size may be clinically uninteresting, but is useful for testing model power since such small effect sizes are extremely difficult to detect in the presence of high noise.
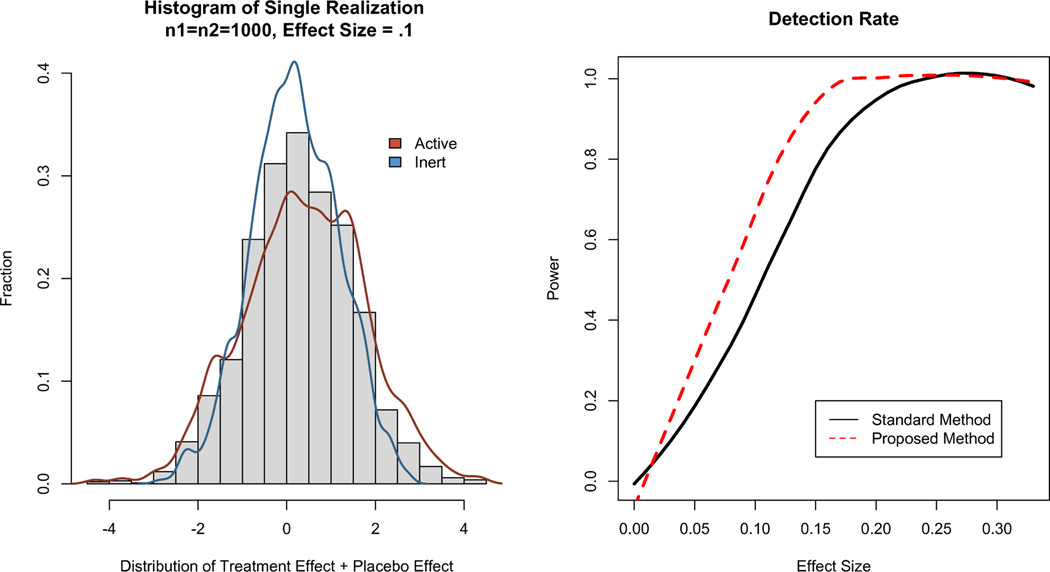
Condition 3: τ = .1, σ = .8, ρ1 = ρ2 = .1. The number of patients in each group, ni, is varied from (30, 31, 32, …, 1000). For each value n of patients, 1000 realizations of size 2n are made, and the two models test the likelihood that τ = 0 given the observations. A single realization of this model with σ = .8, ρ = .1, and τ = .1 is shown in Figure 2 for n1 = n2 = 1000.
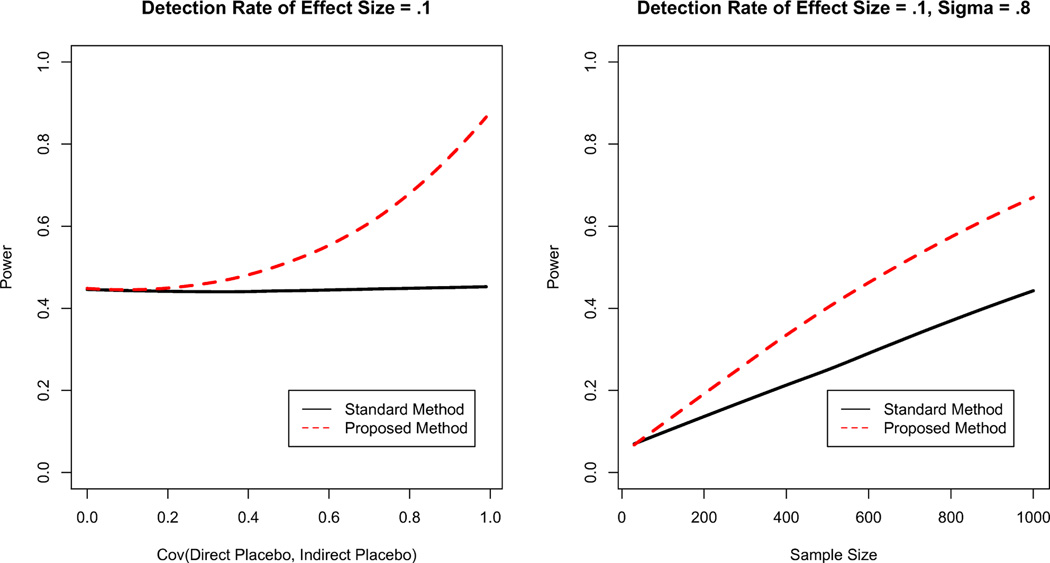
Comparative results for the two models are shown in Figures 2 (Condition 1), 3 (Condition 2), and 3 (Condition 3), using Lowess smoothing for visualization clarity. For all conditions, the proposed method is better able to identify non-zero treatment effects. This identification comes with smaller sample sizes, with greater gains for larger sample sizes. The larger the covariance between the direct and indirect placebo effects Cov(ρ1,ρ2), the greater the difference in detection power.
Figure 2.
(L) A realization of n1 = n2 = 1000, for τ = .1 and ρ = .1. The placebo and treatment groups are separated by τ = .1 (R) Probability of Correctly Rejecting H0 by τ, given ρ = .1, n1 = n2 = 1000. Holding sample size constant, the proposed method is able to detect smaller effect sizes with greater power.
Figure 3.
(L) Probability of Correctly Rejecting H0 by σ = Cov(p1, p2). As the covariance between the direct and indirect placebo effects increases, the power of detection increasingly outperforms conventional analysis. (R) Probability of Correctly Rejecting H0 by Sample Size for effect size τ = .1. The proposed method has more power than the traditional model even for small sample sizes.
The difference in power is even more increased for larger sample sizes, but shows value even for small samples. Given the effect size was strikingly small relative to the variance as shown in Figure 2, this difference in power becomes even more beneficial for implementation in clinical trials. The trivial effect size in itself is not significant, but conclusively determining that the treatment effect is negligibly small distinguishes the weak from the powerful interventions. This echoes the fallacy of classical inference, where any effect size can be found significant with a large enough sample. A “statistically significant” trivial effect size is still trivial. However, establishing the treatment has a trivial effect size using a limited sample size is more informative than merely “failing to reject” the null hypothesis, holding the sample size constant.
4. Discussion
Using existing neuroimaging and treatment data, a prototype has been developed where the placebo network(s) are identified, optimized and measured as described above. The procedure has been applied to isolate and measure placebo networks as defined above. Work was performed within R [4]. This can be considered a completed development that is applicable immediately. Nevertheless, work continues on further optimization. In an implementation, subjects would be scanned pre and post-treatment using a functional brain imaging technique. Using the algorithm described above, the placebo network(s) would be identified and optimized. The relative change in activity in the desired networks between treatment conditions would be a measure of the placebos’ and treatments’ power, and these values would be used to update estimates of the treatment effect and placebo effect(s).
The complementary result is that these methods also allow isolation of the psychologically induced treatment effect, allowing, for example, identification and measurement of regions activated by depression or pain medication. Models testing the effects of the treatment(s) would include as a covariate the placebo imaging effect, and could additionally include as a response the treatment imaging effect. By measuring the treatment effect both psychologically (brain image activity) and physiologically (variable of interest: blood pressure, depression score rating, etc), the analysis becomes a MANOVA where the same effect is captured multiple ways, thus increasing accuracy, decreasing the estimate’s variance, increasing the power, and decreasing the necessary sample size.
5. Conclusion
The general architecture presented herein is a technique that identifies, optimizes and measures placebo effects, giving a quantitative value to the rise in placebo activity during the post-treatment phase. This value then can be used to inform and update estimates of the placebo and treatment effects. Although we did not demonstrate isolating the treatment network here because of brevity, the same models used to isolate and measure the placebo networks can be used to isolate the treatment networks.
If common placebo networks exist in the brain, then the limit of this technology is to turn every patient into its own control, and every trial into a modified cross-over trial. The treatment and placebo effects could be simultaneously measured and contrasted in a single patient, which would be of extreme value not just for regular clinical trials, but also for “orphan drug” trials where it is not possible to recruit large numbers of patients. Because this methodology is new, only future analysis will be able to answer the question of whether a single, or a common, placebo network exists among different disorders and treatments.
Acknowledgments
We thank NIH R21DA26109 to MSC and Burroughs Wellcome Fund Collaborative Research Fellowship to AA for funding this research.
References
- 1.Hyvärinen Aapo, Oja Erkki. Independent component analysis: Algorithms and applications. Neural Networks. 2000;13(4–5):411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 2.Smith Stephen M, Jenkinson Mark, Woolrich Mark W, Beckmann Christian F, Behrens Timothy EJ, Johansen-berg Heidi, Bannister Peter R, De Luca Marilena, Drobnjak Ivana, Flitney David E, Niazy Rami K, Saunders James, Vickers John, Zhang Yongyue, De Stefano Nicola, Brady J Michael, Matthews Paul M. Advances in functional and structural MR image analysis and implementation as fsl. NeuroImage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 3.Ng Andrew Y, Jordan Michael I, Weiss Yair. ADVANCES IN NEURAL INFORMATION PROCESSING SYSTEMS. MIT Press; 2001. On spectral clustering: Analysis and an algorithm; pp. 849–856. [Google Scholar]
- 4.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0. [Google Scholar]