Abstract
Dietary interventions like gluten-free and casein-free diets have been reported to improve intestinal, autoimmune and neurological symptoms in patients with a variety of conditions; however, the underlying mechanism of benefit for such diets remains unclear. Epigenetic programming, including CpG methylation and histone modifications, occurring during early postnatal development can influence the risk of disease in later life, and such programming may be modulated by nutritional factors such as milk and wheat, especially during the transition from a solely milk-based diet to one that includes other forms of nutrition. The hydrolytic digestion of casein (a major milk protein) and gliadin (a wheat-derived protein) releases peptides with opioid activity, and in the present study, we demonstrate that these food-derived proline-rich opioid peptides modulate cysteine uptake in cultured human neuronal and gastrointestinal (GI) epithelial cells via activation of opioid receptors. Decreases in cysteine uptake were associated with changes in the intracellular antioxidant glutathione and the methyl donor S-adenosylmethionine. Bovine and human casein-derived opioid peptides increased genome-wide DNA methylation in the transcription start site region with a potency order similar to their inhibition of cysteine uptake. Altered expression of genes involved in redox and methylation homeostasis was also observed. These results illustrate the potential of milk- and wheat-derived peptides to exert antioxidant and epigenetic changes which may be particularly important during the postnatal transition from placental to GI nutrition. Differences between peptides derived from human and bovine milk may contribute to developmental differences between breastfed and formula-fed infants. Restricted antioxidant capacity, caused by wheat- and milk-derived opioid peptides, may predispose susceptible individuals to inflammation and systemic oxidation, partly explaining the benefits of gluten-free or casein-free diets.
Keywords: Glutathione, casomorphin, gliadin, autism spectrum disorder, schizophrenia, celiac disease, gluten-free/casein-free diet
Introduction
Gluten-free or casein-free diets have been reported to improve intestinal, autoimmune and neurological symptoms in celiac disease (1,2), autism (3,4) and schizophrenia (5,6), but the underlying mechanism of benefit for such diets remains unclear. Emerging evidence from biochemical investigations indicate that neurological disorders like schizophrenia and autism spectrum disorder (ASD) might have underlying defects in pre- and early postnatal neurodevelopment as a contributing factor to the aetiology of these disorders. Especially, systemic oxidative stress has been strongly reported in schizophrenic (7) and ASD (8) patients, associated with significantly lower levels of the antioxidant glutathione (GSH), whose synthesis is limited by cysteine availability. Consistently, brain GSH levels are also reported to be decreased in autism (9) and schizophrenia (10), strongly supporting the proposal that a deficit in this specific antioxidant, namely GSH, might contribute to neurodevelopmental and neuropsychiatric disorders. Notably, intestinal mucosa and plasma levels of GSH are also reduced in pediatric celiac disease patients (11). Moreover, in few cases, patients with neurological conditions (12) like autism (13) and schizophrenia (14) also report a co-morbid problem of celiac disease and/or gut inflammation.
Oxidative stress and DNA methylation changes are metabolically coupled via the transsulfuration pathway and one-carbon metabolism. DNA methylation is carried out by a class of enzymes called DNA methyltransferases DNMTs, which depend on the levels of S-adenosylmethionine (SAM) (15), which in turn are dependent on the action of the enzyme methionine synthase (MS) and the redox status of the cell (16). SAM acts a methyl donor for over 200 methylation reactions and is converted to S-adenosylhomocysteine (SAH), an inhibitor of methylation reactions. Hence, the ratio SAM/SAH is termed as the methylation capacity of the cell. Several studies report decreased levels of SAM and methylation capacity in patients with ASD and schizophrenia. DNA methylation is an important methylation reaction, involving addition of a methyl groups at CpG sites on DNA, and it is a major factor for epigenetic-based changes in gene expression. In recent studies abnormal DNA methylation patterns have been implicated in schizophrenia (17,18) and ASD (19,20), but more prominently in the latter. Rett syndrome, immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome and autosomal dominant cerebellar ataxia are few examples of a heterogeneous group of neurodegenerative and neurodevelopmental disorders caused by changes in proteins that either 'read' or 'write' DNA methylation, resulting in disordered DNA methylation at numerous single-copy genes as well as in specific sequences of repeat DNA in parts of constitutive heterochromatin (17). Similarly, deficiency of B-vitamins, methionine, folic acid, and/or choline can significantly alter DNA methylation by affecting levels of SAM and SAH (21,22). However, while these studies indicate the interaction between nutrients and the pathways regulating DNA methylation, it is still unclear how a specific gene is targeted by nutrients or alternatively, if these changes are stochastic in nature (21,23).
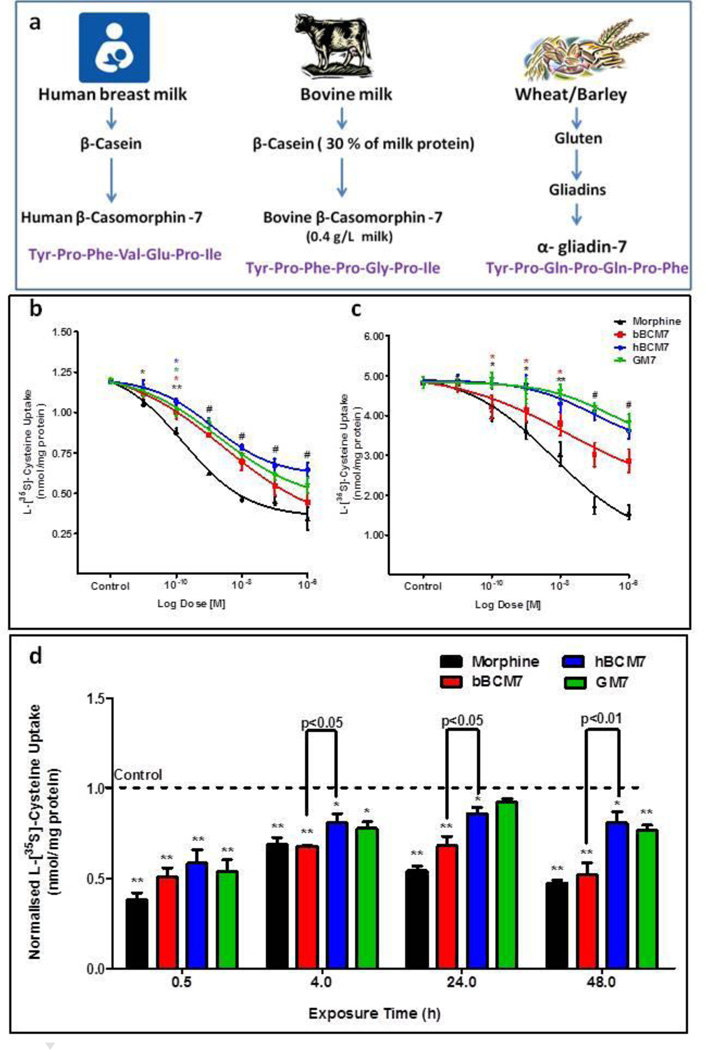
Epigenetic programming during early postnatal development can influence lifelong disease risk (24,25), and may be modulated by nutritional factors, especially factors affecting DNA methylation (26,27). Milk (either human or bovine) or milk-derived products may be the only source of nutrition for infants during the early postnatal development, while wheat-containing foods are commonly introduced after the first year. Casein is an important milk protein and similarly gliadin is a wheat gluten-derived protein, and hydrolytic digestion of both releases peptides with opioid activity (28,29). Enzymatic digestion of human β-casein liberates the heptapeptide hβ-casomorphin-7 (YPFVEPL), designated hBCM7, while bovine β-casein yields bβ-casomorphin-7 (YPFPGPL; bBCM7), and α-gliadin yields gliadinomorphin-7 (YPQPQPF; GM7) (Fig. 1a). Divergent amino acids at positions 3–5 result in distinctive conformations, and the presence of 2 or 3 proline residues makes these peptides resistant to further proteolysis (30). Each of these homologous proline-rich peptides has the capacity to activate opioid receptors (28–31).
Figure 1. Food-derived opioid peptides and morphine inhibit cysteine uptake in SH-SY5Y cells.
a, Beta casein from milk and alpha gliadin from wheat, barley and rye are hydrolysed in the intestine by pepsin, leucine aminopeptidase (LAP) and elastase to release peptides with opioid activity. The 7 aa proline-containing peptides from human and bovine milk, and that from wheat, have homologous sequences, including two or three proline residues. b, Radiolabeled cysteine uptake in SH-SY5Y human neuroblastoma and c, Caco2 colon carcinoma cell lines in the presence of increasing concentrations (0.1 nM to 1 µM) of morphine or opioid peptides. Cells were treated for 30 min. d, Time-dependent effects of opioid peptides and morphine on cysteine uptake by SH-SY5Y cells. SH-SY5Y cells were treated with 1 µM of morphine or opioid peptides for 0.5, 4, 24 and 48 h (n = 6). (*) p < 0.05 and (**) p < 0.01 vs. control.
Endogenous opioid peptides normally found in the body are called endorphins (e.g. beta-endorphins, enkephalins), whereas exogenous opioid peptides (e.g. morphine, hBCM7, bBCM7 and GM7) are called exorphins. Low energy conformations of the food-derived exorphins bear a structural similarity to morphine (Supplementary Fig. 1), particularly at their N-terminal tyrosine residue, which is also a feature of endorphins and is critical for their affinity and efficacy at opioid receptors (32). Studies show that morphine-induced activation of µ opioid receptors induces changes in the expression of EAAT3, which provides the primary pathway for cysteine uptake in both mature neurons (33,34) and GI epithelial cells (33,35), particularly in the distal ileum (35). Morphine can induce oxidative stress and cause a pro-oxidant shift in cell redox status (36–38), which subsequently can cause a decrease in the availability of SAM for more than 200 methylation reactions, including DNA methylation, resulting in subsequent transcriptional changes. Hence, the current study investigates whether morphine and food-derived opioid-peptides can induce alterations in cellular redox status, DNA methylation (39), and transcription process, by altering cysteine uptake in SH-SY5Y cells, and provides biochemical evidence which could potentially support the use of nutritional intervention, i.e. a gluten-free/casein-free diet.
Materials and Methods
Materials
Minimum essential medium, alpha-modification (α-MEM), trypsin, Hank’s balanced salt solution (HBSS) and penicillin/streptomycin/fungizone antibiotic solution were purchased from Mediatech (Manassas, VA). Fetal bovine serum (FBS) was obtained from Hyclone (Logan, UT). [35S]-cysteine was purchased from American Radiolabeled Chemicals (St. Louis, MO). Morphine and other drugs were obtained from Sigma Chemicals (St. Louis, MO). Human and bovine forms of BCM-7, as well as gliadin-7 peptide were custom synthesized by Neopeptide (Cambridge, MA).
Cell Culture
Monolayers of cells were grown in 10 cm tissue culture dishes with 10 mL of α-MEM supplemented with 1% penicillin/streptomycin/fungizone and 10% FBS, in 5% CO2 at 37 °C. For most experiments, cells were plated and incubated for 48 h prior to use.
Cysteine Uptake
The cysteine uptake assay was performed according to a method described previously (39). In brief, SH-SY5Y human neuroblastoma cells plated in six-well plates were pre-treated with drugs and incubated for various times prior to measuring uptake. Media was aspirated and cells were washed with 600 µL of HBSS at 37 °C. Non-radioactive HBSS was aspirated, replaced with 600 µL of 37 °C HBSS containing [35S]-cysteine, (1 µCi/1 mL), 10 µM unlabelled cysteine and 100 µM DTT, and cells were incubated for 5 min. The [35S]-cysteine/HBSS mixture was aspirated and treatment was terminated by two washes with ice-cold HBSS. Cells were then lysed with 600 µL of dH2O, scraped, collected in 1.5 mL microcentrifuge tubes, and sonicated for 10 s. 100 µL of each sample was aliquoted for protein assay. 200 µL of each sample (in triplicate) was aliquoted into scintillation vials with 4 mL of scintillation fluid, vortexed, and counted for radioactivity, normalized against protein content.
Thiol Metabolite Assay
SH-SY5Y neuroblastoma cells were grown to confluence in α-MEM. Media was aspirated and the cells were washed twice with 1 mL of ice cold HBSS. HBSS was aspirated and 0.6 mL ice cold dH2O was added to the cells. Cells were scraped from the flask/dish and suspended in dH2O. The cell suspension was sonicated for 15 s on ice and 100 µL was used to determine protein content. The remaining lysate was added to a microcentrifuge tube and an equal volume of 0.4 N perchloric acid was added, followed by incubation on ice for 5 min. Samples were centrifuged at 5,000 × g and the supernatant transferred to new microcentrifuge tubes. 100 µL of sample was added to a conical micro-autosampler vial and kept at 4 °C in the autosampler cooling tray. 10 µL of this sample was injected into the HPLC system.
The separation of redox and methylation pathway metabolites was accomplished using an Agilent Eclipse XDB-C8 analytical column (3 × 150 mm; 3.5 µm) and an Agilent Eclipse XDB-C8 (4.6 × 12.5 mm; 5 µm) guard column. Two mobile phases were used: Mobile Phase A was 0% acetonitrile, 25 mM sodium phosphate, 1.4 mM 1-octanesulfonic acid, adjusted to pH 2.65 with phosphoric acid. Mobile Phase B was 50% acetonitrile. The flow rate was initially set at 0.6 mL/min and a step gradient was used: 0–9 min 0% B, 9–19 min 50% B, 19–30 min 50% B. The column was then equilibrated with 5% B for 12 min prior to the next run. Temperature was maintained at 27 °C. The electrochemical detector was an ESA CoulArray with BDD Analytical cell Model 5040 and the operating potential was set at 1500 mV. Sample concentrations were determined from the peak areas of metabolites using standard calibration curves and ESA-supplied HPLC software. Sample concentrations were normalized against protein content. In some cases samples were diluted in mobile phase as needed or up to 50 µl of sample were injected to assure that thiol levels were within the range of the standard curve.
DNA Methylation Analysis
Genomic DNA was extracted from samples with the Easy DNA kit (Invitrogen K1800-01) using the appropriate protocol for cell lines. For a full overview of the MethylCap-Seq protocol, refer to De Meyer, T. et al (2013)(40). In summary, fragmentation was performed on Covaris S2 with following settings: duty cycle 10%, intensity 5, 200 cycles per burst during 200 sec, to obtain fragments with an average length of 200 bp. The power mode is frequency sweeping, temperature 6–8° C, water level 12. A maximum of 5 µg is loaded in 130 µl Tris-EDTA in a microtube with AFA intensifier. For samples with less DNA input (down to 500 ng) we dilute the DNA in 1:5 diluted TrisEDTA. DNA with an input from 5–3 µg is analyzed on the Agilent 2100 on a DNA 1000 chip. DNA with an input lower than 3 µg is concentrated in a rotary evaporator to 25 µl and the fragment distribution is checked on a high sensitivity DNA chip. Methylated DNA was captured using the MethylCap kit (Diagenode, Belgium), according to the manufacturer’s protocol; starting concentration was 200 ng. The yield was typically between 0.5 and 8 ng total captured DNA. Fragments were subsequently sequenced using the Illumina Genome Analyzer II. The concentrations of fragmented and captured DNA were determined on a Fluostar Optima plate reader with the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen P7589) at 480/520nm.
To prepare the DNA library we used the DNA Sample Prep Master Mix Set 1 (NEB E6040) in combination with the Multiplexing Sample Preparation Oligo Kit (96 samples, Illumina PE-400-1001). We use the entire fragmented DNA and followed the NEB protocols. NEBNext End Repair Module Protocol: Purify with a Qiaquick PCR Purification Kit (Qiagen 28104) and elute in 37 µl Elution Buffer (EB). NEBNext dA-tailing Module Protocol: Purify on a Minelute PCR Purification Kit (Qiagen 28004) and elute in 25 µl EB. NEBNext Quick Ligation Module Protocol: Purify on a Minelute PCR Purification Kit (Qiagen 28004) and elute in 30 µl EB, using the multiplexing sequencing adapters provided in the Multiplexing Sample Preparation Oligo Kit. Size selection of the library was done on a 2% agarose gel (Low Range Ultra Agarose Biorad 161-3107). We used a 1Kb Plus ladder (Invitrogen 10787-018) and ran the gel at 120 V for 2 hrs. A fragment of 300 bps +/− 50bps was excised and eluted on a Qiagen Gel Extraction Kit column (Qiagen 28704) and eluted in 23 µl EB.
We followed the Illumina library amplification index protocol with the following alterations: We used 22 µl DNA and performed 21 cycles. The sample was purified on a Qiaquick PCR Purification column (Qiagen 28101) and eluted in 50 µl EB, 1:5 diluted, concentrated in a rotary evaporator to 10 µl and 1 µl was applied to a Agilent 2100 HS DNA chip and the concentration was determined by smear analysis on the Agilent 2100. The samples were diluted to 10 nM. After denaturation with NaOH we diluted the samples to 16 pM. The Paired-End flow cell was prepared according to the Cluster Station User Guide. Sequencing was performed according to the HiSeq user guide (performing a Multiplexed PE Run), with 2 × 51 cycles for the paired end runs.
Statistical Methods
Statistical analyses were carried out using Graph Pad Prism® version 5.01. Student’s t-test for independent means was used to test for significant differences between control and experimental groups. Data were expressed as mean ± standard error of the mean (SEM). Best-fit values were calculated using non-linear and linear regression models. Non-linear regressions used a two-phase exponential decay function. Comparisons between multiple groups of data were conducted using one-way analysis of variance (ANOVA), and Tukey’s post-hoc test was used to determine the differences between individual groups.
Results
We previously characterized EAAT3 expression in SH-SY5Y human neuroblastoma cells and indicated that it makes a major (>90%) contribution to [35S]-cysteine uptake.(39) To investigate whether morphine and opioid peptides could alter EAAT3 activity via µ opioid receptors, we carried out a radiolabelled [35S]-cysteine uptake assay in SH-SY5Y cells. Pre-treatment of SH-SY5Y human neuroblastoma cells with either morphine or casein/gluten-derived exorphin peptides for 30 min inhibited the uptake of radiolabelled [35S]-cysteine in a concentration-dependent manner, with an efficacy order of morphine > bBCM7 > hBCM7 > GM7 and IC50 values of 0.16, 1.31, 4.34 and 1.94 nM, respectively (Fig. 1b). Morphine and exorphin peptides inhibited cysteine uptake with similar efficacy but lower potency in Caco2 cells, a cell line derived from human GI epithelial cells, with IC50 values of 6.38, 15.95, 90.08 and 301 nM, respectively (Fig. 1c). Inhibition of cysteine uptake was fully developed at 30 min and was sustained through 48 h of morphine or exorphin exposure, although the extent of inhibition was modestly decreased in a time-dependent manner. Inhibition by bBCM7 was greater than that of hBCM7 at 4, 24 and 48 h (Fig. 1d). Cysteine uptake effects of morphine and exorphin peptides were blocked by naltrexone, an antagonist of both µ and delta opioid receptors (Supplementary Fig. 2), as well as by D-Phe-Cys-Tyr-D-Trp-Arg-Thr-Pen-Thr (CTAP), a selective µ-antagonist, but not by the delta antagonist naltrindole (NTI) (Supplementary Fig. 3).
As mentioned previously, cysteine is the rate limiting precursor for GSH synthesis. Other source of cysteine involves the transsulfuration pathway via homocysteine (HCY). However, in neuronal cells, this pathway is partially blocked, which emphasizes the role of EAAT3–mediated cysteine uptake for GSH homeostasis. HCY can also be converted to methionine via the activity of MS. To investigate whether decreases in cysteine uptake translate into changes in metabolic intermediates involved in transsulfuration and methionine methylation cycle pathways, as illustrated in Supplementary Fig. 4, we measured their levels in SH-SY5Y cells after treatment with morphine and opioid peptides for different times. Morphine and opioid peptides caused time-dependent decreases in cysteine, GSH and methionine, while homocysteine and cystathionine levels increased (Supplementary Fig. 5). These changes are consistent with decreased GSH synthesis, decreased activity of methionine synthase, and increased transsulfuration of homocysteine to cystathionine.
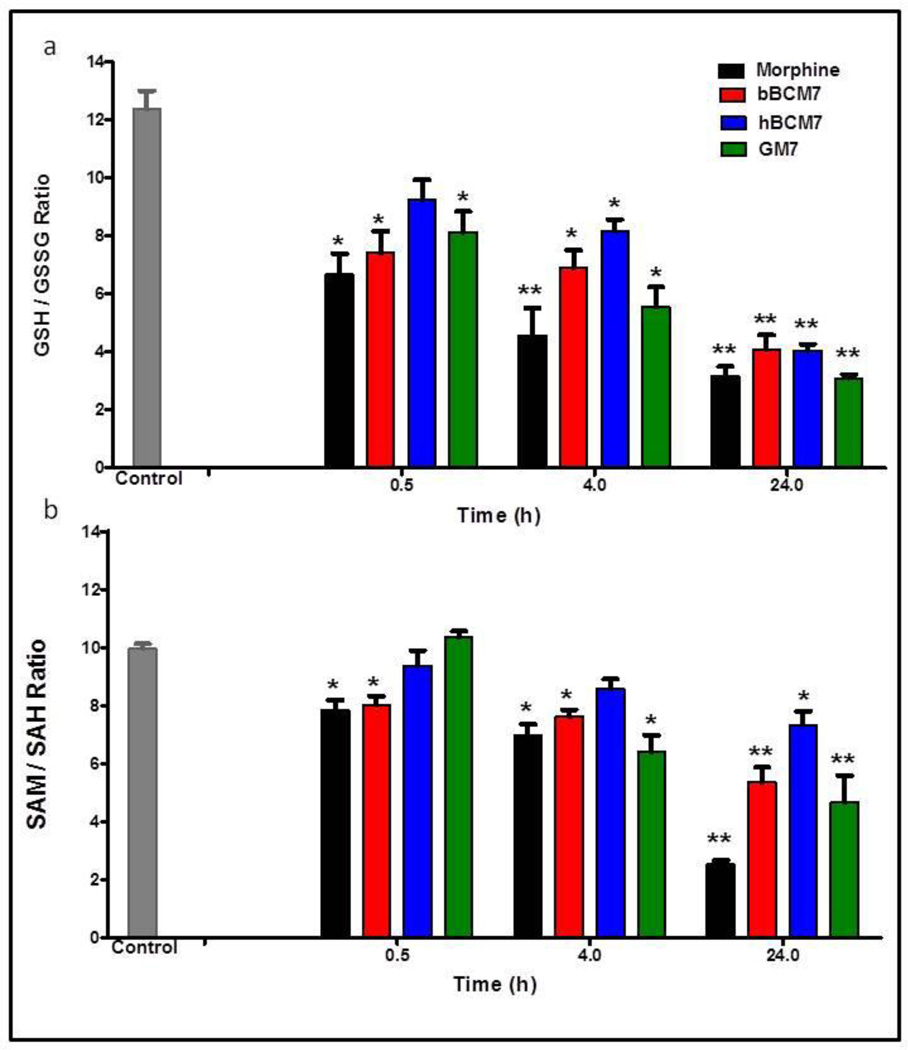
Redox status is reflected by the ratio of reduced GSH to oxidized GSSG (GSH/GSSG), whereas the ratio of SAM to SAH (SAM/SAH) reflects methylation capacity. Morphine and exorphin peptides caused progressive decreases in GSH/GSSG, reaching more than 3-fold at 24 h (p < 0.01, Fig. 2A). Morphine, bBCM7 and GM7, but not hBCM7, transiently decreased SAM/SAH, with a 2–3 fold reduction of SAM/SAH levels observed with morphine, bBCM7 and GM7 treatments at 24 h (p < 0.01, Fig. 2B). Thus the decrease in cysteine uptake caused by morphine and opioid peptides translates into downstream changes affecting redox status and methylation capacity in SH-SY5Y cells.
Figure 2. Effect of opioids on GSH/GSSG and SAM/SAH ratios.
Ratios for a, GSH / GSSG and b, SAM / SAH were calculated after treatment with opioid peptides and morphine for 0.5, 4 and 24 h, at a concentration of 1 µM (n = 4). (*) p < 0.05 and (**) p < 0.01 vs. control.
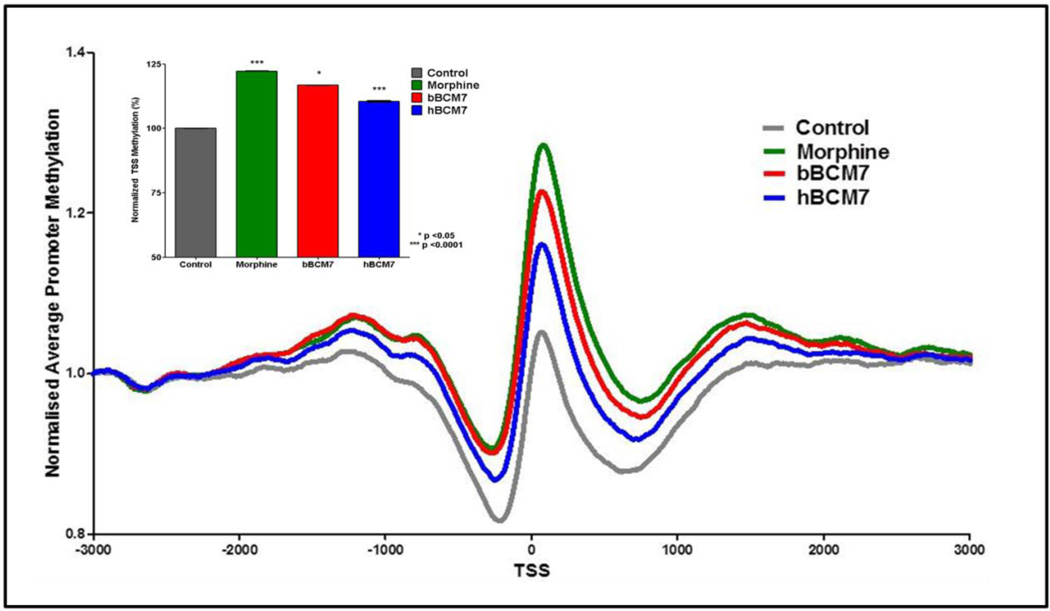
DNA methylation is an important reaction, representing one of the key mechanisms for epigenetic regulation of gene expression, which has particular significance for postnatal development. We used methyl-CpG binding domain (MBD) protein-enriched genome sequencing (MBD-seq) to evaluate the influence of bBCM7 and hBCM7 on global DNA methylation status in SH-SY5Y cells, compared to the effect of morphine. To evaluate DNA methylation status in the promoter region, 53,561 genes were aligned at their transcription start site (TSS) and the level of methylation was calculated for 3,000 bp in either direction, as normalized to the level at the beginning of the region (−3000 bp). As previously described for brain frontal cortex (41), the most prominent feature in the resulting methylation density landscape was an asymmetric peak occurring at about +80 bp, flanked by less methylated valley regions (Fig. 3). A 4 h treatment with morphine or milk-derived opioid peptides (1 µM) caused a significant shift toward increased methylation (i.e., promoter hypermethylation) in the immediate TSS region. Morphine was most effective, increasing genome-wide methylation at the TSS peak to 22% above control levels, followed by bBCM7 and hBCM7, representing increases of 17% and 10%, respectively, a rank order that mirrors their inhibition of cysteine uptake. Morphine treatment significantly altered methylation status of a total of 5,366 genes in this TSS region, bBCM7 3,891 and hBCM7 1,627 (p < 0.05). Thus, opioid-induced changes in cysteine uptake, redox status as well as the SAM/SAH ratios, are associated with significant genome-wide changes in DNA methylation levels at the TSS.
Figure 3. Morphine and milk-derived opioid peptides increase genome-wide promoter methylation.
SH-SY5Y cells were treated with 1 µM morphine, bBCM7 or hBCM7 for 4 h (n = 5) and genome-wide DNA methylation was analysed by MBD-seq. 53,561 genes were aligned at their transcription start site (TSS) and average methylation between −3000 bp and +3000 bp was computed and normalized to values at −3000 bp. Inset: Methylation of all genes was evaluated at the +80 bp peak for each group and expressed as a percentage of the control group level, yielding p-values for morphine, bBCM7 and hBCM7 of 1.8e-25, 0.016 and 3.2e-40, respectively.
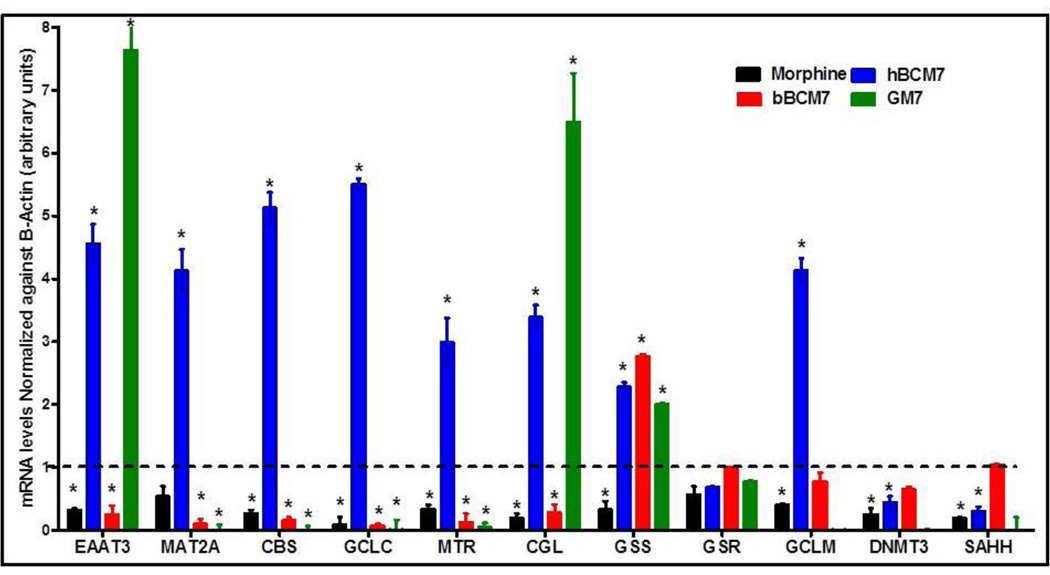
We have previously showed changes in transcription of a panel of 11 redox and methylation pathway genes in association with decreased EAAT3-mediated cysteine uptake in response to the oligomeric Abeta amyloid peptide, implicated in causation of Alzheimer’s disease (39). Hence, we wanted to investigate whether morphine and opioid peptides could induce similar changes in the transcriptional levels of these genes. For this purpose, we used qRT-PCR to evaluate changes in mRNA expression levels after treatment of SH-SY5Y cells with morphine or opioid peptides (4 hrs, 1 µM). hBCM7 significantly increased expression of nine genes in the panel, including EAAT3, GCLC, GCLM, CGL, CBS, MTR, MAT2A, DRD4, and LINE-1 (Fig. 4). In contrast, morphine and bBCM7 decreased the expression of these genes (with the exception of GCLM), while GM7 produced a mixed pattern of increased and decreased gene expression. Thus, food-derived opioid peptides affect redox/methylation gene expression, with hBCM7 and bBCM7 showing opposite effects. These results indicate that morphine and food derived opioid peptides induce changes in the redox and methylation capacity of the cells by modulating cysteine uptake via EAAT3 and subsequent changes in DNA methylation and gene expression levels were observed. However, the changes in the DNA methylation status might not be directly responsible for changes in the transcriptional status of genes involved in the regulation of redox and methylation pathways, and other mechanisms might be involved in regulating mRNA levels (e.g. transcription factors or mRNA stability).
Figure 4. Morphine and opioid peptides alter redox/methylation gene expression.
SH-SY5Y cells were treated with opioid peptides or morphine (1 µM) for 4 h, RNA was isolated and mRNA levels were probed by qRT-PCR with primers designed for a panel of redox and methylation-linked genes. Values were normalized to β-actin levels and to control gene expression (n = 4). The Y-axis indicates fold changes in mRNA levels. (*) indicates significant difference (p < 0.05) from untreated control cells.
Discussion
Food-derived opioid peptides are produced in the gut, where they can exert effects on the gut intestinal epithelium, but they can also enter the systemic circulation (42) and may be able to cross the blood-brain barrier (43). Recently, it has also been revealed that casein-derived opioid peptides are present in the urine of autistic and schizophrenic subjects, but not of controls (44,45). It has been suggested that increased blood levels of bBCM7, in conjunction with lower activity of dipeptidylpeptidase IV (DPPIV), the enzyme responsible for its hydrolysis, may be associated with apnea in sudden infant death syndrome (SIDS) (45), reflecting µ-receptormediated respiratory depression. The current study supports previous results indicating the biochemical effects of the food-derived peptides specifically on the µ-opioid receptor.
Intolerance to casein or gliadin can contribute to inflammation of the gastrointestinal (GI) tract as exemplified by celiac disease, as well as systemic effects including neurological manifestations. While true IgE-based allergy can be triggered by gluten in some individuals, intolerance associated with actions of gluten-derived peptides such as GM7, may affect a larger population, and there is evidence that the number of people suffering from gluten intolerance is increasing (46). A gluten-free (GF) diet usually relieves the symptoms of celiac disease. As mentioned previously, plasma and brain GSH is decreased and other thiol metabolites are abnormal in subjects suffering from autism (8) schizophrenia (7), and celiac disease. In our study, we observed similar pattern of decreased GSH as well as altered thiol metabolites after exposure to morphine and opioid peptides. In light of the above clinical evidence and metabolic relationships, data from the current study suggest that the salutary effects of a GF/CF diet might reflect actions of the opioid peptides released from milk and wheat on biochemical pathways via modulating the cellular redox status.
Only anecdotal evidence is available for the nutritional and dietary interventions recommended for treatment of ASD, schizophrenia and celiac disease patients, and supporting mechanistic or biochemical investigation is rarely performed. The current study is the first to describe the ability of food-derived opioid peptides and morphine to modulate cysteine uptake, with subsequent effects on cellular redox and methylation status leading to global changes in DNA methylation and gene transcription. As summarized in Fig. 5, these actions constitute a biochemical pathway, namely a redox/methylation signalling pathway, which extends our current understanding of the physiological and pharmacological effects of food-derived opioid peptides. In individuals who are sensitive to milk or wheat, these actions of food-derived opioid peptides may contribute to inflammation and, in the case of milk-derived hBCM7 and bBCM7, they may influence epigenetic programming during early postnatal development period. Systemic effects on redox, methylation and epigenetic status could potentially result either from an inhibitory influence of opioid peptides on cysteine absorption from the GI tract. Additionally, distinctive epigenetic effects of hBCM7 vs. bBCM7 might also aid to explain the well-recognized health benefits of breast feeding vs. formula feeding.
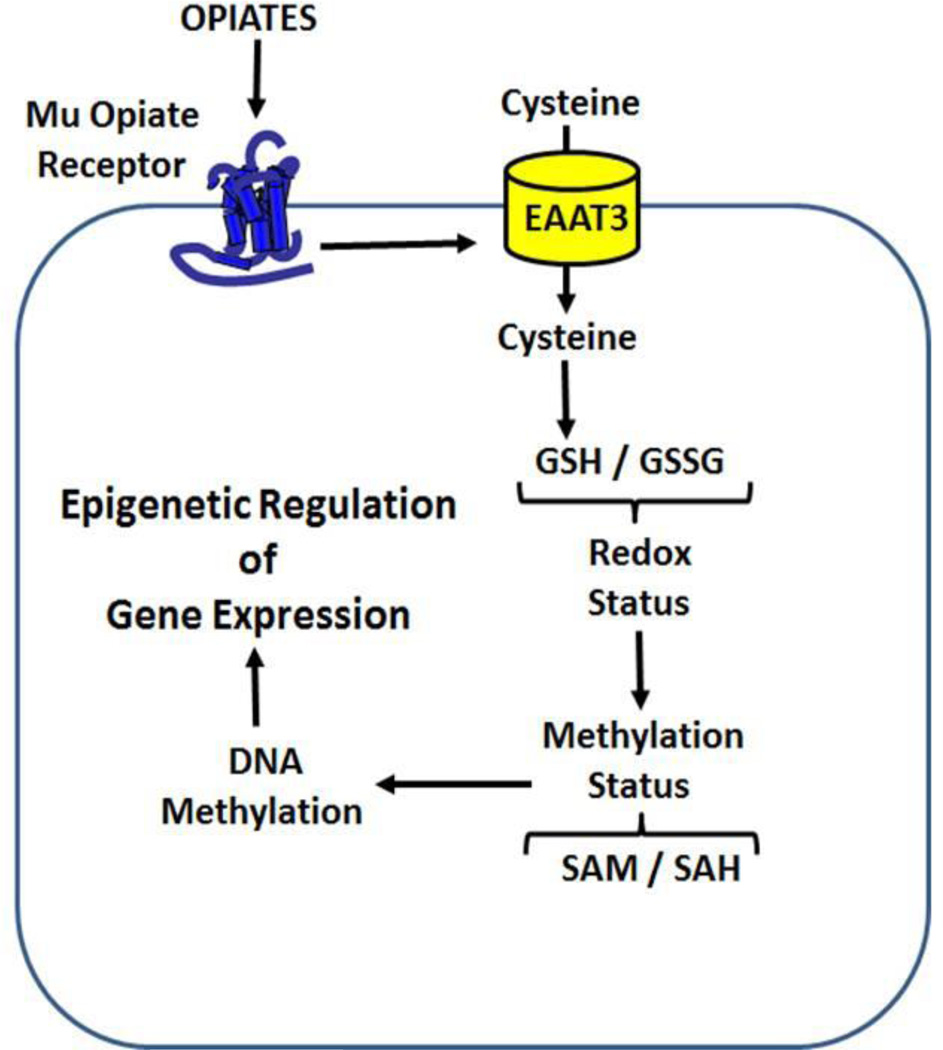
Figure 5. Summary diagram of redox-dependent epigenetic regulation by µ-opioid receptors.
Changes in epigenetic marks such as DNA methylation are crucial for adaptive changes in gene transcription, and early-life experience; for example nutrition, can influence the life-long risk of diseases such as cancer, type 2 diabetes mellitus, obesity, inflammation and neurocognitive disorders (21–23,47). Our findings provide a candidate mechanism by which nutritional and dietary components can affect biochemical pathways and induce changes in epigenetic status and subsequently modulate disease risk. Since EAAT3 is expressed in T-cells, including T-regulatory cells (48), this novel µ-receptor-mediated redox-based signalling pathway may also be important for the immunomodulatory effects of opiate antagonists, such as naltrexone (49). While the GI tract may be the primary locus for opiate peptide effects, their influence can extend throughout the body, by virtue of their influence over systemic antioxidant resources. It should be noted that bovine form of BCM7 is only released from cows with the A1 genotype and not A2 genotype cows.
While the mechanism underlying inhibition of cysteine uptake by food-derived opiate peptides is not fully established, we previously found that protein kinase A (PKA) and/or Mitogen activated protein kinase kinase (MEKK) might be involved in mediating the effects of morphine on EAAT3 (50). The inhibitory effects on cysteine uptake are also observed at the levels of redox status and methylation capacity as well as at the DNA methylation status and the transcription levels of genes involved in maintaining the redox methylation homeostasis. However, the current results do not propose a direct causal relationship between the changes in DNA methylation and the resultant transcriptional state, and several contributing factors, other than the global DNA methylation status, might be affected by opioid peptides. Thus changes in redox and methylation could potentially influence a wide array of factors affecting mRNA levels, including the availability and binding of transcriptional factors or polycomb group proteins, histone modifications or changes in mRNA stability.
It should be noted that the current study was carried out using SH-SY5Y and Caco2 cancer cell lines, which might not reflect responses in non-transformed cells. However, these cell lines have been used previously for epigenetic studies (50,51) and they provide a useful preliminary indication of opiate peptide effects, which need to be replicated in other systems before they can be confidently extrapolated to clinical manifestations. However, the current study does provide the first mechanistic explanation through which nutritional factors might impinge on the epigenetic and transcriptional status of genes. The relationship between gluten- and casein-derived opiate peptide-induced redox and methylation changes and downstream effects of these changes still remains to be elucidated. Taken together, our findings provide a novel antioxidant-based biochemical pathway linking gut and brain function via the diet.
Supplementary Material
Acknowledgements
The authors are appreciative of the encouragement from Drs. Jaqueline McCandless and Karl Reichelt to initiate this research project.
Funding Source:
This work was supported by research grants to RD from the Autism Research Institute, A2 Corporation Limited, and the National Institute for Drug Abuse (R21DA030225). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murray JA, Watson T, Clearman B, Mitros F. Effect of a gluten-free diet on gastrointestinal symptoms in celiac disease. Am J Clin Nutr. 2004 Apr 1;79(4):669–673. doi: 10.1093/ajcn/79.4.669. [DOI] [PubMed] [Google Scholar]
- 2.Benson GD, Kowlessar OD, Sleisenger MH. Adult celiac disease with emphasis upon response to the gluten-free diet. Medicine (Baltimore) 1964 Jan;43:1–40. doi: 10.1097/00005792-196401000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Whiteley P, Rodgers J, Savery D, Shattock P. A Gluten-Free Diet as an Intervention for Autism and Associated Spectrum Disorders: Preliminary Findings. Autism. 1999 Mar 1;3(1):45–65. [Google Scholar]
- 4.Whiteley P, Haracopos D, Knivsberg A-M, Reichelt KL, Parlar S, Jacobsen J, et al. The ScanBrit randomised, controlled, single-blind study of a gluten- and casein-free dietary intervention for children with autism spectrum disorders. Nutr Neurosci. 2010 Apr;13(2):87–100. doi: 10.1179/147683010X12611460763922. [DOI] [PubMed] [Google Scholar]
- 5.Singh MM, Kay SR. Wheat gluten as a pathogenic factor in schizophrenia. Science. 1976 Jan 30;191(4225):401–402. doi: 10.1126/science.1246624. [DOI] [PubMed] [Google Scholar]
- 6.Okusaga O, Yolken RH, Langenberg P, Sleemi A, Kelly DL, Vaswani D, et al. Elevated gliadin antibody levels in individuals with schizophrenia. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2013 Sep;14(7):509–515. doi: 10.3109/15622975.2012.747699. [DOI] [PubMed] [Google Scholar]
- 7.Yao JK, Leonard S, Reddy R. Altered glutathione redox state in schizophrenia. Dis Markers. 2006;22(1–2):83–93. doi: 10.1155/2006/248387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frustaci A, Neri M, Cesario A, Adams JB, Domenici E, Dalla Bernardina B, et al. Oxidative stress-related biomarkers in autism: systematic review and meta-analyses. Free Radic Biol Med. 2012 May 15;52(10):2128–2141. doi: 10.1016/j.freeradbiomed.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, et al. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012 Mar;42(3):367–377. doi: 10.1007/s10803-011-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do KQ, Trabesinger AH, Kirsten-Krüger M, Lauer CJ, Dydak U, Hell D, et al. Schizophrenia: glutathione deficit in cerebrospinal fluid and prefrontal cortex in vivo. Eur J Neurosci. 2000 Oct;12(10):3721–3728. doi: 10.1046/j.1460-9568.2000.00229.x. [DOI] [PubMed] [Google Scholar]
- 11.Stojiljković V, Pejić S, Kasapović J, Gavrilović L, Stojiljković S, Nikolić D, et al. Glutathione redox cycle in small intestinal mucosa and peripheral blood of pediatric celiac disease patients. An Acad Bras Ciênc. 2012 Mar;84(1):175–184. doi: 10.1590/s0001-37652012000100018. [DOI] [PubMed] [Google Scholar]
- 12.Jackson JR, Eaton WW, Cascella NG, Fasano A, Kelly DL. Neurologic and psychiatric manifestations of celiac disease and gluten sensitivity. Psychiatr Q. 2012 Mar;83(1):91–102. doi: 10.1007/s11126-011-9186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genuis SJ, Bouchard TP. Celiac disease presenting as autism. J Child Neurol. 2010 Jan;25(1):114–119. doi: 10.1177/0883073809336127. [DOI] [PubMed] [Google Scholar]
- 14.Cascella NG, Kryszak D, Bhatti B, Gregory P, Kelly DL, Mc Evoy JP, et al. Prevalence of celiac disease and gluten sensitivity in the United States clinical antipsychotic trials of intervention effectiveness study population. Schizophr Bull. 2011 Jan;37(1):94–100. doi: 10.1093/schbul/sbp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng X, Blumenthal R. S-adenosylmethionine-dependent methyltransferases: structures and functions. World Scientific. 1999:422. [Google Scholar]
- 16.Hondorp ER, Matthews RG. Oxidative Stress Inactivates Cobalamin-Independent Methionine Synthase (MetE) in Escherichia coli. PLoS Biol. 2004 Oct 5;2(11):e336. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flight MH. Epigenetics: Methylation and schizophrenia. Nat Rev Neurosci. 2007 Dec;8(12):910–911. [Google Scholar]
- 18.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012 Aug;18(8):1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006 Oct 15;15(Spec No 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 20.Millan MJ. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology. 2013 May;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Park LK, Friso S, Choi S-W. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2012 Feb;71(1):75–83. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 22.Choi S-W, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr Bethesda Md. 2010 Nov;1(1):8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tammen SA, Friso S, Choi S-W. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013 Aug;34(4):753–764. doi: 10.1016/j.mam.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, et al. Early Life Programming and Neurodevelopmental Disorders. Biol Psychiatry. 2010 Aug 15;68(4):314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds LP, Caton JS. Role of the pre- and post-natal environment in developmental programming of health and productivity. Mol Cell Endocrinol. 2012 May;354(1–2):54–59. doi: 10.1016/j.mce.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mischke M, Plösch T. More than just a gut instinct-the potential interplay between a baby’s nutrition, its gut microbiome, and the epigenome. Am J Physiol Regul Integr Comp Physiol. 2013 Jun 15;304(12):R1065–R1069. doi: 10.1152/ajpregu.00551.2012. [DOI] [PubMed] [Google Scholar]
- 27.McGowan PO, Meaney MJ, Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res. 2008 Oct 27;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brantl V, Teschemacher H, Henschen A, Lottspeich F. Novel Opioid Peptides Derived from Casein (β-Casomorphins). I. Isolation from Bovine Casein Peptone. Hoppe-Seyler´s Z Für Physiol Chem. 1979 Jan;360:1211–24. doi: 10.1515/bchm2.1979.360.2.1211. [DOI] [PubMed] [Google Scholar]
- 29.Koch G, Wiedemann K, Teschemacher H. Opioid activities of human beta-casomorphins. Naunyn Schmiedebergs Arch Pharmacol. 1985 Dec;331(4):351–354. doi: 10.1007/BF00500818. [DOI] [PubMed] [Google Scholar]
- 30.Kamiński S, Cieslińska A, Kostyra E. Polymorphism of bovine beta-casein and its potential effect on human health. J Appl Genet. 2007;48(3):189–198. doi: 10.1007/BF03195213. [DOI] [PubMed] [Google Scholar]
- 31.Huebner FR, Lieberman KW, Rubino RP, Wall JS. Demonstration of high opioid-like activity in isolated peptides from wheat gluten hydrolysates. Peptides. 1984 Dec;5(6):1139–1147. doi: 10.1016/0196-9781(84)90180-3. [DOI] [PubMed] [Google Scholar]
- 32.Heyl DL, Schullery SE, Renganathan K, Jayamaha MN, Rodgers DW, Traynor JR. pKa and volume of residue one influence delta/mu opioid binding: QSAR analysis of tyrosine replacement in a nonselective deltorphin analogue. Bioorg Med Chem. 2003 Aug 15;11(17):3761–3768. doi: 10.1016/s0968-0896(03)00329-8. [DOI] [PubMed] [Google Scholar]
- 33.Kanai Y, Hediger MA. Primary structure and functional characterization of a high-affinity glutamate transporter. Nature. 1992 Dec 3;360(6403):467–471. doi: 10.1038/360467a0. [DOI] [PubMed] [Google Scholar]
- 34.Aoyama K, Watabe M, Nakaki T. Regulation of neuronal glutathione synthesis. J Pharmacol Sci. 2008 Nov;108(3):227–238. doi: 10.1254/jphs.08r01cr. [DOI] [PubMed] [Google Scholar]
- 35.Erickson RH, Gum JR, Jr, Lindstrom MM, McKean D, Kim YS. Regional expression and dietary regulation of rat small intestinal peptide and amino acid transporter mRNAs. Biochem Biophys Res Commun. 1995 Nov 2;216(1):249–257. doi: 10.1006/bbrc.1995.2617. [DOI] [PubMed] [Google Scholar]
- 36.Bhat RS, Bhaskaran M, Mongia A, Hitosugi N, Singhal PC. Morphine-induced macrophage apoptosis: oxidative stress and strategies for modulation. [[cited 2011 Sep 19]];J Leukoc Biol [Internet] 2004 Mar 23; doi: 10.1189/jlb.1203639. Available from: http://www.jleukbio.org/content/early/2004/03/23/jlb.1203639.short. [DOI] [PubMed] [Google Scholar]
- 37.Guzmán DC, Vázquez IE, Brizuela NO, Alvarez RG, Mejía GB, García EH, et al. Assessment of oxidative damage induced by acute doses of morphine sulfate in postnatal and adult rat brain. Neurochem Res. 2006 Apr;31(4):549–554. doi: 10.1007/s11064-006-9053-7. [DOI] [PubMed] [Google Scholar]
- 38.Goudas LC, Langlade A, Serrie A, Matson W, Milbury P, Thurel C, et al. Acute decreases in cerebrospinal fluid glutathione levels after intracerebroventricular morphine for cancer pain. Anesth Analg. 1999 Nov;89(5):1209–1215. [PubMed] [Google Scholar]
- 39.Hodgson N, Trivedi M, Muratore C, Li S, Deth R. Soluble Oligomers of Amyloid-β Cause Changes in Redox State, DNA Methylation, and Gene Transcription by Inhibiting EAAT3 Mediated Cysteine Uptake. J Alzheimers Dis JAD. 2013 Apr 11; doi: 10.3233/JAD-130101. [DOI] [PubMed] [Google Scholar]
- 40.De Meyer T, Mampaey E, Vlemmix M, Denil S, Trooskens G, Renard J-P, et al. Quality Evaluation of Methyl Binding Domain Based Kits for Enrichment DNA-Methylation Sequencing. PLoS ONE. 2013 Mar 15;8(3):e59068. doi: 10.1371/journal.pone.0059068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sati S, Ghosh S, Jain V, Scaria V, Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012 Aug;25 doi: 10.1093/nar/gks776. gks776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichelt KL, Tveiten D, Knivsberg A-M, Brønstad G. Peptides’ role in autism with emphasis on exorphins. Microb Ecol Health Dis. 2012;23 doi: 10.3402/mehd.v23i0.18958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Z, Cade R. Findings in normal rats following administration of gliadorphin-7 (GD-7) Peptides. 2003 Feb;24(2):321–323. doi: 10.1016/s0196-9781(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 44.Ermisch A, Rühle HJ, Neubert K, Hartrodt B, Landgraf R. On the blood-brain barrier to peptides: [3H]beta-casomorphin-5 uptake by eighteen brain regions in vivo. J Neurochem. 1983 Nov;41(5):1229–1233. doi: 10.1111/j.1471-4159.1983.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 45.Wasilewska J, Sienkiewicz-Szłapka E, Kuźbida E, Jarmołowska B, Kaczmarski M, Kostyra E. The exogenous opioid peptides and DPPIV serum activity in infants with apnoea expressed as apparent life threatening events (ALTE) . Neuropeptides. 2011 Jun;45(3):189–195. doi: 10.1016/j.npep.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Gasbarrini G, Miele L, Malandrino N, Grieco A, Addolorato G, Gasbarrini A, et al. Celiac disease in the 21st century: issues of under- and over-diagnosis. Int J Immunopathol Pharmacol. 2009 Mar;22(1):1–7. doi: 10.1177/039463200902200101. [DOI] [PubMed] [Google Scholar]
- 47.Gräff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci. 2009 Jul;30(1):1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- 48.Waly MI, Hornig M, Trivedi M, Hodgson N, Kini R, Ohta A, et al. Prenatal and Postnatal Epigenetic Programming: Implications for GI, Immune, and Neuronal Function in Autism. [[cited 2013 Dec 20]];Autism Res Treat [Internet] 2012 Jun 19; doi: 10.1155/2012/190930. 2012. Available from: http://www.hindawi.com/journals/aurt/2012/190930/abs/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouvard MP, Leboyer M, Launay JM, Recasens C, Plumet MH, Waller-Perotte D, et al. Low-dose naltrexone effects on plasma chemistries and clinical symptoms in autism: a double-blind, placebo-controlled study. Psychiatry Res. 1995 Oct 16;58(3):191–201. doi: 10.1016/0165-1781(95)02601-r. [DOI] [PubMed] [Google Scholar]
- 50.Trivedi M, Shah J, Hodgson N, Byun H-M, Deth R. Morphine induces redox-based changes in global DNA methylation and retrotransposon transcription by inhibition of excitatory amino acid transporter type 3-mediated cysteine uptake. Mol Pharmacol. 2014 May;85(5):747–757. doi: 10.1124/mol.114.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Höbaus J, Fetahu IS, Khorchide M, Manhardt T, Kallay E. Epigenetic regulation of the 1,25-dihydroxyvitamin D3 24-hydroxylase (CYP24A1) in colon cancer cells. J Steroid Biochem Mol Biol. 2013 Jul;136:296–299. doi: 10.1016/j.jsbmb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.