Abstract
Improper regulation of signaling in vascular smooth muscle cells (VSMCs) by angiotensin II (AngII) can lead to hypertension, vascular hypertrophy and atherosclerosis. The extent to which the homeostatic levels of the components of signaling networks are regulated through microRNAs (miRNA) modulated by AngII type 1 receptor (AT1R) in VSMCs is not fully understood. Whether AT1R blockers used to treat vascular disorders modulate expression of miRNAs is also not known. To report differential miRNA expression following AT1R activation by AngII, we performed microarray analysis in 23 biological and technical replicates derived from humans, rats and mice. Profiling data revealed a robust regulation of miRNA expression by AngII through AT1R, but not the AngII type 2 receptor (AT2R). The AT1R-specific blockers, losartan and candesartan antagonized >90% of AT1R-regulated miRNAs and AngII-activated AT2R did not modulate their expression. We discovered VSMC-specific modulation of 22 miRNAs by AngII, and validated AT1R-mediated regulation of 17 of those miRNAs by real-time polymerase chain reaction analysis. We selected miR-483-3p as a novel representative candidate for further study because mRNAs of multiple components of the renin angiotensin system (RAS) were predicted to contain the target sequence for this miRNA. MiR-483-3p inhibited the expression of luciferase reporters bearing 3′-UTRs of four different RAS genes and the inhibition was reversed by antagomir-483-3p. The AT1R-regulated expression levels of angiotensinogen and Angiotensin Converting Enzyme 1 (ACE-1) proteins in VSMCs are modulated specifically by miR-483-3p. Our study demonstrates that the AT1R-regulated miRNA expression fingerprint is conserved in VSMCs of humans and rodents. Furthermore, we identify the AT1R-regulated miR-483-3p as a potential negative regulator of steady-state levels of RAS components in VSMCs. Thus, miRNA-regulation by AngII to affect cellular signaling is a novel aspect of RAS biology, which may lead to discovery of potential candidate prognostic markers and therapeutic targets.
Keywords: Angiotensin II, AT1 receptor, miRNA, genome-wide expression, renin angiotensin system, vascular smooth muscle
1. Introduction
Angiotensin II (AngII), the octapeptide hormone produced by the circulating renin angiotensin system (RAS), coordinates fundamental physiological processes such as regulation of normal blood pressure, electrolyte/volume balance, vascular tone, aldosterone secretion and catecholamine release from nerve endings [1–3]. Tissue RAS regulates responses to locally produced AngII in a variety of tissues, including the heart, kidneys, and vasculature [4]. Functioning independently, the tissue RAS provides homeostatic control of growth and movement of cells, such as vascular smooth muscle cells (VSMCs) in the vessel wall. If tissue RAS is not properly regulated, chronic actions of AngII can induce VSMC proliferation or hypertrophy and proinflammatory changes, causing cardiovascular disorders including hypertension, renal disease, coronary artery disease, cardiac hypertrophy, vascular restenosis, and atherosclerosis [1, 2, 5, 6].
Cellular effects of AngII are mediated by two pharmacologically distinct receptors, the AngII type 1 receptor (AT1R) and the AngII type 2 receptor (AT2R). The AT1R mediates the major cardiovascular effects of AngII while the effects of the AT2R are largely unknown, but are believed to be antagonistic to that of the AT1R [7]. AngII activated AT1R leads to the initiation of several key signaling pathways, including activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-related kinase (ERK), and Janus kinase (JAK) [8–10]. These cytoplasmic signaling cascades induce the expression of genes for cytokines, the extracellular matrix and the components of RAS by activating distinct transcription factors (e.g., STAT, NFAT, NFkB and GATA) [8, 9]. VSMCs are adversely affected by increased local RAS due to the high density of AT1R, driving the differentiation of quiescent VSMCs into the metabolically active, proliferative and migratory phenotype [11–13]. These pathophysiological states respond favorably to therapy with AT1R blockers (ARBs) [14]. Growing evidence suggests that miRNAs play critical roles in cardiovascular development and disorders [15]. However, AngII-mediated regulation of micro-RNAs (miRNAs) playing a role in VSMC dysfunction remain incompletely understood. Therefore, we investigated whether a common set of miRNAs are specifically regulated by the AT1R in AngII responsive cells in humans and rodents.
More than one thousand annotated miRNAs are conserved in human, rat and mouse. They mediate post-transcriptional control of >60% of protein encoding genes regulating cell behavior, including proliferation, differentiation, contractility, inflammation, and fibrosis [16, 17]. Biogenesis, maturation and mode of action of a miRNA is conserved between species. Transcription of a miRNA gene by RNA polymerase II or III produces a stem-loop primary miRNA, followed by its nuclear cleavage to generate ∼70 nucleotide long precursor miRNA (pre-miR). The pre-miR hairpin is transported and further processed in the cytosol to single-stranded 18–25 nucleotide functional miRNAs. These miRNAs are incorporated into the RNA-induced silencing complex [16, 18, 19]. The seed sequence of a miRNA in these protein complexes pairs with complementary sites in the 3′-untranslated region (3′-UTR) of target mRNAs, repressing translation or degrading mRNAs to fine-tune gene regulation under diverse pathophysiological conditions [16, 19]. Specific and global alteration of miRNA expression profiles has been shown in heart, kidney and vasculature [15, 20–23]. Functional roles for various miRNAs in modulating VSMC contractility [24], proliferation [25–27], and proinflammatory changes [28] have been reported. A recent, small RNA deep-sequencing study lead to a profile of AngII-regulated miRNAs in rat VSMCs [29]; however, a global pattern of miRNAs that change in AngII-activated VSMCs has not yet been elucidated.
We have previously shown that a single miRNA can affect multiple mRNA targets, potentially providing simultaneous regulation of the genes involved in a physiological pathway and accounting for a complex phenotype, such as human heart failure [30]. In the current study, we hypothesized that AT1R regulated miRNA expression profile in VSMCs is conserved since the functional response of blood vessels to AngII is highly conserved in different species. We profiled miRNAs upon AT1R activation by AngII across 23 samples. We discovered a distinct AngII-regulated miRNA expression pattern in human and rat VSMCs, which was validated in independent samples. Our focus on the insulin-like growth factor 2 gene (IGF2) encoded miR-483-3p provided evidence that this VSMC-specific miRNA is important in the homeostatic regulation of tissue RAS components. We demonstrated that 3′-UTRs of RAS components, angiotensinogen, ACE-1, ACE-2, and AT2R are targets of miR-483-3p and angiotensinogen is derepressed upon AngII-induced down regulation of miR-483-3p in human VSMCs. In the context of mechanisms for de-repressing local RAS, the AngII-regulated miRNA-483-3p will likely have a strong influence on VSMCs in disease. Thus, miRNAs, such as miRNA-483-3p could be novel targets for therapeutic intervention.
2. Materials and Methods
2.1. Samples for miRNA profiling
Characteristics and treatment conditions for twenty-three samples included in the current study are shown in Table 1. The human embryonic kidney (HEK-293) clonal cell lines stably expressing rat HA-AT1R (5.1 pmol/mg) and rat HA-AT2R (5 pmol/mg) were established by geneticin selection (600 µg/ml) as described previously (31). VSMCs were represented by two examples. The primary human aortic smooth muscle cells (HASMC, a gift from A. Majors, Department of Pathobiology, Lerner Research Institute, Cleveland Clinic) [31] were used between passage number 3 and 6. The rat aortic smooth muscle cell (RASMC) line (SV40-LT, CRL-2018) was obtained from American Type Culture Collection. The RASMC cell lines stably expressing rat HA-AT1R and rat HA-AT2R were established by geneticin selection (700 µg/ml). The rat AT1R–expressing (0.9 pmol/mg) mouse atrial cardiomyocyte clonal line (HL-1-AT1R) was established by geneticin selection (800 µg/ml) of HL-1 cells [32]. Hearts were obtained from age and gender matched transgenic (TG) mice over-expressing the human AT1R under the mouse α-MHC promoter and corresponding littermate non-transgenic (NT) mouse lines in C3H and C57BL/6 genetic backgrounds (n = 3 each for TG and NT), which were previously obtained from Paradis et al [33] and heart failure phenotype characterized [34].
Table 1.
Characteristics of biological and technical replicates* utilized in the microarray analysis
| Sample ID |
Cell/Tissue Type | Experimental Condition |
Measure of AT1R Activation/Inhibition |
|---|---|---|---|
| SK-1 | HEK-AT1R | Untreated | Control |
| SK-2 | HEK-AT1R | AngII | ERK1/2, JAK, STAT3 phosphorylation |
| SK-3 | HEK-AT2R | Untreated | Control |
| SK-4 | HEK-AT2R | AngII | – |
| SK-5 | HASMC | Untreated | Control |
| SK-6 | HASMC | AngII | ERK1/2 phosphorylation |
| SK-7 | HASMC | Losartan | ERK1/2 phosphorylation |
| SK-8 | HASMC | Untreated | Control |
| SK-9 | HASMC | AngII | ERK1/2 phosphorylation |
| SK-10 | HASMC | Losartan | ERK1/2 phosphorylation |
| SK-11 | HL1-AT1R | U ntreated | Control |
| SK-12 | HL1-AT1R | AngII | ERK1/2, JAK, STAT3 phosphorylation |
| SK-13 | RASMC | Untreated | Control |
| SK-14 | RASMC | AngII | ERK1/2 phosphorylation |
| SK-15 | RASMC | Losartan | ERK1/2 phosphorylation |
| | SK-16 | RASMC-AT1R | Untreated | Control |
| SK-17 | RASMC-AT1R | AngII | ERK1/2, STAT3 phosphorylation |
| SK-18 | RASMC-AT2R | Untreated | Control |
| SK-19 | RASMC-AT2R | AngII | – |
| SK-21 | Whole heart (C3H NT) |
Non- transgenic |
Control |
| SK-22 | Whole heart (C3H TG) |
Cardiac- specific AT1R transgene overexpression |
ERK1/2, JAK, STAT3 phosphorylation, Cardiac Hypertrophy & HF Phenotype |
| SK-23 | Whole heart (C57BL/6 NT) |
Non- transgenic |
Control |
| SK-24 | Whole heart (C57BL/6 TG) |
Cardiac- specific AT1R transgene overexpression |
ERK1/2, JAK, STAT3 phosphorylation, Cardiac Hypertrophy & HF Phenotype |
Details regarding each RNA sample utilized for miRNA expression profiling are shown. For primary comparison, correlations were made between the untreated and ligand treated cells or between NT and TG heart tissue samples. The samples include HEK-293 cells expressing AT1R or AT2R, an immortalized rat aortic smooth muscle cell (RASMC) line expressing AT1R or AT2R and a mouse atrial cell line (HL-1) expressing AT1R. Two independent human aortic smooth muscle cell (HASMC) samples were profiled to reduce potential bias that may result from the variability in primary cells (passage # 2-3). The heart tissue analyzed was from AT1R overexpressing TG mice in which cardiac hypertrophy and HF phenotype were assessed by 1) measuring the heart weight to body weight ratio, 2) measuring ejection fraction and wall thickness by echocardiography (to monitor ejection fraction and wall thickness), and 3) expression of BNP as an independent marker of heart failure as reported previously [34]. *Biological replicates control for biological diversity of a response, measure a quantity from different sources under the same condition and are often more powerful. Biological replicates in this study are defined as RNA isolated independently from multiple sources, each with a distinct genome under the same condition, AngII-activation of the AT1R. Technical replicates are defined as RNA isolated from an AngII-responsive source with same genome under different treatment conditions.
2.2. Cardiac tissue harvest
NT and TG mice were cared for in accordance with the Guide for the Care and Use of Laboratory Animals (US National Institute of Health publication No. 85–23, revised 1996) in the Biological Resources Unit. All experimental protocols described were reviewed and approved by the Animal Care and Use Committee at the Cleveland Clinic. Parameters of cardiac structure and function in individual mice were determined by echocardiography and correlated to expression of HF marker genes as previously reported [34]. Whole hearts from TG and NT mice were harvested and perfused in 0.1M KCl. Hearts were wrapped in foil and flash-frozen in liquid nitrogen then stored at −80°C until RNA was isolated.
2.3. Cell culture and ligand treatment
The HEK-293, HEK-AT1R, HEK-AT2R, RASMC, RASMC-AT1R, RASMC-AT2R and HASMC were cultured in Dulbecco’s Modified Eagle Media (DMEM, Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) and 100 IU penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA). HL1-AT1R cells were cultured in Claycomb media supplemented with 10% FBS and 1% penicillin/streptomycin solution (Sigma-Aldrich). All cell types were cultivated at 37°C in a humidified atmosphere containing 5% CO2.
When ≈70% confluent, cells were subjected to 4-hour starvation in serum-free DMEM. Cells were then treated with either 1µM [Sar1]AngII or 1µM losartan or candesartan for 24-hour period. Untreated controls were maintained in serum-free DMEM. After 24-hour treatment, cells were trypsinized and pelleted by centrifugation (800 × g). The cell pellets were washed with phosphate buffered saline (PBS), flash-frozen in liquid nitrogen and stored at −80°C until RNA was isolated.
2.4. RNA isolation
Total RNA was isolated from heart tissue (n = 3 each for TG and NT), utilizing the RNeasy Fibrous Tissue Kit (Cat. No. 74704; Qiagen, Valencia, CA, USA) under RNase free conditions, according to manufacturer’s protocol. Briefly, the heart was homogenized in RLT buffer with β-Mercaptoethanol using a Mikro Dismembrator S (Sartorious Mechatronics Corp., Bohemia, NY, USA) for 3 minutes at 1200 RPM. Proteinase K was added to the homogenate, which was then incubated at 55°C for 20 minutes. The mixture was centrifuged at 20–25°C and 5000 × g for 5 minutes. The supernatant was transferred to a clean tube and 100% ethanol was added to each lysate. The sample was transferred to a midi spin column and centrifuged at 20–25 °C and 5000 × g for 5 minutes, washed with the buffer provided then treated with a DNase I mixture at room temperature for 15 minutes. The column was washed and dried by centrifugation (at 20–25°C and 5000 × g) before the RNA was eluted with RNase free water.
Total RNA was isolated from cells using the miRNeasy Mini Kit (Cat. No. 217004; Qiagen) under RNase free conditions, according to manufacturer’s protocol. Briefly, the cell pellet was lysed in QIAzol Lysis Reagent at room temperature for 5 minutes. To the homogenate, chloroform was added and incubated for 3 minutes at room temperature. Following centrifugation for 15 minutes at 12,000 × g at 4°C, the upper aqueous phase was transferred to a new tube. To this 1.5 volumes of 100% ethanol was added and mixed and the mixture was pipetted into an RNeasy Mini spin column in a 2mL collection tube and centrifuged at > 8000 × g for 15 seconds at room temperature. The sample was washed sequentially with buffers RWT and RPE and centrifuged for an additional 15 seconds at > 8000 × g each. The column was placed in a new collection tube and centrifuged at ≥ 14,000 × g for 1 minute. Finally, the column was transferred to a new 1.5mL collection tube and the RNA was eluted by centrifugation at 8000 × g in 50ul of RNase-free water.
A spectrophotometer reading was taken for each sample and a small aliquot was run on a 1% agarose gel to ensure RNA integrity. High-quality RNA preparations (A260/280 ratio >1.8) were submitted to the Genomics Core Facility (http://www.lerner.ccf.org/services/gc/) for RNA processing.
2.5. RNA processing and profiling
Two separate Mouse v2 MicroRNA Expression BeadChip arrays with UDG (Cat. No. MI-202–1124; Illumina, San Diego, CA, USA) were used for each sample, according to the manufacturers recommendations (Illumina MicroRNA Expression Profiling Assay Guide). This miRNA panel contained 656 assays for miRNAs catalogued in the miRBase release 12 database (http://www.mirbase.org), which covers >99% of miRNAs reported to be expressed in different tissues of three species included in our analysis. Rat aortic smooth muscle, mouse cardiomyocyte and cardiac tissue were utilized among other samples. We chose mouse arrays because majority of the samples in our analysis were of rodent origin. The profiling arrays contain probe sets to detect mature miRNAs homologous to mouse miRNAs in rat and human samples. The range of tissues/genes from which these miRNAs are produced in nature is vast and spans the organism’s genome.
The chemistry utilized by the miRNA BeadArray is available at http://www.illumina.com/downloads/MicroRNAAssayWorkFlow.pdf Five hundred nanograms of total RNA was polyadenylated, and then converted into cDNA using a biotinylated oligo-dT primer with a universal primer-tag sequence at its 5′ end. This was followed by annealing of a miRNA-specific oligonucleotide pool (MSO) which consists of three parts: a universal PCR priming site at the 5′ end, an address sequence complementary to a capture sequence on the BeadArray, and a microRNA-specific sequence at the 3′ end. Extension of MSO was facilitated by addition of a polymerase, but only if their 3′ bases were complementary to the cognate sequence in the cDNA template. Common primers were used to amplify the cDNA templates; the primer complimentary to the BeadArray was fluorescently labeled. The single-stranded PCR product was hybridized to the Sentrix Array Matrix (SAM), where the labeled strand binds to the bead on the array containing the complementary address sequence. The SAMs were imaged using an Illumina iScan Reader, which measures the fluorescence intensity at each addressed bead location. Intensity files were analyzed using BeadStudio version 3.1.1. Expression levels were expressed as an average signal value and was compared with untreated samples as baseline.
2.6. Statistical analysis of miRNA microarray data
Raw data were read and preprocessed in BeadStudio and exported for further data processing and analysis in R (www.r-project.org) by a professional statistician (www.lerner.ccf.org/qhs/genetics/). See supplementary information for detailed methodology and graphical representation of the results of statistical analysis. Initially, three datasets were generated for all of the samples: 1) the raw expression dataset exported directly from BeadStudio (containing 656 genes); 2) the normalized dataset, which was obtained based on the raw dataset after false-positive background correction, log2 transformation, and quantile normalization (containing 655 genes); 3) the quality control (QC) filtered dataset, which was obtained based on the normalized dataset by selecting probes using a detection threshold of 0.05 (selected 610 genes). In BeadStudio, we performed one sample to one sample comparisons using differential analysis with parameters set up as ‘quantile normalization, subtract background’. All differential expression was compared with a reference group (Ref Group) with Illumina custom Error Model. Raw p-values were from the Diff Pval column of the Differential Expression Genes results. Fdr p-values were obtained by applying multiple testing corrections using Benjamini and Hochberg False Discovery Rate (a Fdr p-value of <0.05 was considered statistically significant). Following quality control analysis, the expression and p-values were read in R and the results represented graphically in box plots, MA plots, volcano plots and scatter plots (Supplemental Figs. 1A–F). The transformed data were then subjected to unsupervised hierarchical clustering analysis using Euclidean correlations distances as dissimilarity metric and average linkage. Comparisons were made for all the sample pairs (untreated versus treated) and plotted as volcano plots, showing the relationship between –log10 (p-value) and log2 (fold change) of all the genes in each comparison. Raw p-values and fold change were calculated. Genes were categorized as significantly expressed if their raw p-value was <0.001 and the fold change was > threshold or < 1/threshold. The threshold value of 1.2 – 1.3 cut-off was adopted in our analyses, as recommended by Illumina [35].
2.7. Real Time qPCR (RT-qPCR) analysis of independent samples
The miScript PCR system (Cat. No. 218073; Qiagen) was utilized according to the manufacturer’s protocol for validation. Template RNA was mixed with the 5x miScript RT buffer, RNase-free water, and the miScript reverse transcriptase mix. The reaction was incubated for 60 minutes at 37°C followed by heating the samples at 95°C for 5 minutes. For the qPCR, a reaction mix containing 2× QuantiTect SYBR Green PCR master mix, 10× miScript Universal Primer, 10× miRNA specific primer (Supplemental Table 1), and RNase-free water was prepared. The cDNA from the reverse transcription step was dispensed into individual wells of a 96-well PCR plate and appropriate volumes of the reaction mix were added. The plate was briefly centrifuged at 1000 × g to collect the reaction at the bottom of each well and qPCR was started using the Applied Biosystems 7500 real-time cycler. CT values recorded were analyzed, following normalization to RNU6B, an endogenous control, and standardization to the untreated samples.
2.8. Protein isolation and Western immunoblotting analysis
Cells were washed twice with PBS, scraped from the plate, and pelleted by centrifugation at 2200 × g. Each cell pellet was resuspended in 1mL PBS, containing 50µl PopCulture Reagent (Cat. No. 71187; Novagen, Darmstadt, Germany), 100 U of Benzonase Nuclease (Cat. No. 70664; Novagen), 10µl 100X protease inhibitor cocktail (Cat. No. P8340; Sigma-Aldrich), and 10µl 100X phosphatase inhibitor cocktail (Cat. No. 78426; Thermo Scientific, Rockford, IL, USA). The lysates were incubated on ice for one hour then a standard Bradford Assay for protein estimation was performed.
Fifty µg of total protein was used for the Western blotting assay. Fractions were boiled for 5 minutes and separated by SDS-PAGE (10%), and transferred onto a nitrocellulose membrane. After blocking in 150 mM of NaCl buffered with 10 mM of Tris and containing 0.1% Tween 20 and 5% milk powder, the membrane was incubated overnight in primary antibody specific for phospho-p44/42 or phospho-STAT3 (Y705). The membrane was washed and then incubated in 1:5000 diluted IgG secondary antibody labeled with HRP for 1h. Using ECL Plus Western Blotting Reagent (Amersham Biosciences) specific bands were detected. The same blots were subsequently stripped and re-probed for total p44/42 and total STAT3. Densitometry analysis was performed for each experiment. Mean band intensity values (of phosphorylation activity) were determined using the Kodak® 1D Image Analysis Software (Version 3.6.5 K2 – 1B5331010; Eastman Kodak Company, Rochester, NY, USA). Phospho-ERK1/2 and phospho-STAT3 values were normalized to the total protein for each and compared to the untreated controls to determine the percent kinase inhibition.
2.9. Establishment of HEK-293T and RASMC cell lines stably expressing the miRNA miR-483-3p
Pre-miR-483 hairpin sequence complementary DNA sequence was designed to be oriented in the 3′→5′ direction to allow for optimal expression of miR-483-3p in mammalian cells. The 76-nucleotide sequence was synthesized and cloned into the expression vector pRNA U-6.1/Hygro by GenScript (GenScript, NJ, USA). For expression of miR-483-3p, we used the HEK-293T and RASMC cell lines. Transfection of pRNA U6-miR-483-3p plasmid was carried out using the Amaxa Nucleofector™ Technology (Lonza, NJ, USA). Hygromycin-resistant clones were picked and expression of miR-483-3p was analyzed by RNA solution hybridization. For each cell line utilized, a vector control was also made, which was hygromycin resistant.
2.10. 3′-UTR reporter luciferase assay for miR-483-3p predicted targets
The 3′-UTR of Renin (ENSG00000143839), Renin Receptor (ENSG00000182220), AGT (ENSG00000135744), ACE-1 (ENSG00000159640), ACE-2 (ENSG00000130234), Chymase 1 (CMA1; ENSG00000092009), AGTR1 (ENSG00000144891), AGTR2 (ENSG00000180772), AT4 Receptor (ENSG00000113441), and Ang(1–7) Receptor (ENSG00000130368) was uploaded into multiple miRNA target prediction databases (e.g., TargetScan, DIANA-microT 3.0, PITA, Microcosm Targets). MiR-483-3p was predicted to bind to the 3′-UTR of AGT, ACE-1, ACE-2, and AGTR2. The 3′-UTRs of human AGT, ACE-1, ACE-2, and AGTR2 were generated using polymerase chain reaction (PCR) amplification and cloned into the psiCHECK™-2 dual luciferase reporter expression vector, which contains unique Xho1 and Not1 restriction sites (Cat. No. C8021; Promega, WI, USA). Genomic DNA was isolated from human kidney epithelial (HEK-293) cells using a DNeasy Blood and Tissue Kit (Cat. No. 69506; QIAGEN) and served as the DNA template for each individual PCR reaction. Sense and antisense primers were designed to be complementary to each 3′-UTR sequence and harbor nucleotide changes to incorporate restriction sites specific for Xho1 and Not1 enzymes (Supplemental Table 2). The PCR products were fractionated on a 1% agarose gel and isolated using QIAquick Gel Extraction Kit (QIAGEN), digested with Xho1 and Not1, and purified with QIAquick PCR purification Kit (Cat. No. 28104; QIAGEN). The purified products were ligated into psiCHECK-2 and transformed into competent FB10B cells. The plasmids were isolated using a GenElute HP Plasmid Maxiprep Kit (Sigma-Aldrich). The sequence of each 3′-UTR construct was verified using automated DNA sequencing.
Changes in luciferase gene expression were measured using the Dual-Glo Luciferase Assay System (Cat. No. E2920; Promega). HEK-293T cells, overexpressing miR-483-3p (and a vector control), were grown in 6-well culture dishes. Forty-eights hours following transfection of the psiCHECK-2-3′-UTR DNA constructs, cells were trypsinized, re-suspended in culture medium, and counted. Approximately 50,000 cells suspended in culture medium were then seeded onto a poly-L-lysine coated 96-well, white, flat-bottom culture plate. The following day, 75 µl of Dual-Glo Luciferase Assay Reagent was added to each well and incubated with gentle rocking for 1 hour at room temperature. Relative luminescence units (RLUs) for Firefly were read using a FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices, LLC, CA, USA) at 0.5 sec integration and from the bottom of the plate. Immediately following completion of the first luminescence reaction, 75 µl of Dual-Glo Stop and Glo reagent was added to each well. The plate was incubated with gentle rocking for 1 hour at room temperature. Renilla luminescence was then read. The ratio of Firefly to Renilla luminescence was calculated for each well. The ratio of the sample well(s) was normalized to the ratio of the control well(s). In experiments where an antagomir to miR-483-3p was utilized (Cat. No. MIN0002173; QIAGEN), 0.005µM of the antagomir was mixed with the plasmid DNA.
2.11. Analysis of endogenous RAS components
Fifty µg of total protein isolated from RASMC-miR483 cells was used for the Western blotting assay. Fractions were prepared as above. The membrane was incubated overnight in primary antibody specific for AGT (Cat. No. ab108334; Abcam, MA, USA), ACE-1 (Cat. No. ab134709; Abcam), ACE-2 (Cat. No.AF933 R & D Systems, MN, USA and Cat. No. ab108252; Abcam) or AGTR2 (Cat. No. AT21-A and AT22-A; Alpha Diagnostic International, TX, USA). The membrane was washed and then incubated in 1:5000 diluted IgG secondary antibody labeled with HRP for 1h. Using ECL Plus Western Blotting Reagent (Amersham Biosciences) specific bands were detected. The same blots were subsequently stripped and re-probed for actin (Cat. No. MAB1501; Millipore, MA, USA). Densitometry analysis was performed for each experiment. Mean band intensity values were determined using the Kodak® 1D Image Analysis Software (Eastman Kodak Company). Total protein values were normalized to actin and compared to the vector control samples.
2.12. Statistics
Data in the validation experiments are expressed as mean ± SEM (n = 3 experiments performed under identical conditions). Statistical analyses were performed using GraphPad Prism© with an unpaired Student t-test. A p-value <0.05 was considered statistically significant.
3. Results
3.1. AngII-regulated miRNA profiles are distinct in different models
To identify genome-wide differential expression of miRNAs we performed microarray analysis on 23 small RNA libraries prepared from untreated controls, chronic AngII activated samples through either AT1R or AT2R, and AT1R inhibited by losartan (see Table 1 and legend for sample details). To ensure that the cells were in an AngII-activated state when total RNA was isolated for library preparation, ERK1/2 and JAK2/STAT3 pathway responses to AngII were measured in the samples (Table 1) as previously reported from our group (30, 34). All AT1R expressing cell lines were treated with 1µM AngII following serum starvation. The phosphorylation of ERK1/2 and STAT3 were monitored by western immunoblotting with antibodies specific to each protein. Levels of phosphorylation were normalized to total ERK1/2 and STAT3, respectively. Following densitometry analysis of pERK1/2 and pSTAT3 levels, we utilized a 2-fold difference between untreated and AngII treated samples as means to ensure receptor activation. The responses to AngII are robust and highly reproducible in all established cell lines but variable in TG compared to NT heart tissue and primary HASMC cultures. We included multiple samples for these to control for variability (see Table 1 legend).
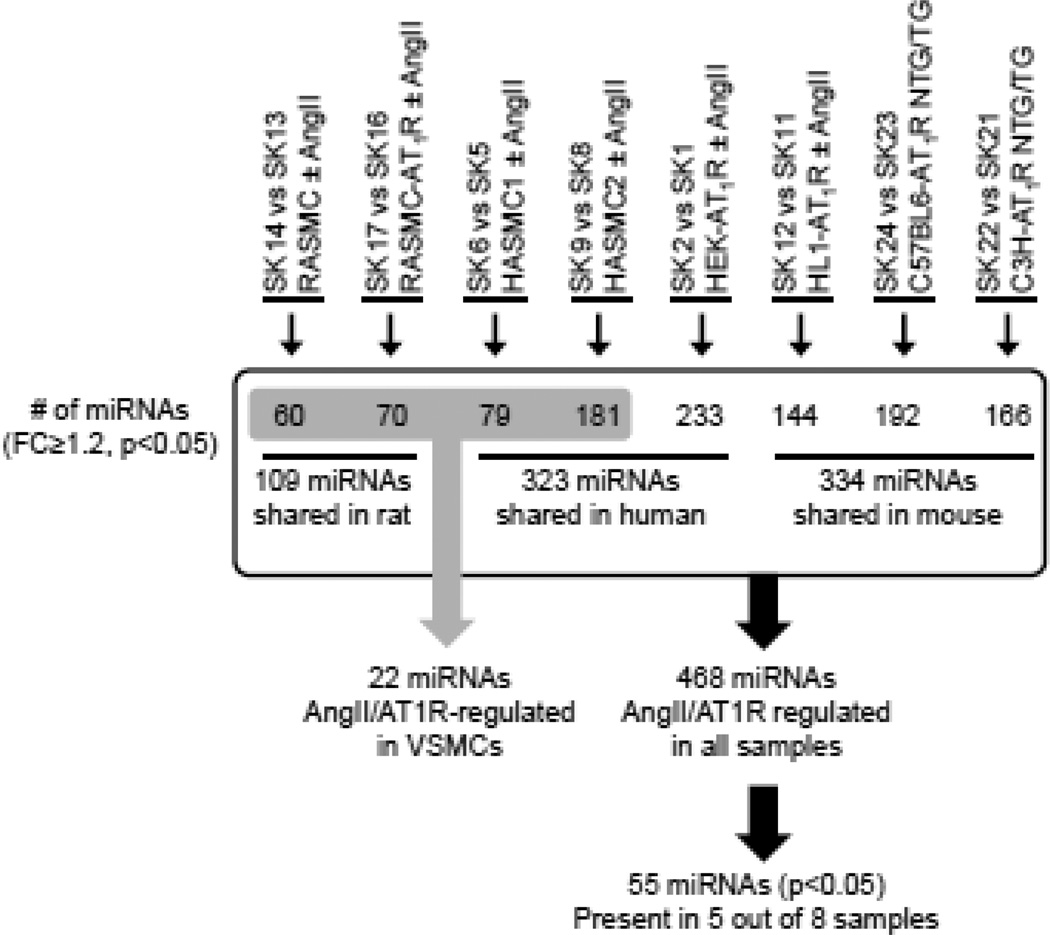
The miRNA bead array used included 656 assays for >99% of miRNA represented in different tissues across mouse genome and the probe sets do detect mature miRNAs homologous to mouse miRNAs across the 24 samples which includes human and rat cells. Expression of 655 miRNAs was detected in quality control assessed microarray data and 610 miRNAs were accepted using the threshold p < 0.05 (see Methods and Supplemental Material). The range of detected miRNAs spanned the organism’s genome in each instance. Risk of not detecting human or rodent specific miRNAs is small because the probes in the array account for conserved miRNAs reported and predicted miRNAs based on cross species reference. Statistical analysis demonstrated that AngII significantly regulated the expression of 468 miRNAs in all samples combined. We employed unsupervised correlation distance and hierarchical clustering analysis of miRNA expression pattern to confirm anticipated relationships between samples. (see Supplemental Figs. 1A–C). For instance, all HEK-293 cell samples clustered together. The heart tissue miRNA profile was closer to the cardiac myocyte, HL1-AT1R miRNA profile than VSMC profile, suggesting that the miRNA expression pattern observed is not random and comparing the data could identify miRNAs common in all samples. Overall strategy applied for comparison between various sample groups is schematized in Fig. 1. AngII regulated the expression of 323 human, 334 mouse, and 109 rat miRNAs. When orthology relationships for these miRNAs were evaluated, 237 of the AngII-regulated miRNA genes expressed in rodents have human orthologs (as determined from miRBase 19). Of these, AngII responsiveness of 168 human orthologs was confirmed in the HEK-293 and HASMC profiles, which demonstrated that genome-wide AngII-regulation of miRNA expression is robust. We identified 30 new miRNAs with unclear functional annotation that have only been predicted in the Solexa sequencing database (www.switchtoi.com). We did not include the Solexa miRNAs in further analyses. (Supplemental Table 3).
Fig. 1. A graphical representation of the comparison of different groups.
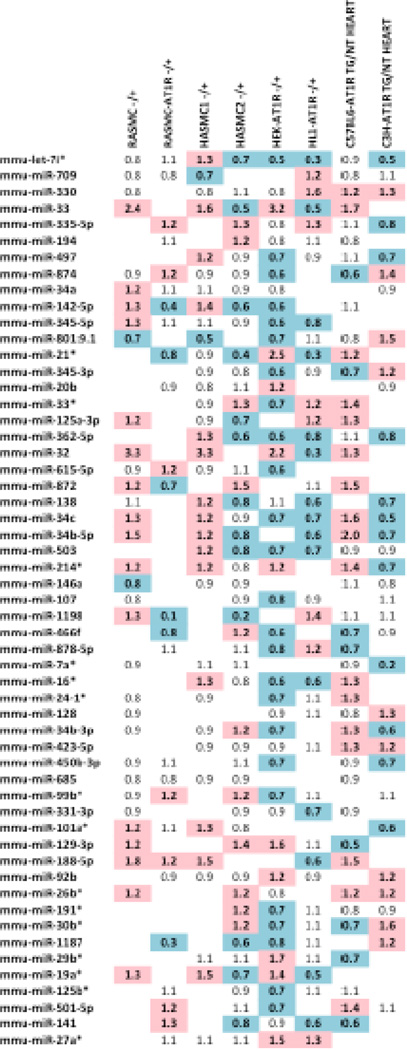
One miRNA, let7i* was expressed in all AngII-activated samples. Fifty-five miRNAs shown in Fig. 2 were present in at least 5 out of 8 data sets that we compared with a p-value ≤ 0.05 for AngII-treated versus untreated conditions. We designated these as most common AT1R-regulated endogenous miRNAs molecules that regulate in vivo responses to AngII in three mammalian genomes in the context of different cell types. These miRNAs may also modulate the effectiveness of AT1R blockade used to treat various pathologies.
Fig. 2. Common miRNAs expressed across AngII activated samples.
Log2 fold change in miRNA gene expression between untreated and AngII/AT1R samples. A blue color indicates that the miRNA is downregulated (< 0.7) in response to the treatment condition (see Table 1 for descriptions of treatments). A red color indicates that the miRNA is upregulated (> 1.2).
3.2. AT1R specificity of miRNA Expression
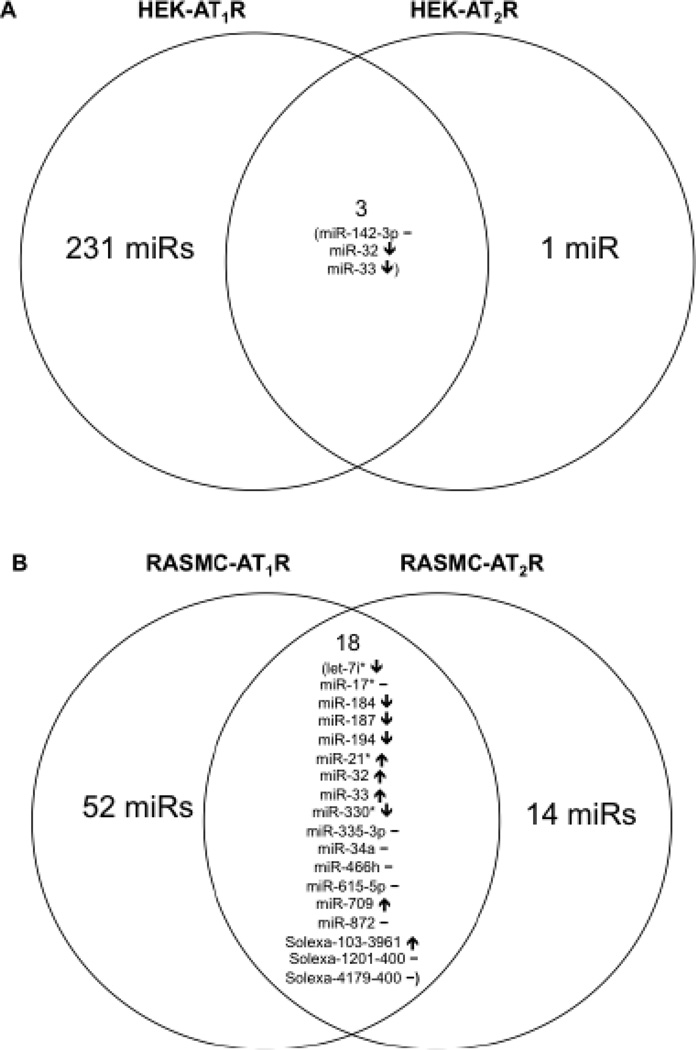
The heat map in Supplemental Fig. 2 depicts whether the same or different miRNAs are regulated in the different samples under different treatment conditions. Genome-wide miRNA expression is robust when AngII activated the AT1R, but only a few miRNAs are expressed when AngII activated the AT2R in both HEK-293 cells and RASMCs. Fig. 3 shows differential response of specific miRNAs to AngII in AT1R-expressing and AT2R-expressing HEK-293 and RASMC cells. In the HEK-AT2R cells four miRNAs are affected, out of which the regulation of three miRNAs was antagonistic to regulation by AT1R (Fig. 3A). In contrast, 32 miRNAs were affected by AngII activation of AT2R in RASMC. The expression pattern of 18 miRNAs common to AT1R and AT2R data (Fig. 3B) was mostly indicative of the well known antagonistic signaling by these AngII receptors [7]. In addition, treatment with the AT1R blocker, losartan, altered 90% of the VSMC miRNAs in a manner opposite from that of AngII treatment. As an additional evidence of specificity, we observed that 24 known miRNA clusters in the human genome, each comprised of several miRNAs and located on 15 different chromosomes [36, 37], respond to AT1R stimulation. Only one or a few specific miRNAs are expressed from each cluster (Table 2), and most of these AngII-regulated miRNAs are evolutionarily conserved in human, mouse and rat species [38–41]. Thus, AT1R specificity of regulation of miRNAs in our analysis is clear.
Fig. 3. Venn diagrams of miRNAs altered in response to AngII treatment.
(A) AngII activation of the AT2R in the human kidney cell line affected only four miRNAs, out of which the regulation of three miRNAs was antagonistic in HEK-AT2R compared to regulation in HEK-AT1R. (B) AngII activation of AT2R in RASMCs affected 32 miRNAs. Eighteen of those miRNAs are common to the expression fingerprint observed in the AT1R RASMCs, albeit the manner of expression is antagonistic. Arrows pointing up (↑) are indicative of an increase in expression while those pointing down (↓) indicate a decrease in expression. A solid line denotes no change. The experimental condition used for obtaining the data in Figure 2A and 2B included 1µM losaratan+1µM AngII, in order to block the endogenous AT1R expressed in the cell lines and to selectively activate transfected AT2R in each case.
Table 2.
Human miRNA clusters differentially regulated upon AngII treatment
| Cluster # |
miRNA Cluster | Chromosome | miRNAs Differentially Regulated in Untreated v. AngII models |
|---|---|---|---|
| 1 | miR-29b-2 ∼ 29c | 1 | miR-29c |
| 2 | miR-200 | 1 | miR-200b, miR-200a |
| 3 | miR-181b-1-181a-1 | 1 | miR-181b-1 |
| 4 | miR-466-467-669 | 2 | miR-466f-1, miR-669d, miR-467b*, miR-466c-1, miR-467e, miR-466a, miR-466d, miR-297a |
| 5 | miR-15b ∼ 16-2 | 3 | miR-15b, miR-16-2 |
| 6 | miR-106b-93-25 | 5 | miR-106a, miR-93, miR-25 |
| 7 | miR-29b-1 ∼ 29a | 6 | miR-29a |
| 8 | miR-290 ∼ 293 ∼ 295 | 7 | miR-290, miR-291a, miR-291b, miR-293 |
| 9 | miR-25 ∼ 93 ∼ 106 | 7 | miR-25, miR-93, miR-106b |
| 10 | miR-23a ∼ 27a ∼ 24-2 | 8 | miR-23a, miR-27a*, miR-24-2* |
| 11 | let-7a-1 ∼ 7f ∼7d | 9 | let-7f* |
| 12 | miR-8 ∼ 141 ∼ 200 | 12 | miR-200c, miR-141 |
| 13 | miR-379 ∼ 411 ∼ 758 | 12 | miR-379, miR-329, miR-667, miR- 668, miR-154, miR-410 |
| 14 | miR-23b-27b-24-1 | 22 | miR-27b*, miR-24-1 |
| 15 | miR-16-1 15a | 13 | miR-15a |
| 16 | miR-17-19a-92a-1 | 13 | miR-17*, miR-18a, miR-19a, miR-20a, |
| 17 | miR-106a-92a-2 | 14 | miR-92a-1*, miR-106a, miR-18b, miR-20b, miR-19b-2*, miR-92a-2*, miR-363 |
| 18 | miR-222 ∼ 221 | X | miR-222, miR-221 |
| 19 | miR-98 ∼ let-7f-2 | X | miR-98 |
| 20 | miR-181c-181d | X | miR-181c, miR-181d |
| 21 | miR-99b ∼ 125 ∼ let-7e | 19 | miR-99b, miR-125, let-7e |
| 22 | miR-23a ∼ 27a ∼ 24-2 | 19 | miR-24-2, miR-23a, miR-27a |
| 23 | miR-99a ∼ let-7c | 19 | miR-99a, let-7c |
| 24 | let-7a-3 ∼ 7b ∼ miR-4763 | 21 | let-7b |
AT1R-responsive human miRNA clusters are defined as regions of the genome harboring two or more miRNAs within 10 kb of each other. These clusters are comprised of several mature miRNAs and located on 15 different chromosomes. Specific MiRNAs within clusters are modulated upon AT1R stimulation with AngII.
3.3. A novel miRNA signature in Human and Rat VSMCs
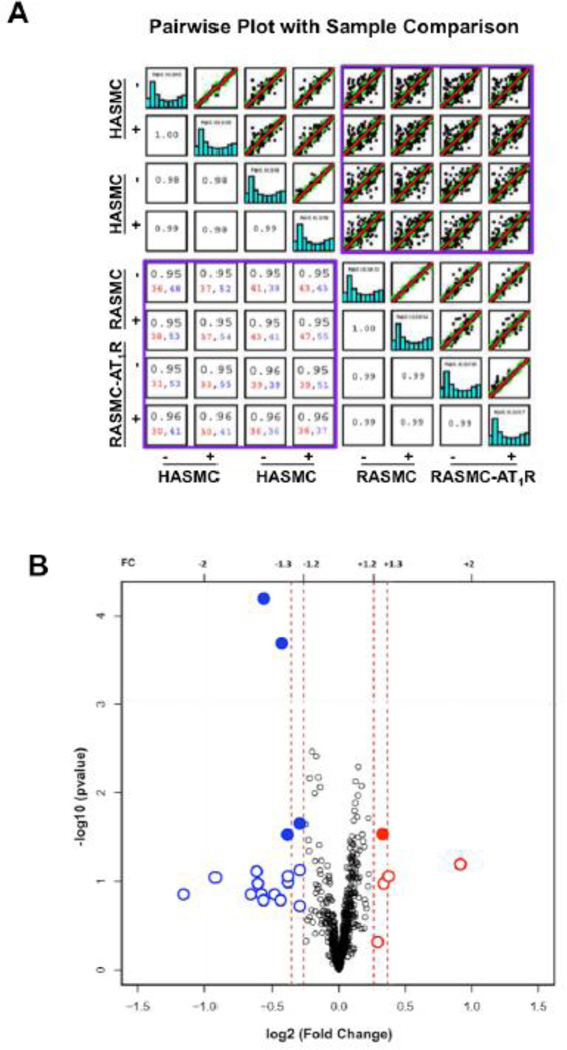
Pearson’s correlation analysis was used to test the hypothesis that the AT1R regulated miRNA profile is conserved in VSMCs from different species. Relationships between expression patterns of miRNAs in VSMC replicates in our analysis are shown by pairwise sample comparisons in Fig. 4A and all replicates in the supplemental Fig. 3. Comparison of profiles between unrelated replicates (e.g., SK-1 and SK-24) produced a scatter plot with weak correlation (r2 <0.7) (Supplemental Fig. 3). The profile of each AngII-treated sample compared to the respective untreated control showed a strong correlation (r2 >0.95). The miRNA expression in human and rodent VSMCs (purple box) yielded correlation coefficients >0.95, indicating a unique pattern of expression (Fig. 4A). This pattern arises from data consisting of >30 upregulated and >30 downregulated miRNAs in all sixteen possible comparisons, demonstrating a robust AngII responsive miRNA signature in VSMCs.
Fig. 4. Characteristics of VSMC-specific miRNAs.
(A) The pairwise plot shows the differential miRNA expression in all AngII activated AT1R VSMC model systems compared. Each blue bar graph represents the overall miRNA profile for a given sample (starting with SK-5 in the upper left hand corner). The unique miRNA expression pattern in the human and rodent VSMC model systems, indicated in purple box. The correlation coefficient is listed for each comparison. In addition, the numbers of miRNAs that are upregulated (>2-fold) are shown in red and the numbers of miRNAs that are downregulated (>2-fold) are shown in blue for the VSMC model systems (lower purple box). (B) The volcano plot shows the log2 fold change in expression of miRNA whose abundance is significantly altered in response to AngII*. Fifteen miRNAs were downregulated (highlighted in blue) and five were upregulated (highlighted in red), following AngII receptor activation. Solid circles represent those miRNAs significantly altered (p<0.05) with a fold change in expression of 1.2 or greater and open circles represent miRNAs altered with a fold change in expression of 1.2 or less.
*Special Note: Conventionally a −1og10 p-value threshold of 1.3 is used for assessing statistical significance. When identifying differentially expressed genes of interest from microarray data, there are multiple methods that can be employed. Researchers can monitor fold change alone as a method to select genes, which is essentially a ratio of the measured intensities from the array. In contrast, a moderated t-test can also be utilized, which compares the difference in mean expression levels and takes into account the variability of the data [56]. We considered both methods when analyzing our data and incorporated the results shown. Some genes, including miRNA genes could have important biological effects even though their change in expression is less than two-fold with p < 0.05. Application of 1.2-fold change is also recommend by the chip manufacture as well as some leading miRNA research groups [57–61].
Expression of 22 miRNAs in VSMCs is highly specific for smooth muscle cells. The volcano plot de-convoluted 5 upregulated and 17 downregulated miRNAs in VSMC signature (Fig. 4B). The genomic locus of each miRNA was identified by aligning the predicted pre-miRNA sequences (miRBase v19) to human, rat, and mouse genome assemblies without allowing mismatches. The known chromosomal locations of the 22 VSMC-specific miRNAs in mouse genome was verified. Many of these miRNAs are encoded within intronic regions in the mouse genome, in which the parent gene is also known. As shown in Table 3, the human genomic complement for 11 of these miRNAs was not found by genome BLAST analysis, however, experimentally detected expression of miRNAs such as miRs-682, −1187, −297c*, −466g, and −1198 in the HASMC samples suggest that gene annotation may have to be updated. Interestingly, 7 miRNAs are clustered to a single region of the genome, the miR-466-467-669 cluster (Table 2).
Table 3.
Genomic characteristics of VSMC-specific AT1R-regulated miRNAs
| miRNA | Mature Sequence | Fold Change (VSMCs) |
Fold Change (HEKs) |
Chromoso me # for Human Homologue |
|---|---|---|---|---|
| miR-675-5p | U GGUG CGGAAAGGGC CC AC AGU | −1.5 | 0.93 | 11 |
| miR-343 | U CU CCCU U C AUGUG C CC AGA | −1.3 | 0.66 | – |
| miR-669c | AUAGUUGUGUGUGGAUGUGUGU | −1.2 | 0.82 | – |
| miR-467b* | AUAUACAUACACACACCAACAC | −1.3 | 0.15 | – |
| miR-218-1* | AAACAUGGUUCCGUCAAGCACC | 1.2 | 0.04 | 4 |
| miR-32 | UAUUGCACAUUACUAAGUUGCA | 1.9 | 0.45 | 9 |
| miR-682 | C U GCAGU C AC AGU GAAGUC U G | −1.2 | 0.99 | – |
| miR-1187 | UAUGUGUGUGUGUAUGUGUGUAA | −1.6 | 0 | – |
| miR-297c*, 297a*, 297b-3p |
UAUACAUACACACAUACCCAUA | −1.9 | 0.81 | – |
| miR-188-5p | CAUCCCUUGCAUGGUGGAGGG | 1.3 | 0.58 | X |
| miR-106a:9.1 | CAAAGUGCUAACAGUGCAGGUA | −1.3 | 0 | X |
| miR-466g | AUACAGACACAUGCACACACA | −1.5 | 0.19 | – |
| miR-325 | UUUAUUGAGCACCUCCUAUCAA | 1.3 | 0.99 | X |
| miR-467a*, 467d* |
AUAUACAUACACACACCUACAC | −1.5 | 1 | – |
| mi R-1198 | UAUGUGUUCCUGGCUGGCUUGG | −2.2 | 0.06 | – |
| miR-301b | CAGUGCAAUGGUAUUGUCAAAGC | −1.5 | 0.12 | 22 |
| miR-21* | CAACAGCAGUCGAUGGGCUGUC | −1.4 | 0.35 | 17 |
| miR-483-3p | U CACU CCU CC CC U C C CGU C U U | −1.5 | 0.93 | 11 |
| miR-297c | AUGUAUGU GUGCAUGUACAUGU | −1.4 | 0.99 | – |
| miR-669f | CAUAUACAUACACACACACGUAU | −1.2 | 0.99 | – |
| miR-208a | AUAAGACGAGCAAAAAGCUUGU | −1.3 | 0.99 | 14 |
| miR-33 | GUGCAUUGUAGUUGCAUUGCA | 1.2 | 0.31 | 22 |
Observed fold change in the levels of AT1R regulated miRNAs is VSMC-specific as evident from comparison to HEK-293. Mouse genome is known to contain genes for the 22 VSMC-specific miRNAs listed; however, the gene assignment for the human equivalent for some of the miRNAs is not yet available (shown with a hyphen). However, the expression of miRs-682, −1187, −297c*, −466g, and −1198 were experimentally detected in the HASMC samples although the genes are not annotated for these miRNAs. The predicted seed region of each miRNA is shown in bold.
3.4. RT-qPCR validation of AT1R-induced miRNA signature in RASMC
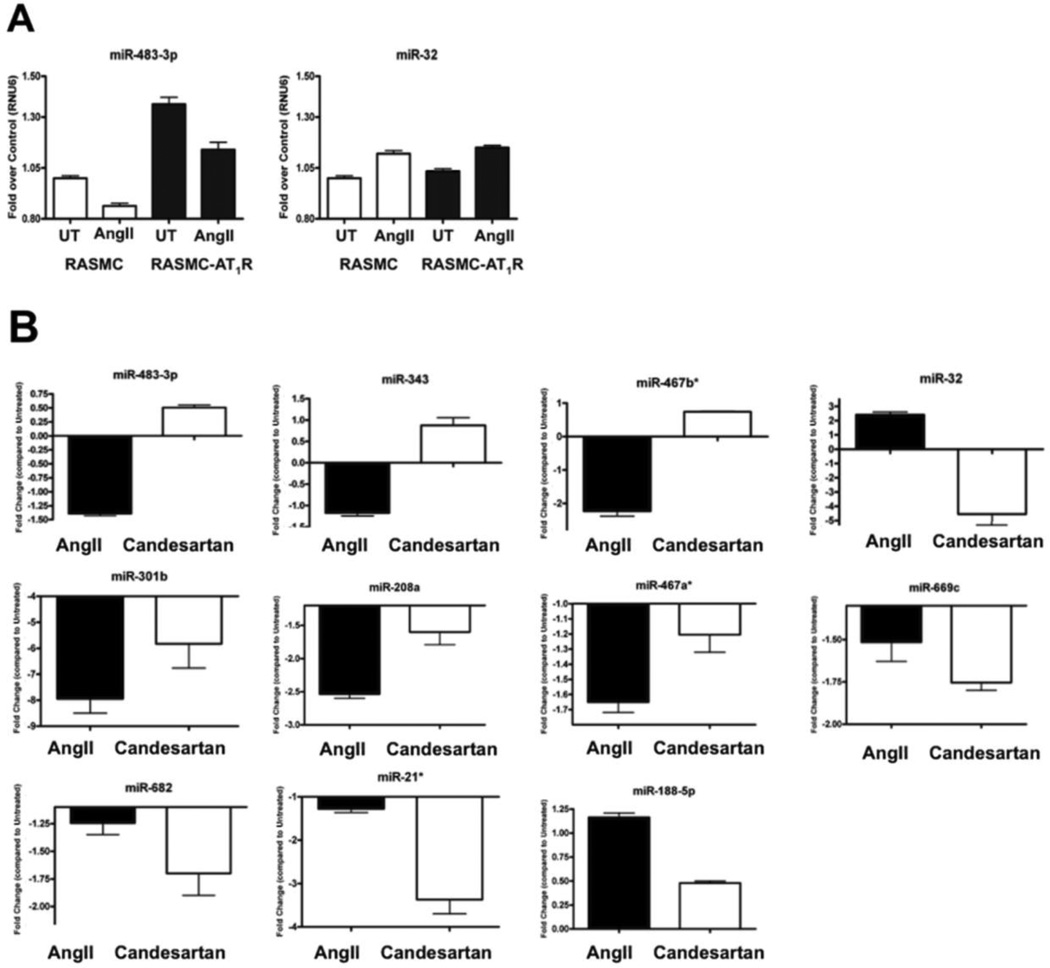
To validate the VSMC-specific miRNA signature predicted by microarray data, the stemloop primer-based, real-time RT-qPCR analysis was performed on independent RNA samples. The basal levels of miR-483-3p, miR-32, miR-669f and miR-1198 in RASMC-AT1R showed small but reproducible change compared to RASMC (Fig. 5A and Supplemental Fig 4A). Similar pattern of change was observed after AngII treatment which suggested that AT1R over expression in the RASMC-AT1R modulates miRNA expression similar to AngII stimulation of RASMC. The time-course for maximal change in the level of each miRNA to AngII treatment may be different, but our focus in this study was on the miRNA levels at 24-hours AngII stimulation.
Fig. 5. RT-qPCR validation of VSMC miRNAs.
(A) Relationship of AT1R cellular levels and miRNA expression analyzed by the stem-loop primer-based real-time RT-qPCR. Steady-state expression of 4 representative miRNAs was monitored in RASMCs and RASMC-AT1R cells (n = 3). Data are presented as fold over U6 RNA internal control following normalization. (B) MiRNA expression modulated by AT1R-specific ligands. Change of expression of 11 miRNAs in the AT1R RASMCs following 1uM AngII or candesartan treatment for 24 hours (n = 3). Data are presented following normalization to U6 RNA, the cellular levels of which did not vary under different experimental conditions and standardization to an untreated control. Also see Supplemental Fig 4 for additional 11 miRNAs.
Fig. 5B shows clear ligand-dependent changes in the levels of 11 different VSMC-specific miRNAs, following treatment with either AngII or Candesartan for 24 hours (n = 3). Supplemental Fig. 4B shows 9 different VSMC-specific miRNAs which responded weakly. Since basal levels of miRNAs were altered in RASMC-AT1R cells due to mere overexpression of the AT1R, the inverse agonist, Candesartan was used to inhibit the basal AT1R-specific effect. The result was then compared to the AngII-induced effect in each case. Of the VSMC candidate miRNAs that were identified from the microarray, 17 showed the same trend in expression (i.e., observed as upregulated or downregulated in Fig. 4) in the independent samples in response to AngII. Candesartan treatment produced distinct response for 11 miRNAs. Either the miRNA expression was reversed or miRNA expression was less diminished compared to treatment with AngII (Fig. 5B). Both types of responses suggest that these miRNAs do respond to AngII activation of the AT1R. The changes observed for 9 miRNAs shown in Supplemental Fig. 4 are small at 24-hours of AngII treatment. Time-course for maximum alterations for these miRNAs may be different. Independent time-course experiments will be needed to find out the functionally effective time point for each of these miRNAs. Taken together, our data demonstrate that AngII activation of AT1R in VSMCs produces a specific miRNA signature that is validated independently.
3.5. Rationale and evidence for a functional role of miR-483-p in VSMCs
The role played by several VSMC signature miRNAs in regulating cellular function is documented as shown in Table 4. MiR-483-3p is one of the 22 VSMC specific miRNAs whose expression is decreased upon chronic AT1R activation but its cardiovascular function is not known. This miRNA is encoded within intron 2 of the IGF2 gene in humans and rodents. IGF2 gene is known to be regulated by AngII signaling and in turn IGF2 signaling affects RAS functions. Therefore we consider miR-483-3p to be an important candidate miRNA for understanding the mechanism by which IGF2 gene regulates RAS functions. As indicated in Table 4, we used bioinformatics analysis to evaluate the potential ability of these 22 miRNAs to target the 3′-UTRs of RAS components. Three independent target prediction programs using multiple databases identified miRNA-483-3p targeting four different components of RAS. Interestingly, miR-483-3p was not predicted to target any components of the ANF system, which is a counter-regulatory system with similar complexity to RAS. The other miRNAs were predicted to target either a single component of RAS (e.g., miR-33 on the AT1R and miR-188-5p on ACE-2) or none of the components (Table 4). In addition, many of the VSMC-specific miRNA have been shown to be involved in cancer, while a few have been implicated in cardiovascular disease. Taken together, miR-483-3p is a very important potential regulator of RAS and thus was selected the candidate for our study.
Table 4.
Mir-483-3p targets multiple RAS components and its functions unknown in VSMC
| miRNA | Predicted RAS Target |
Known Expression and Pathological Context |
|---|---|---|
| miR-675 | – | NA |
| miR-343 | – | NA |
| miR-669c | – | NA |
| miR-297c | – | NA |
| miR-669f | – | NA |
| miR-466g | – | NA |
| mi R-1198 | – | NA |
| miR-33 | AGTR1 | Cholesterol homeostasis in macrophages, atherosclerosis (upregulated) [62] |
| miR-188-5p | ACE-2 | Homocysteine-induced cardiac remodeling (upregulated) [63] |
| miR-483-3p | AGT, ACE-1, ACE-2, AGTR2 |
Wilms’ tumors, hepatocellular carcinoma, colorectal cancer, adrenocortical tumours, pancreatic cancer, proliferation in wounded epithelial cells (upregulated), myocardium of obesity-prone rats, melatonin synthesis in pineal gland (downregulated) [64–70] |
| miR-208a | – | Cardiac hypertrophy/diastolic dysfunction, electrical conductance, acute myocardial infarction (upregulated) [71, 72] |
| miR-218-1 | – | Vascular patterning (upregulated), gastric cancer, oral squamous cell carcinoma, glioma (downregulated) [73– 76] |
| miR-21 | – | Cardiac and pulmonary fibrosis, myocardial infarction, immune response, solid cancers (upregulated) [77] |
| miR-1187 | – | TNFα-mediate hepatic apoptosis (upregulated) [78] |
| miR-297b-3p | – | Lymphoma/carcinogenesis (upregulated) [79] |
| miR-106a | – | Malignant bronchial epithelial, human T-cell leukemia, gastric and pancreatic cancers (upregulated), colon cancer (downregulated) [80–82] |
| miR-301b | – | Colon and pancreatic cancers (upregulated) [83, 84] |
| miR-32 | – | Androgen-regulated prostate, gastric, and colorectal cancers (upregulated) [85–87] |
| miR-325 | – | Stress-induced suppression of luteinizing hormone secretion (upregulated) [88] |
| miR-467a | – | Growth regulation of mouse embryonic stem cells (upregulated) [89] |
| miR-467b* | – | Hepatic steatosis in non-alcoholic fatty liver disease (downregulated) [90] |
| miR-682 | – | Muscle regeneration (upregulated) [91] |
Ten genes known to be components of RAS were analyzed as potential targets of each of the 22 VSMC miRNAs by three target prediction programs. RAS genes predicted as a miRNA target by at least two programs is shown. A solid dash indicates that the miRNA was not predicted to target any components of RAS. NA: Not Available, Reference number is given in parenthesis.
The gene for miR-483-3p is encoded in intron 2 of the IGF2 gene in humans and rodents and AngII regulation of IGF2 gene expression is documented [42]. Mir-483-3p is expressed across muscle and cancer cell lines (Supplemental Fig. 5A). Time-course analysis showed that miR-483-3p levels increased at 4–12h following AT1R stimulation of RASMCs. However, at 24h after stimulation, the level decreased below untreated control levels (Supplemental Fig. 5B, C). Furthermore, pretreatment with the AT1R-blocker Candesartan for 24h significantly increases miR-483-3p in RASMC-AT1R (Fig. 5B, top left), demonstrating a specific role of AT1R in regulating miR-483-3p expression. We also examined whether levels of the predicted transcripts of IGF2 are regulated by AT1R activation (Supplemental Fig. 6). A twofold increase was observed in total IGF2 transcript levels (P total) following treatment of RASMC-AT1R cells with 1 µM AngII for 24 hours (n = 3). Upon AngII stimulation miR-483-3p is decreased when compared to full-length IGF2 transcript (P2 compared to P1), suggesting that steady-state regulation of miR-483-3p may be independent of IGF2 gene transcription. The IGF2 gene contains two promoters, the promoter P-Igf2 for protein expression and the internal promoter P-miR for expressing miR-483 from intronic region of the IGF2 gene (Supplemental Fig. 6).
3.6. MiR-483-3p directly regulates tissue RAS components
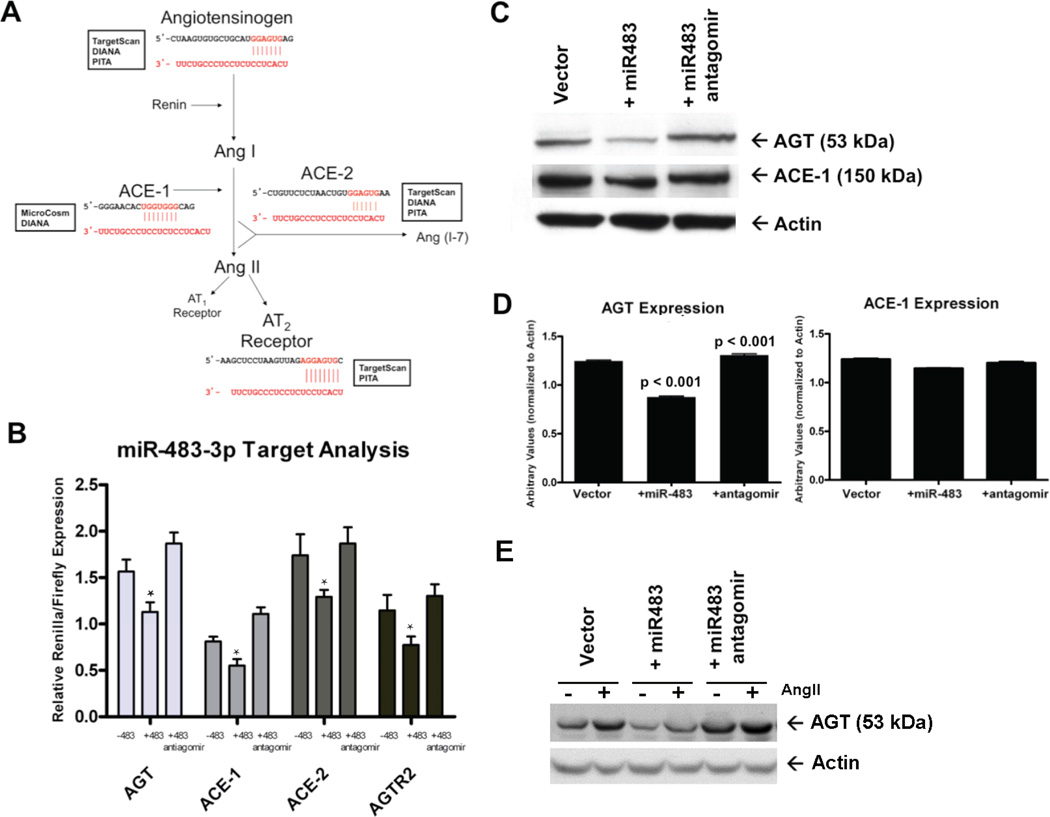
The miR-483-p targets of the components of RAS were predicted using multiple commonly used miRNA target prediction algorithms (Target-Scan, PITA, DIANA and MicroCosm). The criteria we used were theoretically predicted sites by at least two databases and sequence conservation of the predicted site in rodents and human (Fig. 6A). Prediction results showed that 3′-UTRs of angiotensinogen (AGT), angiotensin converting enzyme-1 (ACE-1), angiotensin converting enzyme-2 (ACE-2) and AT2R each contain a single site in the genes’ 3′-UTR for miR-483-3p. We hypothesized that AngII may regulate homeostatic levels of AGT, ACE-1, ACE-2 and AT2R when miR-483-3p targets these components of RAS in VSMCs.
Fig. 6. Effect of miR-483-3p on targeting tissue RAS.
(A) Using multiple target prediction databases, miR-483-3p was predicted to target the 3′-UTR of AGT, ACE-1, ACE-2, and AGTR2 with near complete complementarity. The regions of potential miRNA (red) binding to the 3′-UTR (black) of each gene are shown. (B) HEK-293T-miR483 cells were transfected with a 3′-UTR construct and both firefly and Renilla activity were measured. Symbol * in the bar graphs indicates significant reduction (p>0.05) compared to −483 control. (C) Western immunoblotting for endogenous AGT and ACE-1 in RASMC-pRNA (lane 1), RASMC-miR483 (lane 2), and RASMC-miR483 + antagomir (lane 3) was visualized. (D) Densitometry quantification of AGT and ACE-1 levels are shown. Expression of miR-483-3p significantly reduced the levels of AGT, but had a lesser effect on ACE-1 levels (n = 3; p < 0.0001). Transfection of an antagomir specific for miR-483-3p significantly rescued the levels of AGT, but again had a lesser effect on ACE-1 expression (n = 3; p < 0.001). (E) Effect of 1µM AngII stimulation for 24h on AGT protein expression in RASMC-pRNA (vector), RASMC-miR-483 (+miR483) and RASMC-miR-483+antagomir. The AGT protein expression was visualized by western immunoblotting.
To verify if AGT, ACE-1, ACE-2 and AT2R are direct regulatory targets of miR-483-3p, we cloned the complete 3′-UTR regions containing the predicted miR-483-3p binding sites (see methods for 3′-UTR nucleotide co-ordinates for each gene) downstream of Renilla luciferase expression reporter cassette (Supplemental Fig. 7A). In our dual-luciferase vector system, the expression of Renilla luciferase compared to firefly was stabilized by each of the 3′-UTR cassettes of RAS genes (Supplemental Fig. 7B). We transfected HEK-293 and HEK-293-miR483 cell lines (see methods) with each 3′-UTR luciferase reporter plasmid and measured the ratio of Renilla luciferase to firefly. Renilla luciferase expression was significantly reduced in each case in the presence of miR-483-3p in HEK-293-miR483 cells (Fig. 6B). Co-transfection with inhibitor RNA (antagomir-483-3p) that specifically inhibits miR-483-3p increased the 3′-UTR activity relative to control for all four 3′-UTR luciferase reporters. These experiments clearly demonstrate that miR-483-3p can effectively initiate the RNAi process on the target 3′-UTRs of AGT, ACE-1, ACE-2 and AT2R, suggesting that this miRNA could be a global regulator of tissue RAS.
In addition to the functional readout for the luciferase assay, we monitored levels of endogenous AGT and ACE-1 by immunoblotting in RASMC-AT1R cells stably expressing miR-483-3p. In the miR-483-3p expressing RASMC-AT1R cells, protein levels of these miR-483-3p targets consistently decreased. The suppression of ACE-1 levels was not as dramatic as the effect of miR-483-3p on AGT (p< 0.001), suggesting that miR-483-3p more strongly regulates endogenous AGT. Decreased levels of AGT and ACE-1 in these cells can be rescued by transfection with an antagomir to miR-483-3p (Fig. 6C, D). We monitored effectiveness of miR-483-3p on AGT protein levels in response to chronic AngII-treatment by western immunoblotting (Fig. 6E). Slight increase in AGT expression is observed in the presence of AngII. Both basal and AngII treated AGT expression is decreased in the +miR-483 expression condition. The miR-483 inhibition of AGT expression is reversed following antagomir treatment. Additional effects of this miRNA as a global regulator of RAS in vivo are studies that are currently underway in our laboratory.
Unfortunately, the AGTR2 and ACE-2 primary antibodies utilized were not functional. We also measured mRNA levels of the 4 target genes and observed no change in AGT, ACE-1, ACE-2, or AGTR2 transcripts in the presence of miR-483-3p, thereby suggesting that miR-483-3p did not induce degradation of transcripts of these target genes. MiRNAs primarily act as translational repressors by assembling RNA silencing complex which bind to the 3′-UTR of mRNA targets. The reduction of 3′-UTR-tagged luciferase gene expression in the presence of miR-483-3p in HEK-293-miR483 cells suggested that miR-483-3p acted as inhibitor of 3′-UTR and co-transfection with antagomir-483-3p relieved this inhibition. We found that miR-483-3p is able to decrease endogenous levels of AGT protein (and ACE-1 albeit to a much smaller degree). To examine whether miR-483-3p is affecting levels of transcripts or acting as a repressor of translation of transcripts, we monitored mRNA levels of each of the 4 RAS components. There was no change compared to cells void of miR-483-3p. This allows us to conclude that miR-483-3p is acting on AGT post-transcriptionally and inhibiting protein expression.
4. Discussion
We performed global expression profiling using the Illumina platform in this study to identify AT1R-regulated miRNAs in human and rodent samples. We discovered AT1R-responsive modulation of 55 miRNAs conserved in humans, mice and rats independent of cell context (Fig. 2). These miRNAs are not responsive to the AT2R and AngII-regulation of these miRNAs is blocked by losartan (Fig. 3). We identified VSMC-specific regulation of 22 miRNAs by AT1R for the first time (Fig. 4). Some of these VSMC-signature miRNAs are already known as novel modulators of pathophysiology in VSMCs (Table 4); hence validating their regulation by AT1R (Fig. 5) suggests a significant role for these miRNAs in AngII-mediated VSMC functions. Importantly, we experimentally validated four different predicted RAS components as targets of the AT1R-regulated miR-483-3p (Fig. 6A–B). We demonstrated that miR-483-3p overexpression inhibited expression of AGT and ACE-1 proteins in RASMCs, which is reversed by miR-483-3p antagomir (Fig. 6C–D). Mir-483 was shown to effectively inhibit AngII-induced AGT expression in RASMCs, which is reversed by the miR-483-3p antagomir (Fig. 6E).
Knowledge of genome-wide AngII-regulated miRNAs is envisaged to be exceedingly useful for interpretation of the molecular basis of pathogenesis by AngII, since a single miRNA may orchestrate post-transcriptional regulation of ≈100–200 genes in a cell. The expression profile of miRNAs in cells and heart tissue following chronic AT1R activation was categorized as significant if their raw p-value was <0.001 and the fold change value > 1.2 or < 1/1.2, as recommended by Illumina [35]. The number of modulated miRNAs in this study is relatively higher when compared to other hormone-regulated miRNA profiling studies, but the larger number allowed us to uncover and validate AngII-regulation of many novel miRNAs. The use of human, mouse and rat samples allowed us to identify miRNA response to AngII conserved across species and cell types. Comparing evolutionarily related species or functionally homologous cells from different species as biological replicates in analyzing genome-wide response have been previously reported. For example, Richert et al analyzed the mRNA expression profile in clofibric acid (CLO)-treated and control mouse, rat, and human hepatocytes using expression arrays [43]. In another study, microarray analysis identified temporal expression of a core set of conserved genes across species to 2,3,7,8-Tetrachlorodibenzo-p–dioxin (TCDD) in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells [44]. Comparative gene expression profiles of intestinal transporters in mice, rats and humans have been elucidated in a study of determining interspecies differences and similarities [45]. Based on these examples we hypothesized that if AT1R-regulated miRNAs in human cells are also altered in rodent cells by AngII, then the particular miRNAs should be of interest for further study, not only in the cell type analyzed, but all cell types across the species.
In our study, AT1R-regulated expression changes showed a range between 60 and 234 miRNA genes per sample (Supplemental Fig. 2). This finding is consistent with the miRNA changes reported in studies, which characterized the expression profile in complex tissues, sorted cells and individual cell lines [46, 47]. Authors in these studies reported that on average, change in expression of ≈70–150 miRNA genes per sample is normally observed. Using statistical analysis we inferred a common set of 55 miRNAs (Fig. 2) ubiquitously modulated in AT1R-activated cell lines and tissue samples from rodents and human. The number of miRNA changes we find is consistent with the observation in model studies for basic mechanisms of stem cell maintenance, hormonal differentiation, and malignant transformation to organ systems, in which the authorse stimated ubiquitously expressed miRNAs per sample [46, 47]. Thus, modulating a common set of miRNAs appears to be a critical aspect of the effectiveness of AngII as a wide-ranging agonist in diverse cells. The role played by these miRNAs in different cells may be critical during chronic RAS activation, which increases angiotensinergic risk associated with morbidity and mortality from cardiovascular diseases. These miRNAs may also be important in AT1R-regulated physiological and adaptive tissue remodeling, pro-inflammatory and growth-promoting processes, key functions associated with heart failure, hypertension, restenosis, and atherosclerosis [1]. In many of these disease settings, targeting RAS is an important therapeutic paradigm. Expression of the 55 miRNAs was antagonized by losartan/candesartan blockade of AT1R (Supplemental Fig. 2), suggesting that expression of these miRNAs may also be targeted in most clinical settings. Our study provides insights regarding how RAS-targeted therapies may affect miRNAs, and therefore further experimental studies are needed to elucidate specific functional roles of the AngII-regulated miRNAs.
An unexpected and exciting discovery in this array analysis study is the emergence of a unique AngII-regulated miRNA fingerprint in VSMCs (Fig. 4 and Supplemental Fig. 3) consisting of novel miRNAs and miRNAs with known functions. As shown in Table 4, a significant number of AngII-regulated miRNAs have been implicated in the pathogenesis of other organs and tissues, implying that their role in vasculature may be the basis of pathology of organs and tissues. However, it is important to emphasize that regulation of these miRNAs by AT1R was unknown until now. Our data validated AT1R-modulation of these miRNAs by AngII and also by candesartan (Fig. 5). The amount of a specific miRNA that exists within a cell is determined by its biogenesis and decay. Canonical regulation of miRNA expression through the MEK/ERK1/2 pathway was shown to sequester ERK1/2 in the cytoplasm, thereby inhibiting the ability of ERK1/2 to localize to the nucleus and modulate miRNA gene transcription [48]. In addition, non-canonical pathways of miRNA regulation have been reported in pulmonary artery smooth muscle cells [49, 50]. In future studies we will examine whether miRNAs identified as differentially regulated by the AT1R in this study are able to modulate VSMC phenotypes, such as vascular remodeling and inflammation in response to AngII.
Some miRNAs have been shown to play key roles in regulating different functions of VSMCs. For instance, studies have identified functional roles for various miRNAs; miR-143 and miR-145 may regulate VSMC differentiation, contractility, and hypertension [24, 25], miR-21 and miR-31 [26, 27] may regulate VSMC proliferation, and miR-125b may promote pro-inflammatory responses in VSMCs [28]. However, the involvement of miRNAs in AngII-mediated effects on VSMCs is lacking. A recent study has documented AngII induced expression of the miR-132/212 cluster. Additionally, AngII treatment upregulated miR-132, miR-212, miR-129, miR-21*, and miR-7a [29]. This study, however, was limited to rat vascular smooth muscle. The inclusion of HASMC and RASMC lines in our analysis ensured that the emerging VSMC-specific miRNA fingerprint is conserved in rodents and humans. Also confounded in the whole animal studies, such as heterotypic cell-cell interactions or endocrine, paracrine, autocrine and neuronal influences are absent in our VSMC analysis. Thus, the identified miRNA fingerprint reflects AngII-regulated changes in human and rodent VSMCs through the AT1R.
Our genome-wide screen identified one miRNA whose predicted gene targets are involved in RAS homeostasis. Identifying miR-483-3p as a novel AngII-regulated miRNA has given us some insight into the complexity of miRNA biology. MiRNA expression can certainly change upon acute or short-term agonist treatment in cell culture systems and in vivo, as we observed with miR-483-3p in our VSMC models. The miR-483-3p may down-regulate target genes in early time point. At time points 24h and longer the decreased level of miR-483-3p may have effects opposed to those observed at the earlier time point, which may reflect in vivo scenario under chronic Ang II activation. For our studies, we specifically looked at regulation of miRNAs under chronic AngII stimulation, as those conditions are most relevant to AngII-induced pathological states. We observed that in VSMCs, miR-483-3p is downregulated at 24 hours, whereas, IGF2 expression is upregulated. Analysis of the kinetics of miR-483-3p induction revealed that upon acute treatment with AngII (0.5 h), the miRNA levels increase; however with chronic AngII treatment (24 h), miR-483-3p levels are significantly decreased. Furthermore, VSMCs treated with the AT1R-specific inverse agonist, candesartan showed up-regulation of miR-483-3p. These observations suggested to us that acute and chronic levels of miR-483-3p are regulated independently. Benefits from ARB treatment may involve modulation of miRNAs such as miR-483-3p.
To gain insight into the function of miR-483-3p, we analyzed its potential gene targets, using several miRNA target prediction algorithms [51]. Putative binding sites for miR-483-3p discovered in the 3′UTR of RAS genes, AGT, ACE-1, ACE-2 and AGTR2, suggested that miR-483-3p may coordinate RAS homeostasis. Validation studies in HEK-293T-miR483 cells confirmed repression of all 4 predicted targets by miR-483-3p and reversal by anti-miR-483. In RASMCs, miR-483-3p repressed endogenous protein levels of AGT and to a lesser extent ACE-1. Anti-miR-483 increased the expression of AGT and ACE-1 in RASMCs. The effect of miR-483-3p on endogenous ACE-1 expression in RASMCs is extremely small. Typically, the seed region of the miRNA, which is nucleotides 2–8 of the mature miRNA sequence, is the sequence that is recognized by Ago2 for RISC (Supplemental Fig. 5) loading and strongly pairs with the 3′-UTR of the target. One can see that in the case of ACE-1 the region of complementarity between the miRNA and ACE-1 is not within the 2–8 nucleotide seed region (Fig. 6A), which may be why the inhibitory effect that miR-483-3p has on ACE-1 protein expression is marginal. When we look at AGT, the seed sequence is the region interacting with the 3′-UTR target accounting for significant inhibition of AGT protein expression. These findings showed that miR-483-3p has a physiological role in regulating multiple components of the RAS. Most importantly, the effect of miR-483-3p on AGT is the most convincing and points to a third axis of the RAS that could be a potential therapeutic target. Both ACE-1 inhibitors and ARBs are utilized to date to effectively treat hypertension and related abnormalities caused by over activity of tissue RAS [52]; however, targeting AGT has not been shown until now. Suppressing angiotensinogen would ultimately block production of AngII, similar to ACE-1 inhibitors, but may have fewer side effects. Without in vivo studies, however, we will not know the true capacity of targeting AGT. It is well known that the actions of the AT1R are antagonistic to those of the AT2R. In terms of miR-483-3p, we did not find any significant effect on its ability to decrease AT2R expression in RASMC-AT1R cells, though it is a predicted target of this miRNA. The antagonizing relationship between AT1R and AT2R biology may involve a miRNA-mediated mechanism in some cells, but further studies are needed to tease out this mechanism in vivo.
Regulation of miRNAs in VSMCs by a local increase in AngII can adversely affect vessel functions due to the high density of AT1R in VSMCs. Distinct VSMC phenotypes accumulate within arteries of individuals with disorders such as systemic and pulmonary hypertension, atherosclerosis, and asthma. During insult, VSMCs switch to a proliferative phenotype of poor contractility/excitability, exhibit changes in lipid metabolism, and have high extracellular matrix production leading to vessel remodeling. In contrast, the phenotype of healthy adult VSMCs is restricted cellular plasticity, in which the cells are geared for contraction with a unique repertoire of contractile proteins, agonist-specific receptors, ion channels, and signaling molecules. Thus VSMCs are an interesting model system for studying miRNA-modulated mechanisms of cell maintenance, differentiation, and phenotypic modulation [53, 54].
5. Conclusions
The results presented show that AT1R activation by AngII produces signals that regulate specific miRNA expression. A distinct AngII-regulated miRNA signature emerged in VSMCs, which was validated in independent samples. Further insight into how miRNAs modulate the phenotype of cells in different tissues will be valuable for a greater understanding of AngII biology, as well as in determining the intrinsic regulatory influence of RAS on cardiovascular disease. Following our previous profiling study in human heart failure [30, 55] it is increasingly apparent that understanding the complex network involving miRNAs and their targets, which leads to a coordinated pattern of gene expression, will undoubtedly provide important tools to develop novel therapeutic strategies. This will enhance knowledge of physiological regulation, dysregulated vascular remodeling, and mechanisms for atherosclerotic disease progression. Having focused our efforts on understanding the functional role and mechanism by which miR-483-3p is regulated is only one example of an AngII-responsive miRNA having a substantial impact. Selective regulation of particular miRNAs targeting vascular diseases is a promising prospect for future therapy.
Supplementary Material
Highlights.
Microarray profiling revealed an AT1R–regulated miRNA fingerprint in VSMCs.
The AT1R–blocker, Losartan antagonized AT1R–regulated changes in >90% of miRNAs.
AngII-activated AT2R did not modulate VSMC miRNA expression.
MiR-483-3p targets multiple RAS components, specifically AGT and ACE-1.
Acknowledgements
We thank members of the Karnik lab for constructive suggestions. This work was supported by National Institutes of Health RO1 grants, HL115964 and HL57470 to SK; Graduate Student assistance from the CCF-CSU BGES Graduate Studies program to JK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure summary: No disclosures
References
- 1.Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. American journal of physiology Cell physiology. 2007;292:C82–C97. doi: 10.1152/ajpcell.00287.2006. [DOI] [PubMed] [Google Scholar]
- 2.Lavoie JL, Sigmund CD. Minireview: overview of the renin-angiotensin system--an endocrine and paracrine system. Endocrinology. 2003;144:2179–2183. doi: 10.1210/en.2003-0150. [DOI] [PubMed] [Google Scholar]
- 3.Taubman MB. Angiotensin II: a vasoactive hormone with ever-increasing biological roles. Circulation research. 2003;92:9–11. doi: 10.1161/01.res.0000052920.70316.ae. [DOI] [PubMed] [Google Scholar]
- 4.Bader M, Peters J, Baltatu O, Muller DN, Luft FC, Ganten D. Tissue renin-angiotensin systems: new insights from experimental animal models in hypertension research. Journal of molecular medicine. 2001;79:76–102. doi: 10.1007/s001090100210. [DOI] [PubMed] [Google Scholar]
- 5.Lijnen P, Petrov V. Renin-angiotensin system, hypertrophy and gene expression in cardiac myocytes. Journal of molecular and cellular cardiology. 1999;31:949–970. doi: 10.1006/jmcc.1999.0934. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Iwao H. Molecular and cellular mechanisms of angiotensin II-mediated cardiovascular and renal diseases. Pharmacological reviews. 2000;52:11–34. [PubMed] [Google Scholar]
- 7.de Gasparo M, Catt KJ, Inagami T, Wright JW, Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacological reviews. 2000;52:415–472. [PubMed] [Google Scholar]
- 8.Hunyady L, Catt KJ. Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Molecular endocrinology. 2006;20:953–970. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 9.Saito Y, Berk BC. Angiotensin II-mediated signal transduction pathways. Current hypertension reports. 2002;4:167–171. doi: 10.1007/s11906-002-0042-1. [DOI] [PubMed] [Google Scholar]
- 10.Yin G, Yan C, Berk BC. Angiotensin II signaling pathways mediated by tyrosine kinases. The international journal of biochemistry & cell biology. 2003;35:780–783. doi: 10.1016/s1357-2725(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 11.Ohtsu H, Suzuki H, Nakashima H, Dhobale S, Frank GD, Motley ED, et al. Angiotensin II signal transduction through small GTP-binding proteins: mechanism and significance in vascular smooth muscle cells. Hypertension. 2006;48:534–540. doi: 10.1161/01.HYP.0000237975.90870.eb. [DOI] [PubMed] [Google Scholar]
- 12.Touyz RM, Schiffrin EL. Signal transduction mechanisms mediating the physiological and pathophysiological actions of angiotensin II in vascular smooth muscle cells. Pharmacological reviews. 2000;52:639–672. [PubMed] [Google Scholar]
- 13.Bell L, Madri JA. Influence of the angiotensin system on endothelial and smooth muscle cell migration. The American journal of pathology. 1990;137:7–12. [PMC free article] [PubMed] [Google Scholar]
- 14.Aplin M, Christensen GL, Hansen JL. Pharmacologic perspectives of functional selectivity by the angiotensin II type 1 receptor. Trends in cardiovascular medicine. 2008;18:305–312. doi: 10.1016/j.tcm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Molecular and cellular pharmacology. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. American journal of physiology Renal physiology. 2011;300:F602–F610. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elton TS, Sansom SE, Martin MM. Cardiovascular Disease, Single Nucleotide Polymorphisms; and the Renin Angiotensin System: Is There a MicroRNA Connection? International journal of hypertension. 2010:2010. doi: 10.4061/2010/281692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Current drug targets. 2010;11:913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 23.Scalbert E, Bril A. Implication of microRNAs in the cardiovascular system. Current opinion in pharmacology. 2008;8:181–188. doi: 10.1016/j.coph.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. The Journal of clinical investigation. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circulation research. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Cheng Y, Chen X, Yang J, Xu L, Zhang C. MicroRNA-31 regulated by the extracellular regulated kinase is involved in vascular smooth muscle cell growth via large tumor suppressor homolog 2. The Journal of biological chemistry. 2011;286:42371–42380. doi: 10.1074/jbc.M111.261065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59:2904–2915. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin W, Reddy MA, Chen Z, Putta S, Lanting L, Kato M, et al. Small RNA sequencing reveals microRNAs that modulate angiotensin II effects in vascular smooth muscle cells. The Journal of biological chemistry. 2012;287:15672–15683. doi: 10.1074/jbc.M111.322669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naga Prasad SV, Duan ZH, Gupta MK, Surampudi VS, Volinia S, Calin GA, et al. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. The Journal of biological chemistry. 2009;284:27487–27499. doi: 10.1074/jbc.M109.036541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Majors AK, Sengupta S, Willard B, Kinter MT, Pyeritz RE, Jacobsen DW. Homocysteine binds to human plasma fibronectin and inhibits its interaction with fibrin. Arteriosclerosis, thrombosis, and vascular biology. 2002;22:1354–1359. doi: 10.1161/01.atv.0000023899.93940.7c. [DOI] [PubMed] [Google Scholar]
- 32.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. American journal of physiology Heart and circulatory physiology. 2004;286:H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 33.Paradis P. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proceedings of the National Academy of Sciences. 2000;97:931–936. doi: 10.1073/pnas.97.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue H, Li W, Desnoyer R, Karnik SS. Role of nuclear unphosphorylated STAT3 in angiotensin II type 1 receptor-induced cardiac hypertrophy. Cardiovascular research. 2010;85:90–99. doi: 10.1093/cvr/cvp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Lozach J, Garcia EW, Barnes B, Luo S, Mikoulitch I, et al. Highly sensitive and specific microRNA expression profiling using BeadArray technology. Nucleic acids research. 2008;36:e87. doi: 10.1093/nar/gkn387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Azhar G, Wei JY. The expression of microRNA and microRNA clusters in the aging heart. PloS one. 2012;7:e34688. doi: 10.1371/journal.pone.0034688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochemical and biophysical research communications. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 38.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 39.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 40.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 41.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 42.Lee SD, Chu CH, Huang EJ, Lu MC, Liu JY, Liu CJ, et al. Roles of insulin-like growth factor II in cardiomyoblast apoptosis and in hypertensive rat heart with abdominal aorta ligation. American journal of physiology Endocrinology and metabolism. 2006;291:E306–E314. doi: 10.1152/ajpendo.00127.2005. [DOI] [PubMed] [Google Scholar]
- 43.Richert L, Lamboley C, Viollon-Abadie C, Grass P, Hartmann N, Laurent S, et al. Effects of clofibric acid on mRNA expression profiles in primary cultures of rat, mouse and human hepatocytes. Toxicology and applied pharmacology. 2003;191:130–146. doi: 10.1016/s0041-008x(03)00231-x. [DOI] [PubMed] [Google Scholar]
- 44.Dere E, Lee AW, Burgoon LD, Zacharewski TR. Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC genomics. 2011;12:193. doi: 10.1186/1471-2164-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HR, Park SW, Cho HJ, Chae KA, Sung JM, Kim JS, et al. Comparative gene expression profiles of intestinal transporters in mice, rats and humans. Pharmacological research : the official journal of the Italian Pharmacological Society. 2007;56:224–236. doi: 10.1016/j.phrs.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romano G, Acunzo M, Garofalo M, Di Leva G, Cascione L, Zanca C, et al. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16570–16575. doi: 10.1073/pnas.1207917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Molecular cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis-Dusenbery BN, Hata A. Smad-mediated miRNA processing: a critical role for a conserved RNA sequence. RNA biology. 2011;8:71–76. doi: 10.4161/rna.8.1.14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritter JM. Dual blockade of the renin-angiotensin system with angiotensin converting enzyme (ACE) inhibitors and angiotensin receptor blockers (ARBs) British journal of clinical pharmacology. 2011;71:313–315. doi: 10.1111/j.1365-2125.2011.03918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rensen SS, Doevendans PA, van Eys GJ. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Netherlands heart journal : monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation. 2007;15:100–108. doi: 10.1007/BF03085963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daugherty A, Cassis L. Angiotensin II-mediated development of vascular diseases. Trends in cardiovascular medicine. 2004;14:117–120. doi: 10.1016/j.tcm.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Naga Prasad SV, Karnik SS. MicroRNAs--regulators of signaling networks in dilated cardiomyopathy. Journal of cardiovascular translational research. 2010;3:225–234. doi: 10.1007/s12265-010-9177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarca AL, Romero R, Draghici S. Analysis of microarray experiments of gene expression profiling. American journal of obstetrics and gynecology. 2006;195:373–388. doi: 10.1016/j.ajog.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 58.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calin GA, Trapasso F, Shimizu M, Dumitru CD, Yendamuri S, Godwin AK, et al. Familial cancer associated with a polymorphism in ARLTS1. The New England journal of medicine. 2005;352:1667–1676. doi: 10.1056/NEJMoa042280. [DOI] [PubMed] [Google Scholar]
- 60.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and biophysical research communications. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 61.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 62.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishra PK, Tyagi N, Kundu S, Tyagi SC. MicroRNAs are involved in homocysteine-induced cardiac remodeling. Cell biochemistry and biophysics. 2009;55:153–162. doi: 10.1007/s12013-009-9063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer research. 2010;70:3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veronese A, Visone R, Consiglio J, Acunzo M, Lupini L, Kim T, et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4840–4845. doi: 10.1073/pnas.1101734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gingras B, Rodier G, Giasson E, Coulombe P, Chassagne C, Meloche S. Expression of angiotensin type II receptor downregulates Cdk4 synthesis and inhibits cell-cycle progression. Oncogene. 2003;22:2633–2642. doi: 10.1038/sj.onc.1206346. [DOI] [PubMed] [Google Scholar]
- 67.Bertero T, Gastaldi C, Bourget-Ponzio I, Mari B, Meneguzzi G, Barbry P, et al. CDC25A targeting by miR-483-3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell death and differentiation. 2013 doi: 10.1038/cdd.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertero T, Gastaldi C, Bourget-Ponzio I, Imbert V, Loubat A, Selva E, et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:3092–3105. doi: 10.1096/fj.10-168401. [DOI] [PubMed] [Google Scholar]
- 69.Bertero T, Bourget-Ponzio I, Puissant A, Loubat A, Mari B, Meneguzzi G, et al. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell cycle. 2013:12. doi: 10.4161/cc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, David H, Warner M, Vaag AA, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell death and differentiation. 2012;19:1003–1012. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. The Journal of clinical investigation. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer research. 2013;73:6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 74.Li C, Tu K, Zheng X, Zhang J, Tuo H, Gao J, et al. [MicroRNA-218 expression and its role in hepatocellular carcinoma] Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2013;33:1127–1131. [PubMed] [Google Scholar]
- 75.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circulation research. 2010;107:1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao C, Zhang Z, Liu W, Xiao S, Gu W, Lu H. Reduced microRNA-218 expression is associated with high nuclear factor kappa B activation in gastric cancer. Cancer. 2010;116:41–49. doi: 10.1002/cncr.24743. [DOI] [PubMed] [Google Scholar]
- 77.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. Journal of cellular and molecular medicine. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu DS, An FM, Gong BD, Xiang XG, Lin LY, Wang H, et al. The regulatory role of microRNA-1187 in TNF-alpha-mediated hepatocyte apoptosis in acute liver failure. International journal of molecular medicine. 2012;29:663–668. doi: 10.3892/ijmm.2012.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang T, Luo Y, Wang T, Yang JY. MicroRNA-297b-5p/3p target Mllt3/Af9 to suppress lymphoma cell proliferation, migration and invasion in vitro and tumor growth in nude mice. Leukemia & lymphoma. 2012;53:2033–2040. doi: 10.3109/10428194.2012.678005. [DOI] [PubMed] [Google Scholar]
- 80.Li P, Xu Q, Zhang D, Li X, Han L, Lei J, et al. Upregulated miR-106a plays an oncogenic role in pancreatic cancer. FEBS letters. 2014 doi: 10.1016/j.febslet.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 81.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer research. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 82.Jiang Y, Wu Y, Greenlee AR, Wu J, Han Z, Li X, et al. miR-106a-mediated malignant transformation of cells induced by anti-benzo[a]pyrene-trans-7,8-diol-9,10-epoxide. Toxicological sciences : an official journal of the Society of Toxicology. 2011;119:50–60. doi: 10.1093/toxsci/kfq306. [DOI] [PubMed] [Google Scholar]
- 83.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. Journal of digestive diseases. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 84.Funamizu N, Lacy CR, Parpart ST, Takai A, Hiyoshi Y, Yanaga K. MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells. International journal of oncology. 2014;44:725–734. doi: 10.3892/ijo.2014.2243. [DOI] [PubMed] [Google Scholar]
- 85.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–471. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 86.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, et al. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Molecular cancer. 2013;12:30. doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, Kuai X, Song M, Chen X, Yu Z, Zhang H, et al. microRNA-32 inhibits the proliferation and invasion of the SGC-7901 gastric cancer cell line. Oncology letters. 2014;7:270–274. doi: 10.3892/ol.2013.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lazar L, Nagy B, Molvarec A, Szarka A, Rigo J., Jr. Role of hsa-miR-325 in the etiopathology of preeclampsia. Molecular medicine reports. 2012;6:597–600. doi: 10.3892/mmr.2012.954. [DOI] [PubMed] [Google Scholar]
- 89.Zheng GX, Ravi A, Gould GM, Burge CB, Sharp PA. Genome-wide impact of a recently expanded microRNA cluster in mouse. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15804–15809. doi: 10.1073/pnas.1112772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ahn J, Lee H, Chung CH, Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochemical and biophysical research communications. 2011;414:664–669. doi: 10.1016/j.bbrc.2011.09.120. [DOI] [PubMed] [Google Scholar]
- 91.Chen Y, Gelfond J, McManus LM, Shireman PK. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiological genomics. 2011;43:621–630. doi: 10.1152/physiolgenomics.00136.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.