Abstract
Conjugation of a peptide related to the human immunodeficiency virus type 1 Tat represents a novel method for delivery of antisense morpholino-oligomers. Conjugated and unconjugated oligomers were tested to determine sequence-specific antiviral efficacy against a member of the Coronaviridae, Mouse hepatitis virus (MHV). Specific antisense activity designed to block translation of the viral replicase polyprotein was first confirmed by reduction of luciferase expression from a target sequence-containing reporter construct in both cell-free and transfected cell culture assays. Peptide-conjugated morpholino-oligomers exhibited low toxicity in DBT astrocytoma cells used for culturing MHV. Oligomer administered at micromolar concentrations was delivered to >80% of cells and inhibited virus titers 10- to 100-fold in a sequence-specific and dose-responsive manner. In addition, targeted viral protein synthesis, plaque diameter, and cytopathic effect were significantly reduced. Inhibition of virus infectivity by peptide-conjugated morpholino was comparable to the antiviral activity of the aminoglycoside hygromycin B used at a concentration fivefold higher than the oligomer. These results suggest that this composition of antisense compound has therapeutic potential for control of coronavirus infection.
Coronaviruses are medically and economically important pathogens of humans, livestock, and birds. Coronavirus infections cause a wide range of symptoms, including upper respiratory illness, gastroenteritis, myocarditis, nephritis, central nervous system demyelination, encephalitis, hepatitis, and peritonitis, and can have a debilitating effect on the host immune system, leading to secondary microbial infection. In humans a coronavirus infection marked by aerosol transmissibility and a high degree of immunopathology-related mortality has been linked with the outbreak of severe acute respiratory syndrome (52, 64). Other human coronaviruses cause approximately one-third of all cases of common colds (20, 29, 56).
Coronaviruses have the largest genome among positive-stranded RNA viruses and are the largest known replicating RNA molecules. The coronavirus genome is replicated and packaged by viral proteins produced from a series of mRNAs that have identical 5′ and 3′ terminal regions and that are transcribed by a discontinuous process (66). All coronaviruses have the same genomic arrangement of coding sequence for a basic set of proteins, including a large replicase polyprotein that is processed by two or three virus-encoded proteinases, a helicase which is fused to a zinc-binding domain, an attachment and fusion transmembrane glycoprotein believed to be a class I fusion protein (S), a triple-pass integral membrane protein thought to organize the viral envelope (M), a short hydrophobic protein involved in membrane reorganization and budding (E), and a phosphorylated nucleoprotein that contains numerous positively charged amino acids (N). The 2A and HE genes of mouse hepatitis virus (MHV) are present on a subset of group II coronaviruses and encode luxury functions (61, 67, 68). Production of the viral mRNAs is controlled by specific sequence elements located both at the 3′ terminus of the viral leader and at each mRNA body junction site in a process most carefully dissected in the closely related Arteriviridae (79). Coronaviruses also encode a number of group- or strain-specific genes, some of which are ultimately translated from functionally bicistronic or tricistronic mRNAs and whose products have little similarity to other known proteins.
Treatment of coronavirus infection in vitro and in vivo has proved problematic. Treatments reported in the literature include a wide variety of small molecules and immunomodulators. Some success has been reported for immunomodulatory treatments designed to indirectly block virus growth through enhanced virus-specific immunity. Small molecule viral inhibitors including ribavirin have proved to be largely ineffective at inhibiting coronavirus growth in cell culture and animal models. The only consistently effective inhibitor of coronavirus infection reported is hygromycin B (HYG), an aminoglycoside inhibitor of translation in prokaryotic and eukaryotic ribosomes (25), which is not currently approved for therapeutic use in humans or animals. The specific antiviral effects of HYG are reportedly due to the increased availability of the drug in the cytoplasm of infected cells, leading to inhibition of translation and accelerated destruction of that subset population of cells (9, 12, 17).
Antisense agents have been used to interfere with the gene expression of several human viral pathogens, including vesicular stomatitis virus (63), influenza virus (54), respiratory syncytial virus (42), human papillomavirus (2), herpes simplex virus (7), and human immunodeficiency virus (HIV) (39, 81). The first antisense compound to receive FDA approval, a treatment for cytomegalovirus (CMV) retinitis, targets the IE-2 gene of CMV (51). Two antisense studies using phosphorothioate oligonucleotides alone or complexed with carrier molecules against bovine and murine coronavirus infection have been described (1, 26). Each demonstrated a varying degree of non-sequence-specific antiviral activity at micromolar concentrations, commonly found associated with RNase H-activating antisense agents, although this was more rigorously tested by Hayashi and coworkers (26).
Phosphorodiamidate morpholino-oligomers (PMO) are a class of DNA-like antisense agents typically synthesized to a length of about 20 subunits and contain purine or pyrimidine bases attached to a backbone composed of six member morpholine rings joined by phosphorodiamidate intersubunit linkages (75). They are uncharged, water soluble, and nuclease resistant (28). PMO bind to mRNA by Watson-Crick base pairing and prevent translation by steric blocking (23, 75). When duplexed to RNA, they do not form a substrate for RNase H (72). Regions of eukaryotic mRNA sequence that are likely to be effective targets for PMO antisense agents can be predicted with relative ease and usually involve either pre-mRNA splice sites or the region comprising the 5′-untranslated region, the AUG translational start codon, and the first 20 or so bases of protein coding sequence (24, 74, 77). PMO treatment has resulted in efficacious and specific reduction of target protein levels in a number of tissue culture (40, 43, 45) and in vivo systems (6, 31, 58). PMO have been demonstrated to inhibit translation of hepatitis C virus RNA (32, 53), and studies with two calicivirus strains demonstrated a >80 percent reduction of viral titers in tissue culture (73). In vivo preclinical studies with antisense PMO have demonstrated inhibition of gene expression in rat liver (5) and in the coronary blood vessels of rabbit (37) and pig (38). Further, these studies have established PMO as essentially nontoxic agents with favorable pharmacokinetic properties that remain undegraded following intravenous or oral administration in rats (5). A number of studies have confirmed the ability of PMO to effectively and specifically reduce the translation of target mRNA in vivo via intravenous, intraperitoneal, or transdermal administration (30).
Conjugation of short positively charged peptides resembling a putative protein translocation domain of the HIV type 1 Tat protein was recently shown to increase the efficacy of PMO against specific targets in cell culture systems when compared to PMO without peptide conjugate (57). We have therefore utilized an arginine-rich “Tat-like” peptide, tested a number of peptide-conjugated or unconjugated PMO for antiviral effects against a model coronavirus, MHV, in cell culture and cell-free systems and compared the observed effects with the antiviral effects of HYG.
MATERIALS AND METHODS
Cells and viruses.
Murine astrocytoma DBT were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 0.01 M HEPES, penicillin, and streptomycin or in serum-free medium (VP-SFM; Invitrogen) supplemented with l-glutamine, penicillin, and streptomycin. Serum-free culture conditions were used for all experiments in this report except where stated. Infectious virus in cell culture medium from MHV-A59-infected DBT cells was titrated by plaque assay and used for all inoculations.
PMO design, synthesis and quality control.
PMO were designed to be complementary to the viral (+) strand in regions absolutely conserved in all sequenced MHV strains including the beginning of the nonstructural open reading frame (ORF) 1a polyprotein gene covering two possible initiation codons in favorable Kozak consensus environments (PP), the initiation codon (H1), and an internal region (H2) of the hemagglutinin-esterase gene and the initiation codon of the nonstructural protein 2a gene (2A). A scrambled version of the PP PMO (SC) and a second PMO with irrelevant sequence (FT) were selected for synthesis (Table 1). Antisense and negative-control PMO sequences were screened with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) against murine mRNA sequences in order to preclude unintentional gene-silencing effects. PMO were synthesized with or without a covalently linked peptide, NH2-RRRRRRRRRFFC-CONH2, designated R9F2. Additionally, fluoresceinated R9F2-PP PMO and PP PMO were produced by conjugation of fluorescein to the 3′ end of the PMO. PMO were synthesized at AVI BioPharma Inc. (Corvallis, Oreg.) by a method described previously (75). The conjugation, purification, and analysis of R9F2-PMO compounds were similar to the methods described elsewhere (59).
TABLE 1.
PMO, R9F2-PMO, or oligonucleotide sequences
| Name | Target or purpose | Sequence (5′-3′) |
|---|---|---|
| PP | Over AUG of polyprotein gene (ORF1a) | GCCCATCTTTGCCATTATGC |
| H1 | Hemagglutinin-esterase gene internal (ORF2-1) | CGAGCTTTATTGCCCATCCATC |
| H2 | Hemagglutinin-esterase gene internal (ORF2-1) | CAGCATGTTTAGATTATGCC |
| 2A | ORF 2a | GACATTTACTAGGCTATCG |
| SC | Scrambled PP control | GCACTCTCTGTCTCATAGTC |
| 705 | Human β-globin splice correction control | CCTCTTACCTCAGTTACA |
| FT | Nonsense sequence control | CTCCCTCATGGTGGCAGTTGA |
| MHVL− | (−) Sense MHV target linker | CTAGAGCAGCCCACCCATAGGTTGCATAATGGCAAAGATGGGCAAATACGG |
| MHVL+ | (+) Sense MHV target linker | TCGACCGTATTTGCCCATCTTTGCCATTATGCAACCTATGGG0GGGCTGCT |
Cell-free PMO assays.
The coding sequence for firefly luciferase was subcloned into the multiple cloning site of plasmid pCi-Neo (Promega) at the SalI and NotI sites. Subsequently, complementary oligonucleotides MHVL+ and MHVL− were duplexed and subcloned into the NheI and SalI sites. This replaced the start codon of the luciferase gene with sequence-encoding bases −22 to +21 relative to the A of the AUG initiation translation codon of the MHV polyprotein gene, including the complete −5 to +15 target site for the PP antisense PMO. RNA was produced from this plasmid (pCNM1luc) after linearization with NotI, using the T7 polymerase-based Megascript kit and protocol (Ambion). In vitro translations were carried out by using transcribed RNA at a final concentration in each reaction of 1 nM, with 12 μl of nuclease-treated rabbit reticulocyte lysate (Promega) in addition to PMO, R9F2-PMO, or water. Ten microliters of the translation reaction mixture was mixed with 50 μl of luciferase assay reagent (Promega) according to the manufacturer's instructions, and light emission was read on an FLx800 microplate luminometer (BIO-TEC Instruments). Reactions were assayed for relative light units with the KC Junior program (BIO-TEC) by using the luminescence function and a sensitivity setting of 125. Twelve reactions were assayed at one time, including water control reactions in the first and last wells of each row. The relative light units produced by each reaction was normalized to the mean of all control reactions and expressed as percent inhibition of luciferase translation.
Luciferase assay in cell culture.
Confluent DBT cells were transiently transfected with pCNM1luc by using Lipofectamine (Gibco BRL) according to the manufacturer's directions. The cells were trypsinized 24 h later and 8 × 105 cells/well were plated in six-well plates. The cells were allowed to adhere overnight and were scrape loaded (22) with water or PMO at different concentrations or treated with water or R9F2-PMO. Briefly, cells were treated in a minimal volume of saline or drug-containing saline and then mechanically removed from the culture surface by a consistent number of passes with a tissue culture cell scraper. Cells were then allowed to readhere in culture medium. Cell lysates were prepared and normalized for protein content 18 h later, and luciferase activity was determined as above. The number of relative light units produced by a cell lysate was normalized to the mean of three control reactions and expressed as percent inhibition of luciferase expression.
R9F2-PMO were tested for nonspecific activity in a standard assay based on splice site correction of a missplicing form of human β-globin intron 2 (70). Briefly, the intron was cloned upstream of a luciferase reporter gene designed so that missplicing would result in out-of-frame read-through of the luciferase gene. DBT cells were transfected with target plasmid and treated with PMO or mock treated as above. Cells were lysed 18 h after treatment, and the number of relative light units was recorded.
Microscopy.
DBT cells which had been treated with R9F2-PMO or scrape loaded with unconjugated PMO were rinsed three times with saline and visualized in culture flasks by using a Zeiss Axiovert 200 inverted microscope. Digital images were recorded by using an AxioCam HRc camera and AxioVision 3.1 software. Images of crystal violet-stained cells and syncytia were visualized on a Nikon Diaphot microscope under phase-contrast illumination and recorded on Kodak Ektachrome P1600 35-mm slide film with a Nikon N2000 camera. Developed slide images were digitized by using an Olympus ES-10S film scanner, and digital images were cropped and labeled in Adobe Photoshop 5.0.
Flow cytometry.
Cells were treated for 6 h with R9F2-PMO conjugated at the 3′ end to fluorescein (Fl) and then rinsed five times with saline and removed from culture flasks by using trypsin-EDTA solution (Invitrogen). Cells were pelleted by brief centrifugation at 1,000 rpm in a Heraeus Megafuge 1.10R centrifuge and resuspended in phosphate-buffered saline. Cells were analyzed for green fluorescence by use of a Becton Dickinson FACScan with CellQuest 3.1f software. The cell population to be analyzed was gated for forward and side scatter to include >98% of healthy, untreated DBT cells, and the Fl-1 fluorescence gate was set to exclude >98% of untreated cells.
Toxicity.
MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assays were used to measure cell viability of PMO or R9F2-PMO-treated cells. DBT cells were cultured in DMEM with 10% FBS (supplemented with Pen/Strep) or cultured in VP-SFM without serum. Cells were treated with specified concentrations of test substance in a total volume of 2 ml and plated at a density of 90% confluence. After 24 h of treatment with PMO, R9F2-PMO, or water, 200 μl of 5-mg/ml MTT (Sigma) was added to each well in a six-well plate. Cells were incubated at 37°C when, at 40 min, blue coloration appeared in the majority of cells. The medium was then aspirated and replaced with 1 ml of dimethyl sulfoxide to solubilize the cells. A 200-μl aliquot of this mixture was transferred to a 96-well plate, and the absorbance was read at 540 nm in a Molecular Devices plate reader and analyzed by the SOFTmax program. PMO toxicity and effects on cell growth were also assessed by measuring growth of DBT cells incubated with cell culture medium alone or medium containing various concentrations of R9F2-PMO for up to 36 h. Trypan blue-excluding cells were counted from 6 to 10 replicate flasks at each time point by using a Neubauer hemacytometer.
Virus growth inhibition assay.
DBT cells were seeded at a density of 106 cells per 25-cm2 tissue culture flask and allowed to adhere overnight at 37°C and 5% CO2. Cells were pretreated with 1 ml of VP-SFM or serum-containing DMEM containing various concentrations of R9F2-PMO for 16 h. Cells were inoculated at a multiplicity of 0.1 PFU/cell by removing culture medium and replacing it with 1 ml of culture medium containing 3 × 105 PFU of MHV-A59, and then they were placed at 37°C for 1 h. The inoculum was removed and replaced with fresh VP-SFM, or with serum-containing DMEM where indicated. Cell culture medium was collected, stored, and replaced with fresh medium at designated time points.
Plaque assay.
DBT cells were seeded in 12-well tissue culture plates at 4 × 105 cells per well and allowed to adhere overnight at 37°C, 5% CO2. Culture medium was removed and replaced with 0.5 ml of serial dilutions of inoculum in culture medium, which was placed at 37°C for 1 h. Inocula were left in place, and cells were overlaid with 0.7% agarose (Doc Frugal) in 1× DMEM with 2% serum (final) and incubated at 37°C with 5% CO2 for 2 days. Cells were fixed with 25% formaldehyde in phosphate-buffered saline; agarose plugs were removed, and cells were stained with 0.1% crystal violet.
Plaque size reduction assay.
A modification of a previously described antibody-mediated coronavirus plaque size reduction assay was used (15). DBT cells in 12-well plates were prepared as before and were pretreated with 0.3 ml of VP-SFM containing the desired concentration of the antibiotic HYG or R9F2-PMO for 16 h. Cells were inoculated as before, overlaid with 0.7% agarose in VP-SFM, and incubated at 37°C with 5% CO2 for 2 days. Cells were fixed and stained as before, and the plaque diameter was measured to the nearest 0.5 mm by using a ruler on a lightbox. Minimal and maximal diameters were averaged for noncircular plaques. Plaques in 8 or 16 replicate wells per treatment were measured. The presence of typical MHV cytopathic effects within a plaque was used to confirm identification of small or indistinct plaques.
Syncytium inhibition assay.
DBT cells were seeded in 25-cm2 flasks, pretreated, and inoculated as before. Infection was allowed to proceed for 24 h before medium was removed, and cells were fixed and stained as before. Multinucleate cells, including virus-induced syncytia, were observed and photographed as described above. Rare multinucleate cells are typically present in DBT culture; thus, relative frequency rather than presence of multinucleate cells discriminates MHV-induced cytopathic effects. All multinucleate cells falling within a 4-mm2 window were counted manually for each flask. Statistical significance of differences between syncytium formation after control or antiviral PMO was tested by using a two-sample t test in Instat 3.0 (Graphpad).
Western blotting.
DBT cells that had been pretreated for 16 h with R9F2-PMO were inoculated with MHV-A59 at a multiplicity of 0.1 PFU/ml or were mock inoculated with saline. Cell lysates were prepared 24 h after inoculation by mechanical scraping in 1.5% sodium dodecyl sulfate lysis buffer (also containing 62.5 mM Tris, 10% glycerol, and 0.01% bromophenol blue) and immediate storage at −80°C. Concentration of lysates was normalized for actin content by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting using actin antibody AC-10 (Sigma) and protocols from Santa Cruz Biotechnology (Santa Cruz, Calif.). Antibody complexes were detected by using a horseradish peroxidase-labeled immunoglobulin G secondary antibody (Santa Cruz) with the ECL western blotting detection kit and protocol (Amersham Bioscience). A duplicate blot was prepared as above and probed with ascites fluid to detect the nucleoprotein of MHV (5b188.2; reactive component, murine immunoglobulin G2a [76]). Densitometric analysis of the Western immunoblot was performed by using Kodak ds 1D (v. 3.5.0) image analysis software (Eastman Kodak Company, New Haven, Conn.). Briefly, the autoradiograph was scanned into the computer by using the HP PrecisionScan LTX software and an HP Scan Jet 3400C scanner (Hewlett Packard, Palo Alto, Calif.). This jpeg file was imported into the Kodak ds ID program. Region-of-interest boxes of equal dimensions were subsequently drawn around each band of interest. The software then computed the net intensity of each box, subtracted out the background intensity, and reported the values in relative arbitrary units.
RESULTS
Cell-free translation assay.
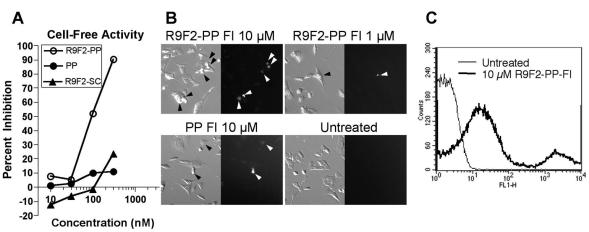
The effects of various PMO on translation of RNA in vitro transcribed from a plasmid containing bases −22 to +21 of MHV ORF 1a fused directly upstream of the coding region for firefly luciferase were measured by in vitro translation reactions in rabbit reticulocyte lysate. In four replicate experiments with one to three samples per treatment condition, it was found that PP PMO was considerably more active in repressing target protein production than noncomplementary PMO controls (Fig. 1A). The R9F2 conjugate was found to increase the effectiveness of antisense PMO sevenfold, based on the concentration required to achieve 50% inhibition of target expression. R9F2 PP PMO was >10-fold more effective at reducing luciferase expression than its base order-scrambled counterpart R9F2 SC PMO.
FIG. 1.
PMO and R9F2-PMO, targeting the MHV polyprotein gene translation initiation codon region, and scrambled control R9F2-PMO were assayed for inhibition of luciferase expression from a chimeric plasmid containing the luciferase gene fused in frame with a 43-base region spanning the MHV polyprotein gene translation initiation codon. The relative percent inhibition by each PMO or R9F2-PMO in one experiment representative of three is shown (A). DBT cells treated with R9F2-PMO-Fl or PMO-Fl were rinsed thoroughly and photographed under mercury vapor illumination to visualize punctate cell-associated fluorescein (B). Fluorescent cells are indicated with black and white arrowheads, respectively, in the phase-contrast and fluorescent image pairs. Fluorescein-positive cells were counted by FACScan. One representative scan from three replicates showing untreated and R9F2-PP PMO-Fl-treated cells is shown (C).
PMO delivery to cultured cells.
Results recently published demonstrate that PMO efficacy in cell culture and cell-free systems could be increased by conjugation of an arginine-rich peptide to the 5′ terminus of the PMO (57). R9F2-PMO-Fl or PMO-Fl were tested for uptake and antiviral effects in cells permissive for MHV growth. Cells were rinsed extensively to remove loosely associated or unassociated PMO. DBT cells scrape loaded with PMO-Fl or incubated for 6 h with R9F2-PMO-Fl displayed punctate green fluorescent labeling (Fig. 1B). Cells took up variable amounts of PMO and formed two populations on flow cytometric analysis (Fig. 1C). The high-fluorescence population corresponded approximately to the percentage of cells with visible PMO staining. Compartmental localization of PMO was not tested, although previous reports indicate that conjugated (57) and unconjugated (22) PMO have some degree of access to the cytoplasm. When the same cells were analyzed by flow cytometry, results from triplicate samples in two independent experiments demonstrated that 80 to 95% of cells treated with R9F2-PMO-Fl and 40 to 90% of cells scrape loaded with PMO-Fl showed more-intense output on the FL-1 channel than 98% of untreated control cells. Independent tests, however, indicated that the in vitro effectiveness of PMO was reduced by approximately 15 to 30% over nonfluoresceinated controls in a standard splice correction assay (H. Moulton, unpublished data), so R9F2-PMO-Fl and PMO-Fl were not pursued for further experiments.
R9F2-PMO toxicity in DBT cells.
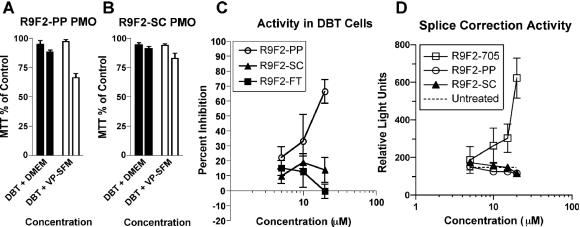
PMO have been reported to have low toxicity both in vitro and in vivo (31). In order to test the R9F2-PMO compounds for toxicity, DBT astrocytoma cells were subjected to the MTT assay, which provides a spectrographic readout of cell viability, in standard cell culture medium and commercially available serum-free culture medium. Cells showed a low toxicity in response to 10 μM R9F2-PP PMO or R9F2-SC PMO but were more sensitive to a 20 μM concentration of either compound (Fig. 2A and B).
FIG. 2.
Cytotoxicity of the R9F2-PP PMO (A) and R9F2-SC PMO (B) was tested by MTT assay at 10 μM (left bar of each pair) or 20 μM (right bar of pair) concentration. The means of three experiments with standard errors are shown. Cells transfected with a plasmid containing a MHV target gene-luciferase fusion were assayed for inhibition of luciferase expression after treatment with R9F2-PMO (C). DBT cells transfected with a plasmid containing a missplicing luciferase reporter gene were assayed in a splice correction assay after treatment in cell culture medium containing R9F2-PMO (D). Relative light units produced from translated luciferase in each monolayer are shown. The dotted line represents the mean of nine controls treated with medium alone. Error bars (C and D) represent the standard errors of the means of three replicates.
In order to measure the effects of R9F2-PMO on growth, cells were treated with 10 to 20 μM R9F2-PP PMO or R9F2-SC PMO and replicate flasks were counted at various time points to construct a cell growth curve. In three experiments using 3 to 10 replicate samples each, no significant difference between the growth of treated and untreated cells was observed (data not shown). Cells incubated with virus and R9F2-PP PMO proliferated to a greater extent than cells inoculated with virus alone (P < 0.05 by Student's t test; n = 10). However, when cells were incubated with R9F2-PMO in the absence of virus for periods of up to 72 h, concentration-dependent thinning of cells was visible after treatment at 20 μM but not 10 μM R9F2-PP PMO or R9F2-SC PMO (data not shown). The R9F2-FT PMO showed no apparent toxicity at a 20 μM concentration over the same period.
Effects on artificial targets in cells.
The pCNM1luc construct was transiently transfected into DBT cells in order to determine PMO effectiveness against an antisense target in cell culture. R9F2-PP-PMO showed 50% inhibition of luciferase expression at concentrations between 10 and 20 μM, and scrambled or irrelevant R9F2-PMO controls showed only background levels of inhibition (Fig. 2C). A positive function test was also performed to test for nonspecific effects in cultured cells. A standard luciferase-based splice correction assay was used to assess R9F2-PMO function in a cellular system other than MHV infection. R9F2-705 PMO-treated DBT cells showed a dose-dependent increase in luciferase production relative to R9F2-PP, R9F2-SC PMO-treated or untreated cells (Fig. 2D). There was a uniform baseline of approximately 150 relative light units in untreated cells that did not vary with R9F2-SC PMO treatment, demonstrating that non-PMO-sequence-specific alteration of an unrelated cellular function was below the threshold of detection in this assay.
Syncytium formation assay.
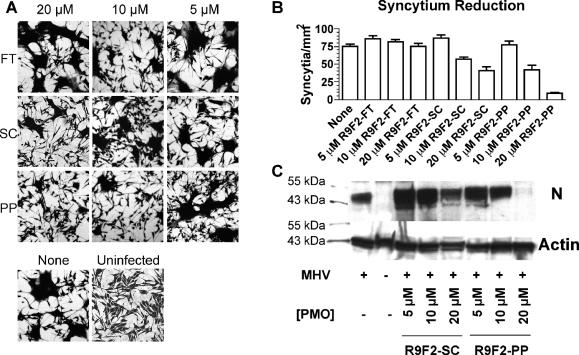
Formation of multinucleate syncytia in mouse astrocytoma DBT cells is a common feature of most murine coronaviruses. Syncytium formation is typically observed at 12 to 14 h after infection, with a majority of cells recruited into syncytia by 24 h. In order to test whether DBT cells could be protected from this highly visible marker of MHV gene expression, cells pretreated with 5 μM, 10 μM, or 20 μM R9F2-PP PMO, R9F2-SC PMO, or R9F2-FT PMO for 12 h were inoculated with MHV-A59, which encodes a highly fusogenic spike protein. Minor toxicity was observed with the PP and SC R9F2-conjugates as measured by a visible decrease in the number of cells present after treatment. However, syncytium formation was reduced in R9F2-PP PMO-treated cells in a dose-dependent fashion. Significantly fewer syncytia were observed for triplicate flasks after treatment with 10 μM or 20 μM R9F2-PP PMO than for cells treated with either R9F2-SC PMO or R9F2-FT PMO (Fig. 3A and B). Treatment with 20 μM R9F2-PP PMO delayed the appearance of MHV cytopathic effects by about 24 h compared to what was observed with untreated infected cells.
FIG. 3.
MHV-infected or mock-infected DBT cells treated with R9F2-PMO or water were fixed and stained with crystal violet 24 h after inoculation (A). Multinucleate syncytia were observed and quantified within three randomly placed 4-mm2 windows per flask of three replicates. The number of multinucleate cells per square millimiter is shown for each treatment type and concentration, with error bars representing the standard errors of the means (B). None, cells were inoculated with MHV in the absence of R9F2-PMO treatment; Uninfected, cells were mock inoculated and not treated with R9F2-PMO. (C) Western blot probed with anti-β-actin or anti-MHV N antibody on samples from treated and untreated MHV-infected or mock-infected cells with loading normalized with respect to actin content. Standard molecular weight markers show that each of the proteins detected corresponds to the expected size of murine β-actin (43 kDa) or MHV N (50 kDa).
Quantification of viral protein synthesis.
Western blotting was performed to confirm that the lack of syncytium formation was due to decreased viral protein expression. Cell lysates were normalized for actin content by Western blotting, and an identical blot was then probed with antiserum to detect the MHV nucleoprotein (N). N expression decreased in a dose-dependent fashion after treatment with R9F2-PP PMO to a much greater extent than what was observed after treatment with R9F2-SC PMO (Fig. 3C). Densitometry analysis indicated that the reduction in expression associated with 10 and 20 μM R9F2-PP PMO corresponded to decreases of 21 and 98%, respectively, in N protein expression, while 20 μM nonsense PMO decreased expression by only 6%.
Reduction of virus growth.
Some preliminary studies in DBT cells showed significant reduction of viral titer with the PP PMO delivered by scrape loading, although these results were not consistent in all experiments. MHV grew to similar titers in serum-free and serum-containing cultures. No apparent inhibition with other anti-MHV or control PMO was detected; therefore, the only antiviral PMO selected for further study and conjugation to the R9F2 peptide was the anti-AUG sequence PP.
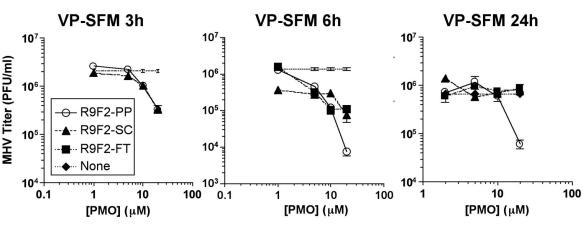
In our hands, with this set of cell culture conditions, the efficacy of PP-R9F2 depended on the absence of serum in the culture medium. Medium containing 10% serum reduced sequence-specific effects to background levels that were identical to those of controls. It was suspected that the highly charged peptide R9F2 might mediate attachment to serum proteins, thus delaying or blocking cellular entry of the PMO. In order to test whether the effects of serum-containing medium were limited to the uptake period, serum-free medium was used for incubations and then replaced by serum-containing medium at different time points. The presence of serum was found to affect R9F2-PP PMO efficacy only during a minimal incubation period. Readdition of serum-containing medium after 3 h but not 6 h of incubation prevented specific antiviral effects of R9F2-PP PMO, indicating that 3 h was insufficient for adequate drug uptake (Fig. 4, left panel). Only nonspecific effects, presumably related to mild toxicity, were present in antisense-treated, serum-pulsed cells. Cells maintained in the absence of serum did not display nonspecific PMO-related antiviral activity. The DBT cells were less viable in serum-free medium by MTT assay, and the stress of serum removal appeared to mask the low-level toxic effects associated with R9F2-SC PMO treatment. When the incubation of R9F2-PP PMO was performed in serum protein-free medium in four independent experiments with three replicate flasks of cells for each treatment, the result was a consistent 10- to 100-fold inhibition of viral growth (Fig. 4, middle and right panels). Time course experiments revealed the inhibition of MHV growth reflected a delay in viral kinetics by about 24 h. Forty-eight-hour titers from R9F2-PP PMO-treated cells were equivalent to 24 h titers in untreated or R9F2-SC PMO-treated cells. Specific antiviral effects remained between 10 and 100-fold for 20 μM R9F2-PP PMO in cells inoculated at a multiplicity of 0.01 to 10 PFU/cell (data not shown).
FIG. 4.
Three MHV growth experiments in which serum-free culture medium was substituted for serum-containing culture medium at different points during incubation of R9F2-PMO with DBT cells or virus growth period were performed. Cells were treated in 1 ml of VP-SFM, and then 10 ml of serum-containing medium was added after 3 h (left panel) or 6 h (middle panel). One treatment group received 10 ml of VP-SFM after inoculation (right panel). Results are presented for a minimum of three replicate flasks per treatment, and cells were inoculated with MHV after 6 h of R9F2-PMO treatment. Infectious MHV present in culture supernatant recovered 18 h after inoculation was titrated by plaque assay. Mean titers in PFU per milliliter are shown with error bars corresponding to the standard errors of the means. An MHV growth experiment was performed with incubation in the absence of serum. Results shown are mean values of three replicate flasks in one of four independent assays, with standard errors indicated by error bars.
Effects on MHV spread.
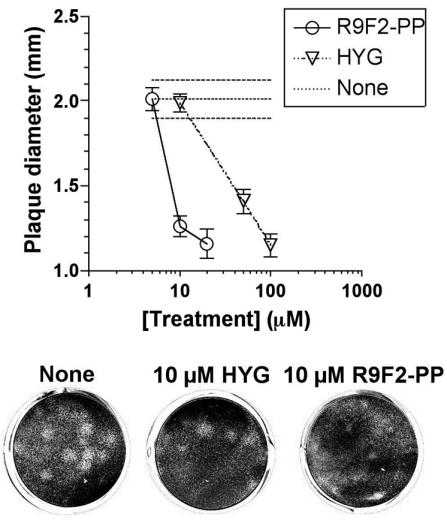
A plaque size reduction assay was performed to assess the effects on MHV spread and to determine the contribution in these assays of potential inhibition of virus entry mentioned in a previous publication (73). This assay assesses the long-term effects on MHV growth and spread in cell culture after a single treatment. Cell-to-cell spread of MHV, unlike viral entry, does not require the presence of a specific MHV receptor on the target cell (21). Treatment of DBT cells with HYG during MHV-A59 plaque assay reduced plaque diameter by a maximum of approximately twofold when HYG concentration was 50 μM or greater, and cells displayed evidence of HYG toxicity with HYG concentration of 100 μM and higher. Plaque size on cells treated with R9F2-SC PMO did not differ significantly from untreated controls, and the plaques on R9F2-PP PMO-treated cells were significantly smaller than the plaques on cells treated with equimolar HYG or R9F2-SC PMO (by Student's t test; P < 0.01). In two determinations with 12 replicate wells, R9F2-PP PMO treatment at 10 μM concentration reduced plaque size approximately twofold, indicating that the antiviral effects of R9F2-PP PMO were approximately fivefold more potent than the effects of HYG treatment (Fig. 5). The correspondence between effects on MHV spread, growth, protein production, and onset of cytopathic effects indicated that possible inhibition of MHV binding made a negligible contribution to antiviral effects.
FIG. 5.
Comparison of HYG and PP-P003. MHV was titrated by plaque assay on DBT cells with R9F2-PMO present in the indicated concentrations in the agarose-VP-SFM overlay. The average plaque diameter, measured to the nearest 0.5 mm ± standard error, is shown for each treatment group (top). Results and a selection of plaques (bottom) from 10 μM concentration treatment groups of one experiment representative of two are shown. MHV plaques are visible as large or small holes in the DBT monolayer that has been degraded to some extent by 72-h incubation with R9F2-PMO or HYG.
DISCUSSION
The literature describes many attempts to control coronavirus infection with pharmaceuticals. Attempts which have been most successful include immunotherapy with various combinations of interferon (14, 19, 60) or with neutralizing antibodies or antibody fragments (41, 62). Limited data indicate mixed anticoronaviral activity for nucleoside analogs including ribavirin (11, 16, 18, 35, 44, 59, 71, 78, 82) and an assortment of other small molecules (8, 10, 13, 55, 69, 80). Corticosteroid treatment has proved generally ineffective against coronaviruses (3, 4). One study demonstrated an antiviral ribozyme that could be generated in situ for control of persistent or acute murine coronavirus infection in cell culture (50). Likewise, phosphorothioate antisense oligomers, while somewhat effective in cell culture (1, 26), are subject to degradation in cells and are most effective against shorter sequences with inherently lower target specificity due to RNase H promiscuity (74). Proteinase inhibitors, including EST (E-64d), have been used to treat coronavirus infection in cell culture (36), but EST was found to achieve a tenfold reduction in titer at a concentration of approximately 1,200 μM. Treatment with HYG reduced viral titers in murine (46-49), feline (8), and bovine (33, 34) coronavirus experimental systems and appears to have the broadest anticoronaviral activity of the small molecule antivirals tested. However, there exist reports of incomplete inhibition of virus growth and strains of bovine coronavirus apparently not susceptible to HYG-mediated inhibition (33, 34). The mechanism of resistance to the antiviral effects of HYG was not elucidated and is difficult to rationalize with the proposed mechanism of HYG specificity for virus-infected cells.
In this study, an antisense strategy was designed to knock down translation of the ORF 1ab replicase polyprotein from the positive-sense viral genome. Coronaviruses do not package a viral polymerase complex; the genomic RNA is translated immediately to produce the ORF 1ab replicase polyprotein. Secondary antiviral effects that might accompany decreased replicase production are loss of replication intermediates (65) and prevention of the downregulation of host gene expression (27). The approach described here was moderately successful, resulting in an approximately 1-log reduction of viral titer when compared with negative controls. One inherent drawback of the targeting strategy used is due to the nature of coronavirus genome replication. Like other positively stranded RNA viruses, coronaviruses produce a low number of negative-strand genome replicates from an original positive strand, and these negative transcripts in turn serve as templates for the production of a relatively high number of positive-strand genome progeny. Should the original positive-sense genome particles escape PMO duplexing and become converted into negative-sense RNA templates, the remaining PMO may be challenged by an amount of positive-sense genome-length RNA far greater than that present from the initial infection. The development and characterization of potentially drug-resistant MHV that may be formed through nonlethal mutation in the ORF 1a AUG region or elsewhere will require further experimentation.
The R9F2 peptide enhanced delivery of PMO in the cell culture systems tested. The mechanism of action of the R9F2 peptide is unclear and merits further study. Antiviral effects of PMO, which were increased upon conjugation with R9F2, indicate that at least a portion of the R9F2-PMO passing through the cell had access to the cytoplasm. PMO directed against the 2A and HE genes of MHV-A59 were expected to bind the genomic RNA and would be expected to have antiviral effects through inhibition of replicase processing. Lack of effect may indicate that the MHV helicase is sufficiently robust to dislodge bound PMO. It would be interesting to learn if these apparently ineffective PMO sequences are conferred detectable antiviral function on conjugation to the cell culture delivery and activity-enhancing R9F2 peptide.
That R9F2-PMO was ineffective in the presence of culture medium containing 10% FBS raises a question about the utility of intravenous or orally administered R9F2-conjugated PMO in vivo. Various experiments with rats, showing that PMO can decrease target gene expression in the liver after topical or intraperitoneal application, indicate that the R9F2 is not necessarily required for systemic PMO function. Thus, we view the peptide conjugate as a developmental tool to increase uptake in cell culture that may be dispensable in vivo. Further experimentation will be required to address efficacy of morpholino-based antisense drugs against coronaviruses in an animal model. Peptide conjugation did not result in antiviral function of the nonsense sequence FT PMO, indicating that the peptide-conjugate itself does not cause but instead enhances antisense activity in cell culture and cell-free translation systems. The SC PMO displayed mild toxicity and limited antiviral effects, both of which appeared also to be enhanced by addition of R9F2 conjugate. The mechanism of low-level nonspecific activity associated with the SC sequence could not be ascertained from these data.
The increased antiviral effects of the PP antisense PMO over antiviral aminoglycoside treatment, together with other qualities including the rapid, complete sequence customization possible, make PMO an attractive candidate for control of coronavirus infection. The key to the effectiveness of PMO as therapeutics would be predicted to reside at the level of target selection and delivery, since this class of compound has proven its potential for precise, sequence-specific down-regulation of target gene expression in numerous experimental systems.
Acknowledgments
We thank Michelle Nelson and The Chemistry Group at AVI BioPharma for synthesis, purification, and analysis of the PMO and R9F2-PMO used in this work and Jennifer Abma, Nicola Benning, and J. Lindsay Whitton for technical assistance.
B.W.N. was supported by NIH training grant NS-41219. This work was supported by funding under NIH grants AI43103 and AI25913 to M.J.B.
REFERENCES
- 1.Abdou, S., J. Collomb, F. Sallas, A. Marsura, and C. Finance. 1997. beta-Cyclodextrin derivatives as carriers to enhance the antiviral activity of an antisense oligonucleotide directed toward a coronavirus intergenic consensus sequence. Arch. Virol. 142:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Salas, L. M., T. E. Arpawong, and J. A. DiPaolo. 1999. Growth inhibition of cervical tumor cells by antisense oligodeoxynucleotides directed to the human papillomavirus type 16 E6 gene. Antisense Nucleic Acid Drug Dev. 9:441-450. [DOI] [PubMed] [Google Scholar]
- 3.Ambali, A. G., and R. C. Jones. 1990. The effects of three reproductive hormones and cortisone on the replication of avian infectious bronchitis virus in vitro. Rev. Roum. Virol. 41:151-156. [PubMed] [Google Scholar]
- 4.Andrew, S. E. 2000. Feline infectious peritonitis. Vet. Clin. North Am. Small Anim. Pract. 30:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora, V., D. C. Knapp, M. T. Reddy, D. D. Weller, and P. L. Iversen. 2002. Bioavailability and efficacy of antisense morpholino oligomers targeted to c-myc and cytochrome P-450 3A2 following oral administration in rats. J. Pharm. Sci. 91:1009-1018. [DOI] [PubMed] [Google Scholar]
- 6.Arora, V., D. C. Knapp, B. L. Smith, M. L. Statdfield, D. A. Stein, M. T. Reddy, D. D. Weller, and P. L. Iversen. 2000. c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J. Pharmacol. Exp. Ther. 292:921-928. [PubMed] [Google Scholar]
- 7.Aurelian, L., and C. C. Smith. 2000. Herpes simplex virus type 2 growth and latency reactivation by cocultivation are inhibited with antisense oligonucleotides complementary to the translation initiation site of the large subunit of ribonucleotide reductase (RR1). Antisense Nucleic Acid Drug Dev. 10:77-85. [DOI] [PubMed] [Google Scholar]
- 8.Barlough, J. E., and B. L. Shacklett. 1994. Antiviral studies of feline infectious peritonitis virus in vitro. Vet. Rec. 135:177-179. [DOI] [PubMed] [Google Scholar]
- 9.Benedetto, A., G. B. Rossi, C. Amici, F. Belardelli, L. Cioe, G. Carruba, and L. Carrasco. 1980. Inhibition of animal virus production by means of translation inhibitors unable to penetrate normal cells. Virology 106:123-132. [DOI] [PubMed] [Google Scholar]
- 10.Bermejo Martin, J. F., J. L. Jimenez, and A. Munoz-Fernandez. 2003. Pentoxifylline and severe acute respiratory syndrome (SARS): a drug to be considered. Med. Sci. Monit. 9:SR29-SR34. [PubMed]
- 11.Booth, C. M., L. M. Matukas, G. A. Tomlinson, A. R. Rachlis, D. B. Rose, H. A. Dwosh, S. L. Walmsley, T. Mazzulli, M. Avendano, P. Derkach, I. E. Ephtimios, I. Kitai, B. D. Mederski, S. B. Shadowitz, W. L. Gold, L. A. Hawryluck, E. Rea, J. S. Chenkin, D. W. Cescon, S. M. Poutanen, and A. S. Detsky. 2003. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 289:2801-2809. [DOI] [PubMed] [Google Scholar]
- 12.Cameron, J. M., M. J. Clemens, M. A. Gray, D. E. Menzies, B. J. Mills, A. P. Warren, and C. A. Pasternak. 1986. Increased sensitivity of virus-infected cells to inhibitors of protein synthesis does not correlate with changes in plasma membrane permeability. Virology 155:534-544. [DOI] [PubMed] [Google Scholar]
- 13.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361:2045-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Treatment of SARS with human interferons. Lancet 362:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Stefano, G., A. Bignamini, C. Busi, F. P. Colonna, and L. Fiume. 1997. Enhanced accumulation of ribavirin and its metabolites in liver versus erythrocytes in mice administered with the liver targeted drug. Ital. J. Gastroenterol. Hepatol. 29:420-426. [PubMed] [Google Scholar]
- 17.Fernandez-Puentes, C. 1984. Permeability to inhibitors of protein synthesis in virus infected cells. Mol. Biol. Rep. 10:65-68. [DOI] [PubMed] [Google Scholar]
- 18.Fingerote, R. J., B. M. Cruz, R. M. Gorczynski, L. S. Fung, H. R. Hubbell, R. J. Suhadolnik, and G. A. Levy. 1995. A 2′,5′-oligoadenylate analogue inhibits murine hepatitis virus strain 3 (MHV-3) replication in vitro but does not reduce MHV-3-related mortality or induction of procoagulant activity in susceptible mice. J. Gen. Virol. 76(Pt 2):373-380. [DOI] [PubMed] [Google Scholar]
- 19.Fuchizaki, U., S. Kaneko, Y. Nakamoto, Y. Sugiyama, K. Imagawa, M. Kikuchi, and K. Kobayashi. 2003. Synergistic antiviral effect of a combination of mouse interferon-alpha and interferon-gamma on mouse hepatitis virus. J. Med. Virol. 69:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagneur, A., J. Sizun, S. Vallet, M. C. Legr, B. Picard, and P. J. Talbot. 2002. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect. 51:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher, T. M., M. J. Buchmeier, and S. Perlman. 1992. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology 191:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, C., and P. L. Iversen. 2000. Intracellular delivery strategies for antisense phosphorodiamidate morpholino oligomers. Antisense Nucleic Acid Drug Dev. 10:263-274. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh, C., D. Stein, D. Weller, and P. Iversen. 2000. Evaluation of antisense mechanisms of action. Methods Enzymol. 313:135-143. [DOI] [PubMed] [Google Scholar]
- 24.Giles, R. V., D. G. Spiller, R. E. Clark, and D. M. Tidd. 1999. Antisense morpholino oligonucleotide analog induces missplicing of C-myc mRNA. Antisense Nucleic Acid Drug Dev. 9:213-220. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez, A., A. Jimenez, D. Vazquez, J. E. Davies, and D. Schindler. 1978. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim. Biophys. Acta 521:459-469. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, M., A. Maeda, T. Watanabe, K. Hanaki, S. Arai, and S. Nozaki. 1995. Inhibition of mouse hepatitis virus multiplication by phosphorothioate analogues of oligodeoxynucleotides complementary to the leader RNA. J. Vet. Med. Sci. 57:1081-1083. [DOI] [PubMed] [Google Scholar]
- 27.Hilton, A., L. Mizzen, G. MacIntyre, S. Cheley, and R. Anderson. 1986. Translational control in murine hepatitis virus infection. J. Gen. Virol. 67(Pt 5):923-932. [DOI] [PubMed] [Google Scholar]
- 28.Hudziak, R. M., E. Barofsky, D. F. Barofsky, D. L. Weller, S. B. Huang, and D. D. Weller. 1996. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 6:267-272. [DOI] [PubMed] [Google Scholar]
- 29.Isaacs, D., D. Flowers, J. R. Clarke, H. B. Valman, and M. R. MacNaughton. 1983. Epidemiology of coronavirus respiratory infections. Arch. Dis. Child. 58:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iversen, P. L. 2001. Phosphorodiamidate morpholino oligomers: favorable properties for sequence-specific gene inactivation. Curr. Opin. Mol. Ther. 3:235-238. [PubMed] [Google Scholar]
- 31.Iversen, P. L., V. Arora, A. J. Acker, D. H. Mason, and G. R. Devi. 2003. Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin. Cancer Res. 9:2510-2519. [PubMed] [Google Scholar]
- 32.Jubin, R., N. E. Vantuno, J. S. Kieft, M. G. Murray, J. A. Doudna, J. Y. Lau, and B. M. Baroudy. 2000. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 74:10430-10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapil, S., C. Chard-Bergstrom, P. Bolin, and D. Landers. 1995. Plaque variations in clinical isolates of bovine coronavirus. J. Vet. Diagn. Investig. 7:538-539. [DOI] [PubMed] [Google Scholar]
- 34.Kapil, S., K. L. Richardson, T. R. Maag, and S. M. Goyal. 1999. Characterization of bovine coronavirus isolates from eight different states in the USA. Vet. Microbiol. 67:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keck, J. G., P. H. Wang, E. J. Lien, and M. M. Lai. 1989. Inhibition of murine coronavirus RNA synthesis by hydroxyguanidine derivatives. Virus Res. 14:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipshidze, N., J. Moses, L. R. Shankar, and M. Leon. 2001. Perspectives on antisense therapy for the prevention of restenosis. Curr. Opin. Mol. Ther. 3:265-277. [PubMed] [Google Scholar]
- 38.Kipshidze, N. N., H. S. Kim, P. Iversen, H. A. Yazdi, B. Bhargava, G. New, R. Mehran, F. Tio, C. Haudenschild, G. Dangas, G. W. Stone, S. Iyer, G. S. Roubin, M. B. Leon, and J. W. Moses. 2002. Intramural coronary delivery of advanced antisense oligonucleotides reduces neointimal formation in the porcine stent restenosis model. J. Am. Coll. Cardiol. 39:1686-1691. [DOI] [PubMed] [Google Scholar]
- 39.Kusunoki, A., N. Miyano-Kurosaki, T. Kimura, K. Takai, N. Yamamoto, H. Gushima, and H. Takaku. 2000. Antisense phosphorothioate oligonucleotides targeted to the human chemokine receptor CXCR4. Nucleosides Nucleotides Nucleic Acids 19:1709-1719. [DOI] [PubMed] [Google Scholar]
- 40.Lacerra, G., H. Sierakowska, C. Carestia, S. Fucharoen, J. Summerton, D. Weller, and R. Kole. 2000. Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc. Natl. Acad. Sci. USA 97:9591-9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamarre, A., and P. J. Talbot. 1995. Protection from lethal coronavirus infection by immunoglobulin fragments. J. Immunol. 154:3975-3984. [PubMed] [Google Scholar]
- 42.Leaman, D. W., F. J. Longano, J. R. Okicki, K. F. Soike, P. F. Torrence, R. H. Silverman, and H. Cramer. 2002. Targeted therapy of respiratory syncytial virus in African green monkeys by intranasally administered 2-5A antisense. Virology 292:70-77. [DOI] [PubMed] [Google Scholar]
- 43.Lemjabbar, H., and C. Basbaum. 2002. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat. Med. 8:41-46. [DOI] [PubMed] [Google Scholar]
- 44.Liu, M. F., Q. Ning, M. Pope, T. Mosmann, J. Leibowitz, J. W. Ding, L. S. Fung, O. Rotstein, R. Gorczynski, and G. A. Levy. 1998. Resistance of naive mice to murine hepatitis virus strain 3 requires development of a Th1, but not a Th2, response, whereas pre-existing antibody partially protects against primary infection. Adv. Exp. Med. Biol. 440:415-423. [DOI] [PubMed] [Google Scholar]
- 45.Luo, T., Y. H. Lee, J. P. Saint-Jeannet, and T. D. Sargent. 2003. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. USA 100:532-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macintyre, G., B. Curry, F. Wong, and R. Anderson. 1991. Hygromycin B therapy of a murine coronaviral hepatitis. Antimicrob. Agents Chemother. 35:2125-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macintyre, G., C. Kooi, F. Wong, and R. Anderson. 1990. On the membrane cytopathology of mouse hepatitis virus infection as probed by a semi-permeable translation-inhibiting drug. Adv. Exp. Med. Biol. 276:67-72. [DOI] [PubMed] [Google Scholar]
- 48.Macintyre, G., F. Wong, and R. Anderson. 1989. A model for persistent murine coronavirus infection involving maintenance via cytopathically infected cell centres. J. Gen. Virol. 70(Pt 3):763-768. [DOI] [PubMed] [Google Scholar]
- 49.Macintyre, G., D. E. Woods, and R. Anderson. 1991. Hygromycin B inhibits synthesis of murine coronavirus RNA. Antimicrob. Agents Chemother. 35:2630-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda, A., T. Mizutani, M. Hayashi, K. Ishida, T. Watanabe, and S. Namioka. 1995. Inhibition of viral multiplication in acute and chronic stages of infection by ribozymes targeted against the polymerase gene of mouse hepatitis virus. Adv. Exp. Med. Biol. 380:399-404. [DOI] [PubMed] [Google Scholar]
- 51.Margraf, S., M. Bittoova, J. U. Vogel, R. Kotchekov, H. W. Doerr, and J. Cinatl, Jr. 2001. Antisense oligonucleotide ISIS 2922 targets IE-expression and prevents HCMV-IE-induced suppression of TSP-1 and TSP-2 expression. Nucleosides Nucleotides Nucleic Acids 20:1425-1428. [DOI] [PubMed] [Google Scholar]
- 52.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 53.McCaffrey, A. P., L. Meuse, M. Karimi, C. H. Contag, and M. A. Kay. 2003. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology 38:503-508. [DOI] [PubMed] [Google Scholar]
- 54.Mizuta, T., M. Fujiwara, T. Hatta, T. Abe, N. Miyano-Kurosaki, S. Shigeta, T. Yokota, and H. Takaku. 1999. Antisense oligonucleotides directed against the viral RNA polymerase gene enhance survival of mice infected with influenza A. Nat. Biotechnol. 17:583-587. [DOI] [PubMed] [Google Scholar]
- 55.Mizutani, T., M. Hayashi, A. Maeda, K. Ishida, T. Watanabe, and S. Namioka. 1994. The inhibitory effects of MgSO4 on the multiplication and transcription of mouse hepatitis virus. Jpn. J. Vet. Res. 42:95-102. [PubMed] [Google Scholar]
- 56.Monto, A. S., and S. K. Lim. 1974. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 129:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moulton, H. M., M. C. Hase, K. M. Smith, and P. L. Iversen. 2003. HIV Tat peptide enhances cellular delivery of antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 13:31-43. [DOI] [PubMed] [Google Scholar]
- 58.Nasevicius, A., and S. C. Ekker. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216-220. [DOI] [PubMed] [Google Scholar]
- 59.Ning, Q., D. Brown, J. Parodo, M. Cattral, R. Gorczynski, E. Cole, L. Fung, J. W. Ding, M. F. Liu, O. Rotstein, M. J. Phillips, and G. Levy. 1998. Ribavirin inhibits viral-induced macrophage production of TNF, IL-1, the procoagulant fgl2 prothrombinase and preserves Th1 cytokine production but inhibits Th2 cytokine response. J. Immunol. 160:3487-3493. [PubMed] [Google Scholar]
- 60.Pei, J., M. J. Sekellick, P. I. Marcus, I. S. Choi, and E. W. Collisson. 2001. Chicken interferon type I inhibits infectious bronchitis virus replication and associated respiratory illness. J. Interferon Cytokine Res. 21:1071-1077. [DOI] [PubMed] [Google Scholar]
- 61.Popova, R., and X. Zhang. 2002. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology 294:222-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramakrishna, C., C. C. Bergmann, R. Atkinson, and S. A. Stohlman. 2003. Control of central nervous system viral persistence by neutralizing antibody. J. Virol. 77:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robbins, I., and B. Lebleu. 2000. Vesicular stomatitis virus as model system for studies of antisense oligonucleotide translation arrest. Methods Enzymol. 313:189-203. [DOI] [PubMed] [Google Scholar]
- 64.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 65.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 67.Schwarz, B., E. Routledge, and S. G. Siddell. 1990. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 64:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shieh, C. K., H. J. Lee, K. Yokomori, N. La Monica, S. Makino, and M. M. Lai. 1989. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J. Virol. 63:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sidwell, R. W., J. H. Huffman, E. W. Call, R. P. Warren, L. A. Radov, and R. J. Murray. 1987. Inhibition of murine hepatitis virus infections by the immunomodulator 2,3,5,6,7,8-hexahydro-2-phenyl-8,8-dimethoxy-imidazo[1,2a]pyridine (PR-879-317A). Antimicrob. Agents Chemother. 31:1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sierakowska, H., M. J. Sambade, S. Agrawal, and R. Kole. 1996. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 93:12840-12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smee, D. F., H. A. Alaghamandan, K. Ramasamy, and G. R. Revankar. 1995. Broad-spectrum activity of 8-chloro-7-deazaguanosine against RNA virus infections in mice and rats. Antivir. Res. 26:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stein, D., E. Foster, S. B. Huang, D. Weller, and J. Summerton. 1997. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 73.Stein, D. A., D. E. Skilling, P. L. Iversen, and A. W. Smith. 2001. Inhibition of Vesivirus infections in mammalian tissue culture with antisense morpholino oligomers. Antisense Nucleic Acid Drug Dev. 11:317-325. [DOI] [PubMed] [Google Scholar]
- 74.Summerton, J. 1999. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489:141-158. [DOI] [PubMed] [Google Scholar]
- 75.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187-195. [DOI] [PubMed] [Google Scholar]
- 76.Talbot, P. J., A. A. Salmi, R. L. Knobler, and M. J. Buchmeier. 1984. Topographical mapping of epitopes on the glycoproteins of murine hepatitis virus-4 (strain JHM): correlation with biological activities. Virology 132:250-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor, M. F., D. D. Weller, and L. Kobzik. 1998. Effect of TNF-alpha antisense oligomers on cytokine production by primary murine alveolar macrophages. Antisense Nucleic Acid Drug Dev. 8:199-205. [DOI] [PubMed] [Google Scholar]
- 78.Tkachuk, Z., L. I. Semernikova, V. V. Tkachuk, L. M. Reshot'ko, L. V. Tkachuk, T. Eremenko, G. K. Matsuka, and I. A. Mikhailopulo. 1998. Antiviral effect of trimeric 2′,5′-oligoadenylic acid and some of its analogues. New Microbiol. 21:141-146. [PubMed] [Google Scholar]
- 79.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warren, R. P., R. W. Sidwell, J. H. Huffman, E. W. Call, M. C. Healey, A. V. Johnston, L. A. Radov, R. J. Murray, and C. R. Kinsolving. 1990. Effect of oxamisole on immune parameters of mice infected with murine hepatitis. Int. J. Immunopharmacol. 12:625-630. [DOI] [PubMed] [Google Scholar]
- 81.Wei, X., M. Gotte, and M. A. Wainberg. 2000. Human immunodeficiency virus type-1 reverse transcription can be inhibited in vitro by oligonucleotides that target both natural and synthetic tRNA primers. Nucleic Acids Res. 28:3065-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weiss, R. C., N. R. Cox, and M. L. Martinez. 1993. Evaluation of free or liposome-encapsulated ribavirin for antiviral therapy of experimentally induced feline infectious peritonitis. Res. Vet. Sci. 55:162-172. [DOI] [PMC free article] [PubMed] [Google Scholar]