Abstract
Eph-ephrin interactions control the signal transduction between cells and play an important role in carcinogenesis and other diseases. The interactions between Eph receptors and ephrins of the same subclass are promiscuous; there are cross-interactions between some subclasses, but not all. To understand how Eph-ephrin interactions can be both promiscuous and specific, we investigated sixteen energy landscapes of four Eph receptors (A2, A4, B2, and B4) interacting with four ephrin ligands (A1, A2, A5, and B2). We generated conformational ensembles and recognition energy landscapes starting from separated Eph and ephrin molecules and proceeding up to the formation of Eph-ephrin complexes. Analysis of the Eph-Ephrin recognition trajectories and the co-evolution entropy of 400 ligand binding domains of Eph receptor and 241 ephrin ligands identified conserved residues during the recognition process. Our study correctly predicted the promiscuity and specificity of the interactions and provided insights into their recognition. The dynamic conformational changes during Eph-ephrin recognition can be described by progressive conformational selection and population shift events, with two dynamic salt bridges between EphB4 and Ephrin-B2 contributing to the specific recognition. EphA3 cancer-related mutations lowered the binding energies. The specificity is not only controlled by the final stage of the interaction across the protein-protein interface, but also has large contributions from binding kinetics with the help of dynamic intermediates along the pathway from the separated Eph and ephrin to the Eph-ephrin complex.
Keywords: Eph receptor tyrosine kinase, conformational selection, induced fit, protein-protein interaction, energy landscape, conformational dynamics
1. Introduction
Three theories were proposed to explain protein-ligand interactions. The ‘lock and key’ mechanism assumes that the protein is rigid and that to form a functional complex the binding site should be an exact match of the ligand. The ‘induced fit’ hypothesis argues that the fact that protein complexes often have different conformations from those of their unbound protein constituents suggests that the bound conformation is ‘induced’ by the binding partner. ‘ Induced fit’ assumes that the protein is flexible around the binding site. However, proteins are dynamic molecules and both binding partners are flexible and exist in conformational distributions. The ‘conformational selection and population shift’ mechanism [1–8] suggests that a ligand selects its most favored preexisting receptor protein conformation, and that binding shifts equilibrium of the conformational ensemble of the receptor toward this favored state. The conformational selection and population shift theory provides not only an explanation of protein recognition, but also a general framework for cellular communication [9], particularly when extended and complemented by induced fit to optimize the interaction [10]. Protein conformational dynamics can encode functional regulation which a static description of molecular structure is unable to do [11]. Intrinsically disordered protein regions which may allow certain functional promiscuity are at the extreme end of the dynamic spectrum [12]. Elucidation of the detailed mechanisms of how conformational energy landscapes can shape dynamic recognition can help in understanding these processes at the molecular level.
Eph (Erythropoietin-producing hepatoma) tyrosine kinase cell surface receptors comprise a large group of receptor tyrosine kinases [13]. These receptors and their ephrin ligands form signaling hubs, orchestrating signal transduction within interacting cells to regulate cell proliferation, differentiation, migration and adhesion [14–16]. The roles of Eph/ephrin have been well characterized in embryogenesis [17–19] and carcinogenesis [20–24], and it is clear that Eph/ephrin signaling can play an important role in the development of novel inhibition strategies [15, 24–26]. Eph-Ephrin interactions also regulate the proliferation of stem and progenitor cells [22]. Eph receptors can have a dual role in both tumor promotion and tumor suppression [27]. Mutations and overexpression of Eph receptor and ephrin can result in tumor growth, invasiveness and metastasis in many human cancers [22, 25, 27–29]. Sixty-one percent of glioblastomas, 76% of ovarian cancers, and 85% of prostate cancers overexpress EphA2, and EphB4 is also upregulated [30]. EphA3 is one of the most frequently mutated proteins in lung cancer. Many point mutations were observed in EphA3 receptor, but the oncogenic potential remains unknown [31]. Eph receptors and their ephrin ligands are also important players in chronic inflammatory diseases and immune function [32]. Viruses can make use of ephrin in evasion of host innate immune responses [33]. Functional recognition of Eph receptors by their ephrin ligands is important to control these complex biological processes.
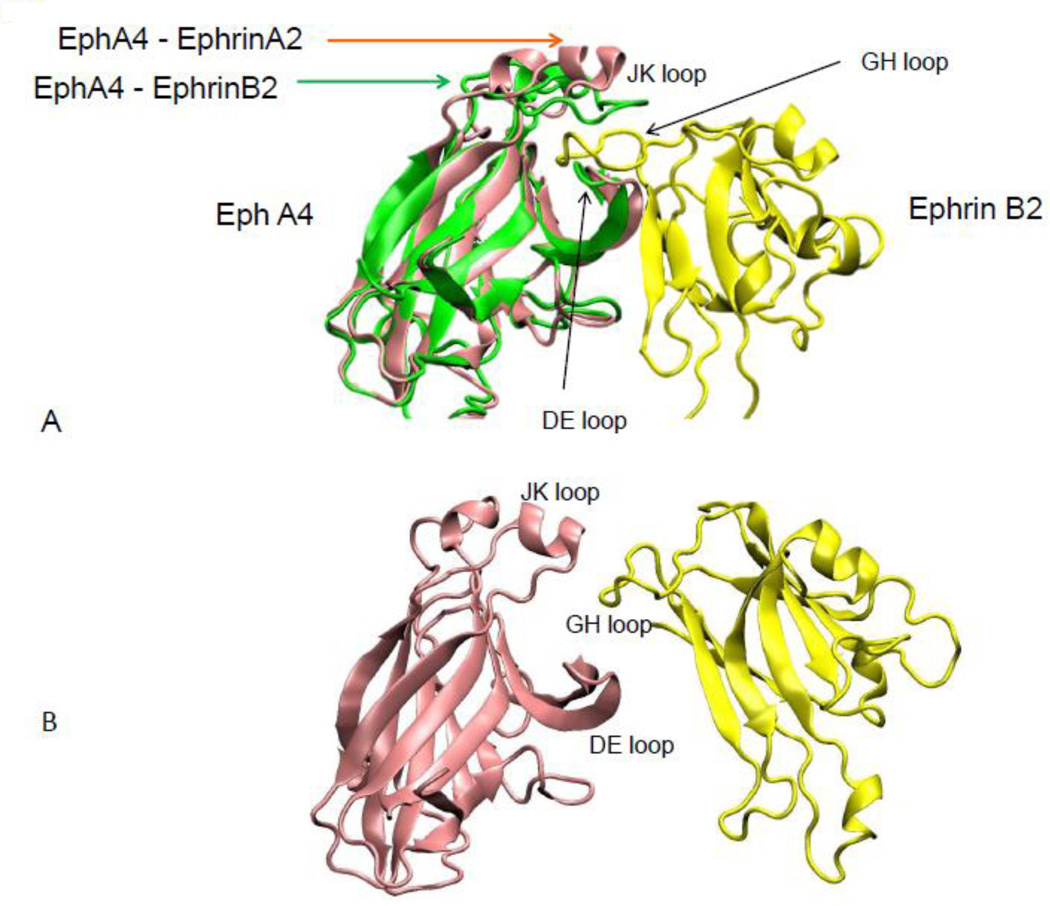
10 EphA and 6 EphB receptors have been observed to interact with 6 ephrin-A and 3 ephrin-B ligands, respectively. The interactions between the Eph receptors and ephrin ligands of the same subclass are promiscuous but cross-binding between subclasses is less observed, except for three receptors, EphA3 can bind ephrin-B2, EphB2 can bind ephrin-A5 and EphA4 interacts with all 9 ephrin ligands. Depending on the environment and ligand types, these interactions can lead to different biological functions. The structures of Eph and ephrin are given in Figure 1. Based on loop conformations near the binding site, Eph ligand-binding domain (LBD) structures can be classified into open and closed states. In the apo state, the Eph receptor binding pocket is closed. A conformational change opening the binding site allows ephrin binding.
Figure 1.
Eph-ephrin complexes are conformationally flexible, as indicated by comparing two Eph-ephrin complexes. (A) Two overlaid conformations of Eph receptors reveal the change in the secondary structure of the JK loop binding to ephrin-A2 and ephrin-B2. ephrin-A2 is not shown for clarity. (B) Starting conformations in the simulations of Eph-ephrin recognition with separated Eph receptor and ephrin ligand.
These considerations underscore the importance of delineating the conformational changes during Eph-ephrins recognition. The combinational interactions between Eph receptors and ephrins raise the question of how promiscuity and specificity are regulated. Eph-ephrins provide a good system for exploring the mechanisms of protein recognition processes with different free energy landscapes. Here we focused on three aspects of Eph-ephrins recognition. First, we investigated the energy landscape of the recognition pathways. The pathways start from the separated Eph and ephrin molecules and proceed up to the formation of the Eph-ephrin complexes. Second, we identified the contact residues responsible for the promiscuous and specific Eph-ephrin recognition at various stages of the recognition pathways. Finally, we studied the evolution entropies of residues involved in specific and non-specific Eph-ephrin recognition. We correctly predicted the promiscuity and specificity of the interaction among the four Eph receptors and four ephrins and provide insights into their recognition. We found that Eph-ephrin recognition can be characterized by progressive conformational selection and population shift [34]. The specificity is not only controlled by the final interaction across the protein-protein interface, but also contributed by dynamic intermediate conformations along the pathways from the separated Eph and ephrin molecules to the Eph-ephrin complex. We observed two dynamic salt bridges between EphB4 and Ephrin-B2 that may contribute to the specific recognition.
2. Materials and Methods
The simulations had three steps. (1) Generation of conformational ensembles to describe the Eph-ephrin recognition. The ensembles were generated by simulating the recognition processes, bringing the separated Eph and ephrin proteins to the final bound state using Structure-Based Models (SBMs). (2) Interaction energy evaluation to characterize the energy landscape of Eph-ephrin recognition. (3) Structure and sequence analysis of recognition residues.
2.1 Dataset
Seven crystal structures of Eph-ephrin complexes (PDB codes: 3HEI, 3MX0, 2WO2, 2WO3, 1SHW, 1KGY, and 2HLE) and one unbound EphA4 structure (PDB code: 2WO1) were downloaded from the PDB website. The structures of the EphA3 wild-type and three point mutants (T37K, S46F, and N85S) in the ligand-binding domain were constructed by homology modeling based on EphA2 in the EphA2 ephrin-A1 complex (PDB codes: 3HEI) as template. We split each complex crystal structure into two individual structures for the corresponding Eph receptor and ephrin (Figure 1B). Based on the available crystal structures, sixteen Eph-ephrin complexes were chosen (Table 1). Of them, seven complex structures have already been included in the downloaded set, as indicated by the PDB codes in Table 1. The structures for the other nine complexes were constructed by replacing the Eph and ephrin structures based on structural alignment of the known Eph-ephrin complex. For example, the structure for the EphA4 ephrin-A1 complex was generated using the individual EphA4 structure extracted from 2WO3 and the ephrin-A1 structure from 3HEI, by aligning the ephrin-A1 structure with respect to the ephrin-A2 in 2WO3. The structures of the wild-type and three point mutants of EphA3-ephrins complexes were constructed based on the EphA4 ephrin-A1 complex, by aligning the EphA3 structure with respect to EphA2 structure in EphA2 ephrins (A1, A2, A5, and B2) complexes.
Table 1.
Averaged interaction free energies between Eph and ephrin reflect their tendencies to form complexes. Complexes with available crystal structures are indicated with their PDB codes.
| Eph-ephrin complex |
Interaction based on classification |
Interaction energies* (kcal/mol) |
Results from simulations |
|---|---|---|---|
| 2wo2(EphA4 ephrin-B2) |
√, promiscuous | −77.5 ± 10.4 | √ |
| 2wo3(EphA4 ephrin-A2) |
√, promiscuous | −33.2 ± 9.3 | √ |
| 3hei(EphA2 ephrin-A1) |
√, specific | −42.7 ± 8.2 | √ |
| 2hle(EphB4 ephrin-B2) |
√, specific | −40.1 ± 9.4 | √ |
| 1kgy(EphB2 ephrin-B2) |
√, promiscuous | −28.0 1± 0.7 | √ |
| 3mx0(EphA2 ephrin-A5) |
√, specific | −4.0 ± 6.8 | √? |
| 1shw(EphB2 ephrin-A5) |
√, promiscuous | −10.4 ± 4.7 | √ |
| 4BKA(EphA4 ephrin-A5) |
√, promiscuous | −7.0 ± 7.9 | √ |
|
EphA4 ephrin-A1 |
√, promiscuous | −30.7 ± 7.9 | √ |
|
EphA2 ephrin-A2 |
√, specific | −14.7 ± 6.2 | √ |
| EphB4 ephrin-A5 | X | 18.3 ± 12.2 | X |
| EphB2 ephrin-A2 | X | 0.4 ± 7.0 | X? |
| EphA2 ephrin-B2 | X | 39.4 ± 9.9 | X |
| EphB2 ephrin-A1 | X | 11.0 ± 6.2 | X |
| EphB4 ephrin-A2 | X | 11.9 ± 11.3 | X |
| EphB4 ephrin-A1 | X | 11.5 ± 3.4 | X |
Based on the trajctories from 300 ps to 500 ps after RMSDs are stablized.
2.2 MD simulation with structure-based models
In the creation of GROMACS topology files, the web server (http://smog-server.org/) uses previously published and validated structure-based Hamiltonians for all-atom models. We simulated four kinds of complex recognition trajectories. In the first, the initial and terminal states were the separated (using VMD software) crystal structures. In the second, constructed complexes were used in the same manner. The conformations of Eph both in the initial and terminal states were open in these two cases. We also simulated recognition trajectories from separated complexes replacing open Eph receptor conformations by closed terminal states. In the third type, complexes were used as terminal states, with the initial states being the separated complexes and the Eph receptors replaced by their closed conformations. Simulations of recognition trajectories of complexes whose Eph receptor was built by homology modeling were carried out. The Eph receptor conformations were open in the initial and terminal states.
The SBM model was implemented using Langevin MD simulations in GROMACS. Simulations were performed at reference temperature T=50. The MD simulations with total time of 0.5 ns were performed to ensure that each complex be formed from separated conformation. Simulation time steps of 0.0001 and 0.0005 ps were used and coordinates were saved every 1000 steps. To ensure as high a quality as possible given the differences between the structure-based models and other commonly used models [35], the temperature and time step used in the structure-based models are in reduced units, smaller than many other MD simulations. Many proteins fold around T=100–120 in the structure-based models.
2.3 Energy landscape analysis
The energy landscape of the conformational change of a protein or complex can be obtained by an appropriate conformational sampling method. Conformations generated by SBM simulations were used for energy analysis in this study. In order to get a more accurate characterization of the energy landscape, we recalculated the interaction energies between the Eph and ephrin molecules using the MM-GBSA in the Amber software. MM-GBSA methods calculate binding free energies for macromolecules by combining molecular mechanics calculations and continuum solvation models. In MM/GBSA, the binding free energy (Δ Gbind) between a ligand (L) and a rector (R) to form a complex RL is calculated as [36]:
Before calculating binding energies, conformations extracted from the simulated trajectories were energy optimized for 100 steps using SANDER from the Amber software. Then, we collected the binding energies of all conformations for each simulation and drew the energy landscapes.
The "complexes" without structural evidence and have repulsive energy could be artificial, and there is no real binding process for their formations. However, in our computational study, we probe their formations in silica to show the different between energetically favored recognition and disallowed binding. The study of the disallowed “binding process” reveals how Eph receptors selectively avoid binding with these ligands.
2.4 Co-evolution and entropt analysis of protein sequence
The Observed Minus Expected Squared (OMES), Mutual Information (MI), and Statistical Coupling Analysis (SCA) covariance algorithms were used to find the conserved residues of Eph receptors and ephrins [37]. Protein families of the Eph receptor ligand-binding domain (accession: PF01404) and ephrin ligand (accession: PF00812) were downloaded from the Pfam database. We obtained a dataset that includes 400 homologous sequences of the Eph receptor ligand-binding domain family and 241 sequences for ephrin ligands. The sequences of each family were aligned using ClustalW. The scores were calculated by the OMES, MI, SCA algorithms for every residue in the proteins, and residues in the top sixteen percent of the scores were defined as conserved.
The protein sequence entropy analysis was performed as described by Fuchs et al. [38] and Shen et al. [39]. The entropy of each residue in the homologous families of the Eph receptor ligand binding domain and eprhin ligand used in co-evolution analysis was calculated. It is defined as
The scores derived from the entropy analysis were divided into four cataogories: highly specific residues with entropy of 0.5 and less, specific residues with entropy of 0.5 < Si ≤ 1.0, variable residues with entropy of 1.0 < Si ≤ 2.0, and non-specific residues with entropy larger than 2.0.
Results
3.1 Eph-ephrin recognition trajectories in the SBM simulations
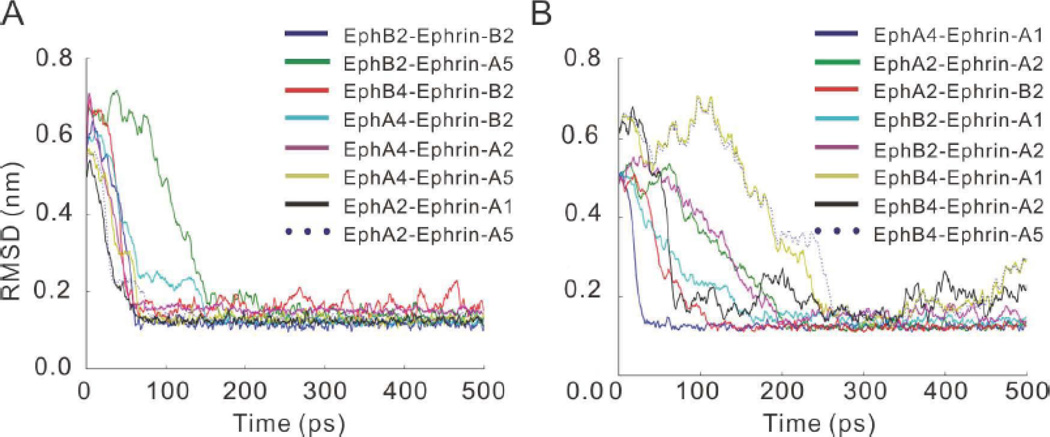
Structure-based models (SBMs) are simplified models of biomolecular dynamics that arise from funneled energy landscapes [40]. They were developed to study long-time scale molecular dynamics simulations. The simplest varieties of SBM are coarse-grained, where each residue is represented by a single bead and only the interactions present in the native state are attractive [41]. Here we use an all-atom SBM, which allows a more explicit connection with experimental observables. SBMs were used to understand the interplay between side-chain and backbone dynamics during folding of proteins and RNAs and protein-protein recognition [35, 40, 42]. We used the all-atom SBMs to simulate conformational changes from the initial unbound state to the terminal bound state. In the terminal bound state, contacts are defined between native atom pairs. Using the final bound state as reference, we examined the root mean square deviation (RMSD) values of the conformational ensembles in the trajectory. The recognition processes observed during the simulation are progressive with initial small changes of RMSD, large drop of RMSD, and final RMSD fluctuations.
As can be seen in Figure 2 and Sup-Figure 1, the RMSDs for the initial unbound states for sixteen Eph-ephrin complexes (Table 1) are more than 0.6 nm. In some complexes, at the beginning of the simulation the RMSD values increase during the simulation, e.g., those of EphB4 ephrin-A5, EphA2 ephrin-A5, EphA4 ephrin-A5 in Figure 2. This suggests that as expected, during the recognition process interface residues between the two proteins adjust their positions. As indicated by the RMSD vs time plots, the RMSDs for all simulated Eph-ephrin pairs decrease to small values around 0.1 nm, indicating that they reach the bound states with conformations similar to those in the crystal structures.
Figure 2.
The RMSD trajectories of Structure-Based Models (SBMs) simulations of Eph-ephrin recognition for (A) complexes with bound crystal structure available, and (B) modeled complexes of bound structures. Each line was averaged from five simulations. Both kinds of complexes can have either large or small RMSD fluctuations after 300 ps, with the modeled complexes having larger fluctuations.
Grouping the RMSD trajectories, we observed three recognition processes. The majority of the complexes can reach near-bound states with small RMSD regions within 100 ps. This group represents Eph-ephrin complexes with fast recognition. EphB2 ephrin-A5 has intermediate recognition kinetics, reaching the bound state at around 150 ps. EphA2 ephrin-A2, EphB2 ephrin-A2, EphB4 ephrin-A1 and EphB4 ephrin-A5 pairs undergo large conformational changes and thus are slow to reach the final bound states. Note that EphB2 ephrin-A2, EphB4 ephrin-A1 and EphB4 ephrin-A5 are disallowed binding pairs.
After reaching the bound states, there are still fluctuations in the RMSD values (Figure 2), suggesting that the two proteins continuously adjust their positions for optimal interactions. However, no significant difference was found between the crystal structures and the modeled complexes. Both kinds of complexes could have either large or small RMSD fluctuations in the final bound states.
3.2 Energy landscapes of Eph-ephrin recognition from open conformations
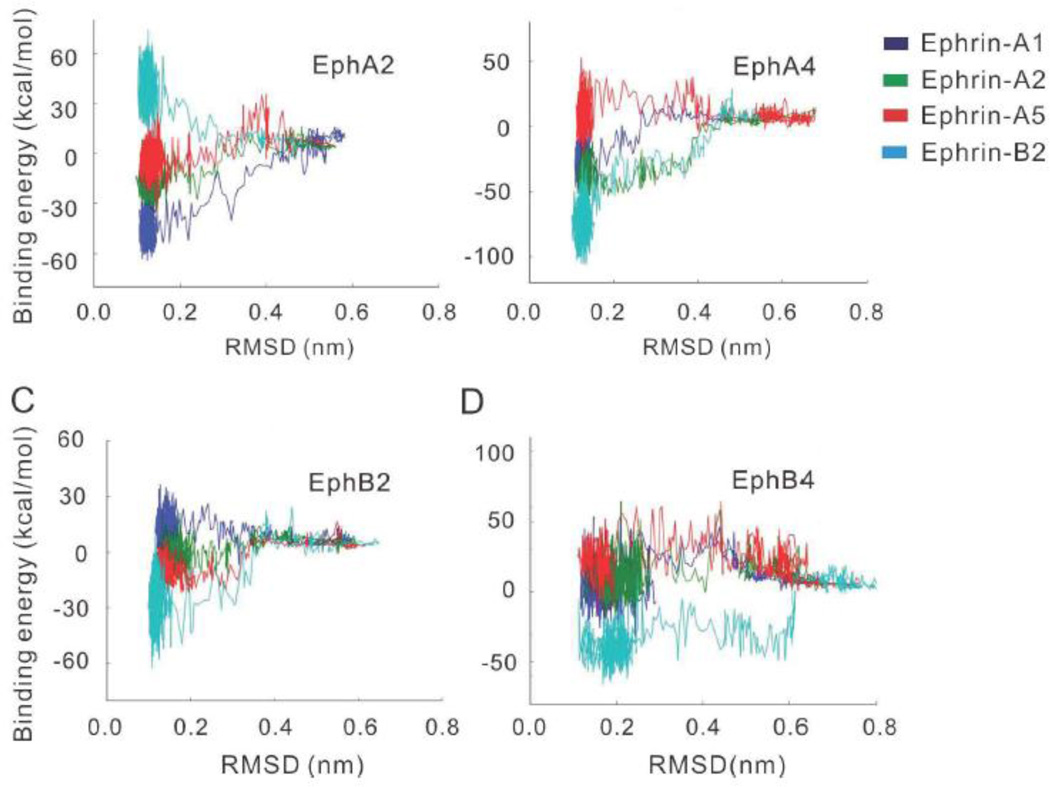
To characterize the energy landscape in Eph-ephrin recognition, we calculated the interaction energies between the Eph receptors and the ephrin ligands. The energy-RMSD plots are displayed in Figure 3 and the binding energy (average binding energy of conformations derived from last 100 frames) for the recognition of 16 Eph-ephrin pairs are listed in Table 1. Starting from initial separated states to the recognized complex states, steps of energy changes of varying extents were observed (Figure 3). Depending on the combinations among the Eph receptors and ephrin ligands, the energy changes can be attractive (energy drop) or repulsive (energy rise) at different recognition times.
Figure 3.
Energy landscapes of four Eph receptors (A2, A4, B2, and B4) interacting with four ephrins (A1, A2, A5, and B2) reflect the binding tendencies between ephrins and their receptors, consistent with the promiscuity and specificity of their interactions. (A) EphA2 is sepcific to ephrin-A; (B) EphA4 is promiscus to both ephrinA and ephrin-B; (C) EphB2 is promiscus to both ephrin-A and ephrin-B; and (D) EphB4 is sepcific to ephrin-B.
As can be seen in Figure 3A, the energy landscapes of EphA2 interacting with ephrin-A1, ephrin-A2 and ephrin-A5 are attractive funnel-like, while that of EphA2 ephrin-B2 is strongly repulsive. The EphA2 ephrin-A2 complex was based on homology modeling, suggesting its potential strong interaction. Even though there is no direct information about whether EphA2 ephrin-A2 is a specific interacting pair [43], recent report suggested their specific interactions in bone-resorbing osteoclasts and bone-forming osteoblasts. For example, it has been shown that bidirectional signaling through EphA2 ephrin-A2 enhances osteoclastogenesis and suppresses osteoblastogenesis [44, 45].
The EphA4 receptor could recognize both A-class and B-class ephrin ligands. EphA4 interactions show that EphA4 ephrin-A1, EphA4 ephrin-A2, and EphA4 ephrin-B2 have attractive energy landscape. In the bound states, the binding free energies of EphA4 to various ephrin ligands (EphA4 ephrin-A1, EphA4 ephrin-A2, and EphA4 ephrin-B2) are attractive, except for the binding free energy to ephrin-A5 (Figure 3B). The EphA4 ephrin-A5 complex was initially modeled based on homology. Recently, a crystal structure of EphA4 ephrin-A5 complex was published [46] (PDB code 4BKA). Using the newly available crystal structure of EphA4 ephrin-A5 complex, we obtained similar results (Sup-Figure 2). While EphA4 ephrin-A5 presents a repulsive energy landscape, the energies in the final stage indicate that there are attractive conformers.
Similarly, EphB2 also binds both ephrin-A and ephrin-B (Figure 3C). It has funnel-like interactions with ephrin-A5 and ephrin-B2. In contrast, two EphAs specific to ephrin-A1 and A2 do not have favorable interactions with EphB2 and the EphB2 ephrin-A1 energy landscape is strongly repulsive. EphB4 only binds to class B ephrin ligands. The crystal structure of EphB4 ephrin-B2 complex is available [47] and there are indications that EphB4 may also interact with ephrin-B1 [43, 45, 48, 49]. Consistently, the energy landscapes of EphB4 ligand interactions reveal attractive interactions with ephrin-B2 and repulsive interactions with ephrin-A1, ephrin-A2, and ephrin-A5 (Figure 3D). Therefore, the EphB4 energy landscape indicates that the specificity of EphB4 is not only controlled by the final interaction across the protein-protein interface, but also contributed by dynamic intermediate stages along the pathway from the separated Eph and ephrin to the Eph-ephrin complex. Detailed analysis of EphB4 interactions from structural dynamics is provided in later sections.
Essentially, the obtained energy landscapes reflect the binding tendencies between ephrins and their receptors, consistent with the promiscuity and specificity of their interactions. Of these, the Eph receptors of 10 Eph-ephrin pairs could recognize the corresponding ephrin ligands, but those of other 6 pairs could not (Table 1). Among the six energetically non-favored pairs, three (EphB4 ephrin-A2, EphB2 ephrin-A2, and EphB4 ephrin-A5) display a large time to reach at 0.1 nm RMSD in the MD simulations (Figure 2B), indicating the coupling of energetic and dynamical control of Eph ephrin recognition processes.
3.3 Energy landscapes of Eph-ephrin recognition from closed conformations
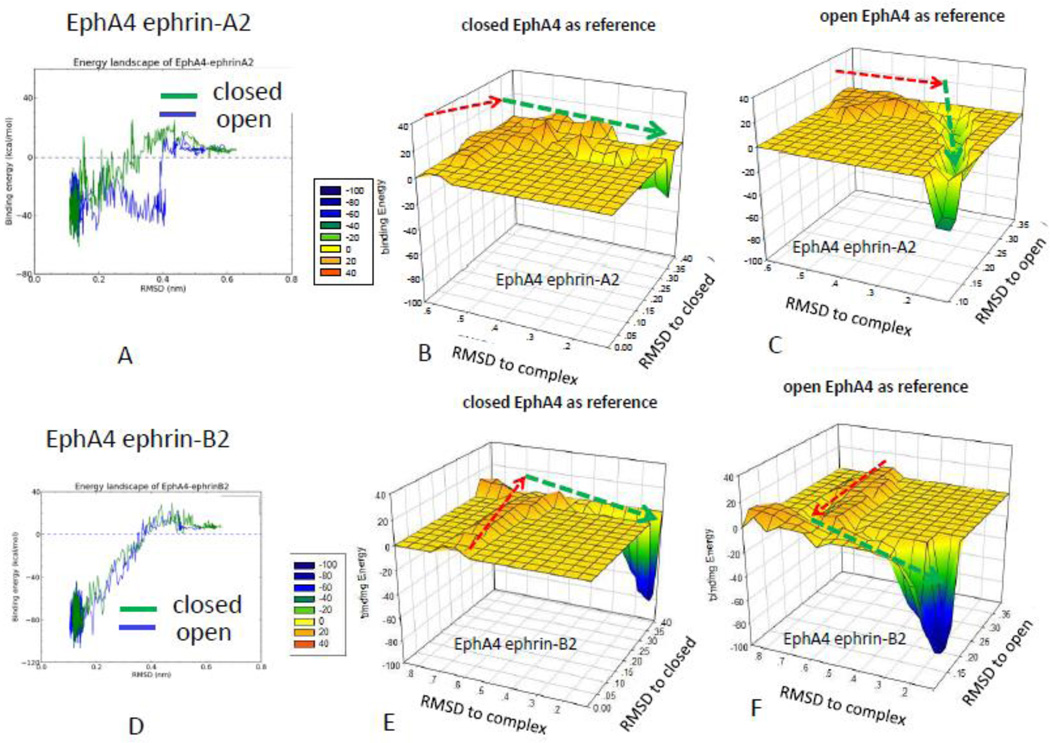
We have investigated the Eph-ephrin recognition assuming that the initial Eph receptor states are in open conformations, which allows fast conformational selection events with the incoming ephrin ligands. This can be seen from the trajectory of the EphA4 ephrin-A2 interaction, which has an attractive funnel-like energy landscape with a sharp energy drop around RMSD = 0.4 nm (Figure 4A). However, for the closed EphA4 conformation, gradual conformational adjustments and energy changes are observed during the recognition process (Figures 4A, 4B and 4C), indicating a progressive conformational selection process. Opening up the closed EphA4 conformation (PDB code: 2WO1) to allow initial EphA4 ephrin-A2 recognition before the RMSDcomplex reaches 0.4 nm, encounters an energy barrier (Figure 4A). Prior to reaching the energy barrier, the EphA4 conformational change has increasing RMSDclosed with respect to the closed conformation (red arrow, Figure 4B). However, the conformer in the energy barrier does not get closer to the final bound conformation (red arrow, Fig 4C), since the RMSDopen with respect to the final open conformation does not decrease. After passing the barrier, the conformations of EphA4 get closer and closer to the final bound open conformer (red arrow, Figure 4C) toward reaching the final recognition with ephrin-A2.
Figure 4.
Comparison of the energy landscape of the Eph-ephrin recognition starting with open and closed Eph conformations reveals that the EphA4 receptor uses a different mechanism to interact with ephrin-A2 and ephrin-B2. (A) Comparison of EphA4 ephrin-A2 interaction starting with closed and open EphA4 conformations. Starting from a closed EphA4 conformation, the RMSD with respect to the closed (B) and open (C) EphA4 conformations suggests simutanous ephrin-A2 recognition and adjustment to the open EphA4 conformation. (D) Comparison of EphA4 ephrin-B2 interaction starting with closed and open EphA4 conformations. Starting from a closed EphA4 conformation, the three-dimensional energy landscapes of the EphA4 ephrin-B2 interaction of the closed (E) and (F) open EphA4 conformations suggests that adjusting to the open EphA4 conformation takes place at the early stage of ephrin-B2 recognition.
Interestingly, the same EphA4 receptor uses a different mechanism to interact with ephrin-B2 (Figures 4D, 4E and 4F). While formation of the EphA4 ephrin-B2 interaction also needs to overcome a barrier (red arrows, Fig 4E and 4F) as in the case of the EphA4 ephrin-A2 interaction, the EphA4 conformation on the barrier reached a state which has a small RMSDopen and is very similar to the final bound open conformation (Figure 4F). Figure 4A and 4D provided insights to understand the specific and promiscuous binding as the pattern of binding energy regarding distance varies depending on type of ligand. While specific recognition within the same class can have either progressive conformational selection or a direct conformational selection with sharp energy drop (as EphA4 ephrin-A2 in Figure 4A and EphB4 eprhin-B2 in Figure 3D), the promiscuous binding need go through progressive conformational selections to reach final recognized state.
When the EphA4 receptor starts the recognition process from the closed conformation, the complex will form as long as EphA4 receptor opens only sufficiently to allow the ligand binding. It seems that complete opening is not a prerequisite for the formation of the final complex. However, when starting from the open EphA4 conformation, the recognition process has a lower energy barrier than when starting from closed conformation. A new x-ray structure revealed that the high affinity ephrin-binding pocket of EphA5 has an open pocket in the unbound state, resembling that of other Eph receptors bound to ephrins [50].
3.4 Contact residues in the Eph-ephrin recognition and their evolution entropies
We analyzed the contact residues between Eph and ephrin ligands for each conformation in several time frames. An interacting residue is defined as one whose distance to other residues across the interface is less than 0.6nm. We listed the interacting residues in different recognition time in Table 2.
Table 2.
Contact residues during Eph-ephrin recognition at different time frame*. The common anchoring residues are highlighted in bold fonts.
| EphA4-ephrinA2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Opened EphA4 | Closed EphA4 | ||||||||||||||
| Snapshot 1 (27 ps) |
Snapshot 2 (39 ps) |
Snapshot 3 (48 ps) |
Snapshot 4 (63 ps) |
Snapshot 1 (49 ps) |
Snapshot 2 (53 ps) |
Snapshot 3 (61 ps) |
Snapshot 4 (73 ps) |
||||||||
| Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin |
| CYS73 | PRO135 | CYS73 | PRO135 | CYS73 | PRO135 | GLN71 | ARG111 | CYS73 | PRO135 | CYS73 | PRO135 | ILE59 | ARG111 | ||
| PHE136 | THR104 | PHE136 | THR104 | PHE136 | CYS73 | PRO135 | THR104 | PHE136 | THR104 | PHE136 | CYS73 | PRO135 | |||
| SER137 | LEU105 | SER137 | LEU105 | SER137 | THR104 | PHE136 | LEU105 | SER137 | LEU105 | SER137 | THR104 | PHE136 | |||
| ARG106 | LEU138 | ARG106 | LEU138 | LEU105 | SER137 | ARG106 | LEU138 | ARG106 | LEU138 | LEU105 | SER137 | ||||
| ARG162 | ARG162 | ARG106 | LEU138 | CYS191 | ARG162 | ARG106 | LEU138 | ||||||||
| CYS191 | CYS191 | ARG162 | ILE192 | CYS191 | ARG162 | ||||||||||
| ILE192 | ILE192 | CYS191 | ALA193 | ILE192 | CYS191 | ||||||||||
| ALA193 | ALA193 | ILE192 ALA193 | ALA193 | ILE192 ALA193 | |||||||||||
| EphA4-ephrinB2 | |||||||||||||||
| Opened EphA4 | Closed EphA4 | ||||||||||||||
| Snapshot 1 (39 ps) |
Snapshot 2 (54 ps) |
Snapshot 3 (64 ps) |
Snapshot 4 (150 ps) |
Snapshot 1 (56 ps) |
Snapshot 2 (60 ps) |
Snapshot 3 (62 ps) |
Snapshot 4 (66 ps) |
||||||||
| Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin | Eph | Ephrin |
| ILE59 | PRO119 | ILE59 | ILE108 | ILE59 | THR96 | ILE59 | THR96 | ILE59 | THR96 | ILE59 | THR96 | ||||
| THR104 | ASN120 | THR104 | LYS109 | GLN71 | LEU97 | LEU97 | THR104 | LEU97 | THR104 | LEU97 | |||||
| LEU105 | LEU121 | LEU105 | PHE110 | CYS73 | ASP107 | ASP107 | LEU105 | ASP107 | LEU105 | ASP107 | |||||
| ARG106 | TRP122 | ARG106 | THR111 | THR104 | ILE108 | ILE108 | ARG106 | ILE108 | ARG106 | ILE108 | |||||
| CYS191 | GLY123 | CYS191 | PRO119 | LEU105 | LYS109 | LYS109 | CYS191 | LYS109 | ARG162 | LYS109 | |||||
| ILE192 | ILE192 | ASN120 | ARG106 | PHE110 | LEU121 | ILE192 | PHE110 | CYS191 | PHE110 | ||||||
| ALA193 | ALA193 | LEU121 | CYS191 | THR111 | TRP122 | ALA193 | THR111 | ILE192 | THR111 | ||||||
| TRP122 | ILE192 | ILE112 | GLY123 | PRO119 | ALA193 | ILE112 | |||||||||
| GLY123 | ALA193 | LYS113 | ASN120 | LYS113 | |||||||||||
| PRO119 | LEU121 | PRO119 | |||||||||||||
| ASN120 | TRP122 | ASN120 | |||||||||||||
| LEU121 | GLY123 | LEU121 | |||||||||||||
| TRP122 | TRP122 | ||||||||||||||
| GLY123 | GLY123 | ||||||||||||||
The snapshots were taken when a complex structure reaches different RMSD stages: snapshot 1: RMSD = 0.45 nm; snapshot2: RMSD = 0.35; snapshot3: RMSD = 0.25; snapshot4: RMSD = 0.15.
When Eph receptor and ephrin contact near binding pocket, and the Eph receptor initiates from an open or a closed conformation, residues in the GH loop of ephrin (Figures 1 and 5) contact residues at the edge of the Eph binding pocket. Additional residues in the Eph binding pocket contact the ephrin GH loop as the RMSDs drop from 0.4nm to 0.3nm. Residues outsides the GH loop begin to contact the Eph binding pocket further decreasing the RMSDs from 0.3 nm to 0.2 nm. The binding conformations become similar to those in the crystal structures when the RMSDs reach 0.1 nm.
Figure 5.
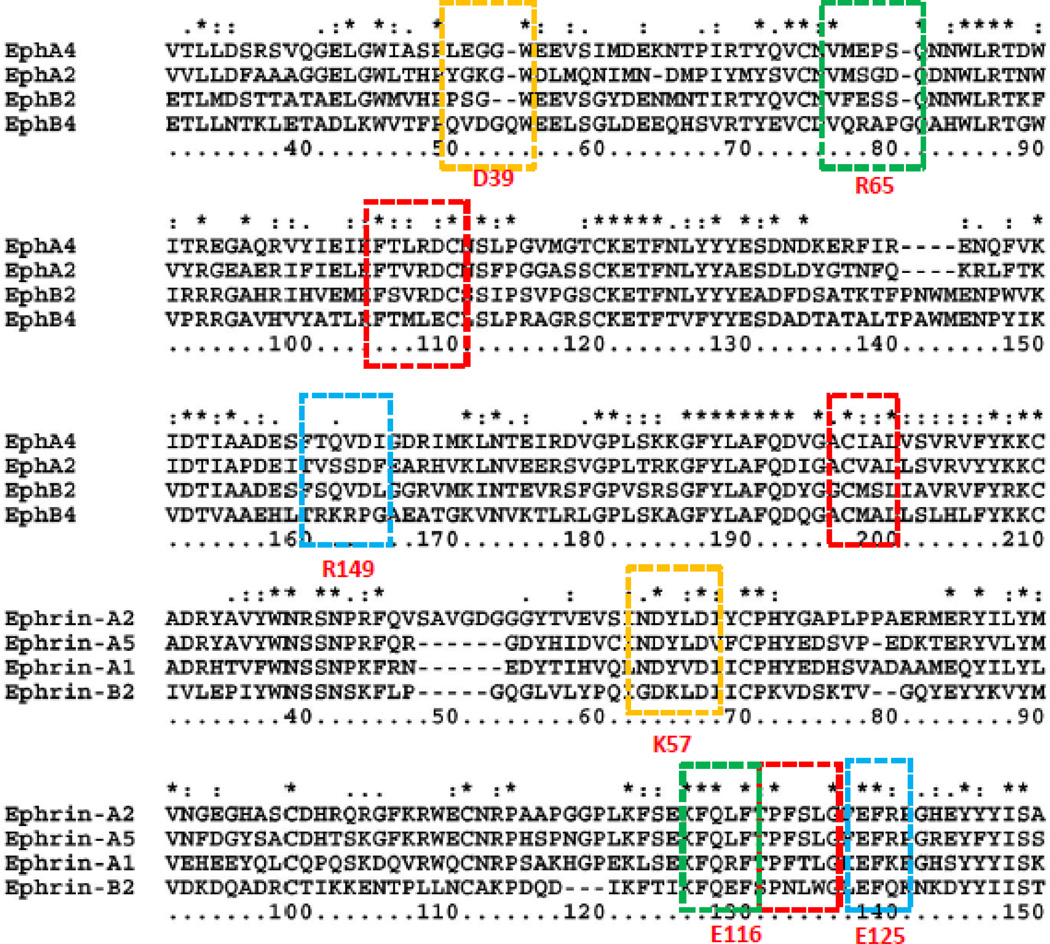
Sequence alignments of several Eph receptors and ephrins indicate that anchoring residues can be conserved across the Eph and ephrin family (red boxes), or unique for specific recongnition (orange and green boxes). The blue box indicates that R149 in EphB4 is unique, but does not contribute to specific ephrin selection since the E125 in ephrins is common. The residue numberings are based on EphB4 and ephrin-B2, respectively.
We showed that Eph-ephrin recognition involves large conformational changes of Eph receptors and the energy landscapes differ across Eph-ephrin pairs. However, when we cross-examined the contact residues in EphA4 ephrin-A2 and EphA4 ephrin-B2 from both the open and closed EphA4 conformations, we found that there are common anchoring residues. In Table 2, we highlight these residues in bold font. These common anchoring residues include Ile59, Cys73, Thr104, Leu105, Arg106, Arg162 and Cys191, Ile192, and Ala193. They mainly interact with the ephrin GH loop residues Pro135Phe136Eer137Leu138. Sequence alignments of Eph receptors and ephrins indicate that these common anchoring residues and GH loop residues are conserved across Eph receptors and ephrins, respectively (red boxes, Figure 5). The existence of common anchoring residues may provide insight into the Eph-ephrin recognition mechanism, especially for the EphA4 in closed conformation. Even though EphA4 presents different conformational dynamics in the recognition of ephrin-A2 and ephrin-B2, the common anchoring residues constitute key interaction sites. Comparing the time when a complex structure reaches different RMSD stages, we can see that early contact of the anchoring residues also differentiates the EphA4 recognition of ephrin-A2 and ephrin-B2 (Table 2). The EphA4 ephrin-A2 complex has faster contact of the anchoring residues than the EphA4 ephrin-B2 complex for snapshots 1 and 2, indicating the importance of dynamics in specific and promiscuous binding.
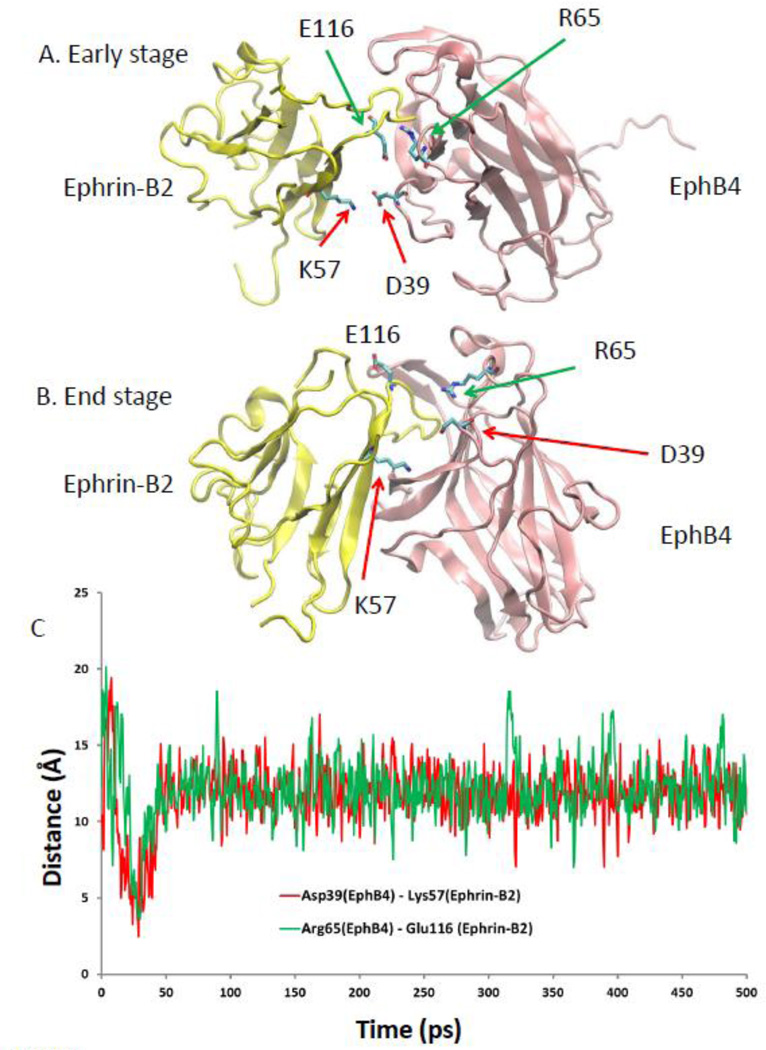
The importance of these common anchoring residues, Thr104Leu105Arg106 and Cys191Ile192Ala193 has been previously noticed from crystal structures [47]. For example Leu90 in EphB4, which is equivalent to Arg106 (in EphA4), was suggested to contribute to the specific EphB4 interaction [47]. However, since these regions are also involved in ephrin-A2 EphA4 recognition, we believe that Leu90 alone cannot determine the specificity between EphB4 and ephrin-B2. Our EphB4 ephrin-B2 simulation trajectories revealed specificity (Figure 6). Two pairs of salt bridges (Asp39 in EphB4 and Lys57 in ephrin-B2, and Arg65 in EphB4 and Glu116 in ephrin-B2, Figure 6A) are formed only during the second stage of EphB4 and ephrin-B2 recognition, which may help guide the entry of ephrin-B2 into the EphB4 binding pocket. These two salt bridges are not located in the binding site and cannot be identified from the crystal structure of the EphB4 and ephrin-B2 complex. As clearly shown in Figure 6C, these two salt bridges form quickly when EphB4 and ephrin-B2 approach each other. The formation of the salt bridges corresponds to the starting decrease of the RMSD (Figure 2A, red line) and energy drop (Figure 3D, blue line). However, the salt bridges break away (Figures 6B and 6C) after the GH loop of ephrin-B2 entered into the EphB4 binding pocket. Sequence alignments of Eph receptors and ephrins revealed that these two pairs of the salt bridges (Asp39 in EphB4 and Lys57 in Ephrin-B2, and Arg65 in EphB4 and Glu116 in ephrin-B2, orange and green boxes in Figure 5) are unique for EphB4 and ephrin-B2. Therefore, our study suggests that the specificity between EphB4 and ephrin-B2 has a dynamic component. The common anchoring residues together with the dynamic component lead to different energetic landscape for binding of Eph receptors and ephrins.
Figure 6.
Dynamic salt bridges contribute to specific EphB4 ephrin-B2 recognition. (A) Illustration of the dynamically formed salt bridges during the early stage of EphB4 ephrin-B2 recognition. (B) The salt bridges did not appear in the final bound structure of the EphB4 ephrin-B2 complex. (C) Distance trajactories during the simualtion of EphB4 ephrin-B2 recognition.
We finally examined the co-evolution of 400 sequences of the Eph receptor family ligand-binding domain and 241 of ephrin ligands. We focused on conserved residues on the interface in the four Eph-ephrin recognition snapshots listed in Table 3 and Sup-Table 2. First we analyze the co-evolution entropy of EphA4 ephrin-A2 and EphA4 ephrin-B2 complexes. Using ‘cleavage entropy’ as a quantitative measure of protease specificity [38], we calculate the specificity. Using 0.5 as stringent specificity, several anchoring residues in EphA4 are highly specific. As shown in Sup-Table 2 and Sup-Figure 3, the contact residues of EphA4 have a broad range of co-evolution entropy, from highly specific residues Gln71/Arg106 to highly variable like Phe154 and Asp61. Similarly for ephrin-A2 and ephrin-B2 both specific and variable residues are involved in binding. Although GH loop residues Pro135Phe136Eer137Leu138 are conserved across human ephrin families, these residues are not necessarily conserved evolutionally. Only proline (Pro135 for ephrin-A2 and its equivalent Pro139 in ephrin-B2) has low co-evolution entropy. ephrin-B2, which can cross-interact with EphA and EphB receptors, has the most highly specific residue Gly123. The high conservation of Gly123 in ephrin-B2 suggests the importance of conformational flexibility of the GH loop.
Table 3.
| receptor | Complex c | highly specific | specific |
|---|---|---|---|
| EphA2 | EphA2 ephrinA | 14.3% (2/14) | 35.7% (5/14) |
| EphA2 ephrin-B2 | 10.5% (2/19) | 15.8% (3/19) | |
| EphA4 | EphA4 ephrinA | 14.3% (2/14) | 42.8% (6/14) |
| EphA4 ephrin-B2 | 10.5% (2/19) | 36.8% (7/19) | |
| EphB2 | EphB2 ephrinA | 14.3% (2/14) | 28.6% (4/14) |
| EphB2 ephrin-B2 | 10.5% (2/19) | 36.8% (7/19) | |
| EphB4 | EphB4 ephrinA | 71.4% (10/14) | 14.3% (2/14) |
| EphB4 ephrin-B2 | 52.9% (9/17) | 17.6% (3/17) | |
| ligand | |||
| ephrinA1 | EphA ephrin-A1 | 0% (0/5) | 40% (2/5) |
| EphB eprhin-A1 | 21.4% (3/14) | 5.9% (1/14) | |
| ephrinA2 | EphA ephrin-A2 | 0% (0/5) | 40% (2/5) |
| EphB ephrin-A2 | 21.4% (3/14) | 5.9% (1/14) | |
| eprhinA5 | EphA ephrin-A5 | 0% (0/5) | 40% (2/5) |
| EphB ephrin-A5 | 21.4% (3/14) | 0% (0/14) | |
| ephrinB2 | EphA ephrin-B2 | 20% (1/5) | 0% (0/5) |
| EphB ephrin-B2 | 21.4% (3/14) | 5.9% (1/14) |
The percentages in the table are based on the counts of with the number of contact residues and their evolution entropies. Highly specific residues : entropy < 0.5 ; specific residues : 0.5 < entropy ≤ 1.0.
Some complexes are artificial. for example, EphB4 EphrinA complex. We mapped the contact residues to see if these complexes are formed, what are the evolution entropies for the residues to be involved in recognition.
We grouped the Eph and ephrin by their classes. EphA includes EphA2 and EphA4; EphB includes EphB2 and EphB4; and ephrinA includes ephrin-A1, ephrin-A2, and ephrin-A5. Therefore, for example EphA2 ephrinA is the group of EphA2 ephrin-A1, EphrA2 Ephrin-A2, and Eph ephrin-A5; and EphA ephrinA1 is the group of EphA2 ephrin-A1 and EphA4 ephrin-A1.
Table 3 and Sup-Table 2 list the classification of residues involved in the sixteen Eph-ephrin complexes based on their evolution entropies. We grouped the Eph and ephrin by their classes. EphA includes EphA2 and EphA4; EphB includes EphB2 and EphB4; and ephrinA includes ephrin-A1, ephrin-A2, and ephrin-A5. Therefore, for example EphA2 ephrinA is the group of EphA2 ephrin-A1, EphrA2 Ephrin-A2, and Eph ephrin-A5; and EphA ephrinA1 is the group of EphA2 ephrin-A1 and EphA4 ephrin-A1. Some complexes are artificial, for example, EphB4 only interacts with ephrin-B2. Still, we mapped the contact residues to see if these complexes can form and what are the evolution entropies for residues to be involved in recognition. Sup-Figures 4 and 5 provide the entropies for Eph receptors residues involved in the recognition of A-type and B-type ephrins, respectively. Sup-Figures 6 and 7 provide the entropies of ephrin residues involved in the interaction with A-type and B-type of Eph receptors, respectively. Table 3 and Sup-Table 2 summarize the evolution entropies for the contact residues in the sixteen Eph-ephrin complexes. One clear feature observed from Sup-Figures 4 and 5 is that the contact residues in EphB4 are highly conserved. We also can see from Table 3 that EphB4 employs highly conserved residues to recognize B-type ephrins, consistent with specific EphB4 ephrin-B2 interactions. However, if EphB4 forms a complex with A-type ephrins, essentially all (or many) interacting residues have to be conserved and specific. The requirement of highly specific interacting residues may disable EphB4 receptor to interact with A-type ephrins prevents cross-recognition of EphB4 with A-type ephrins. A-type ephrins do not use highly specific residues to interact with EphA receptors. However, when interacting with EphB receptors, A-type ephrins exploit specific residues. The above observations reveal a compensatory mechanism to balance between specific and promiscuous recognition, consistent with a previous suggestion that sequence variability among homologs can hold the key to specificity [51].
3.5 The interaction of Eph cancer mutants with ephrin ligands
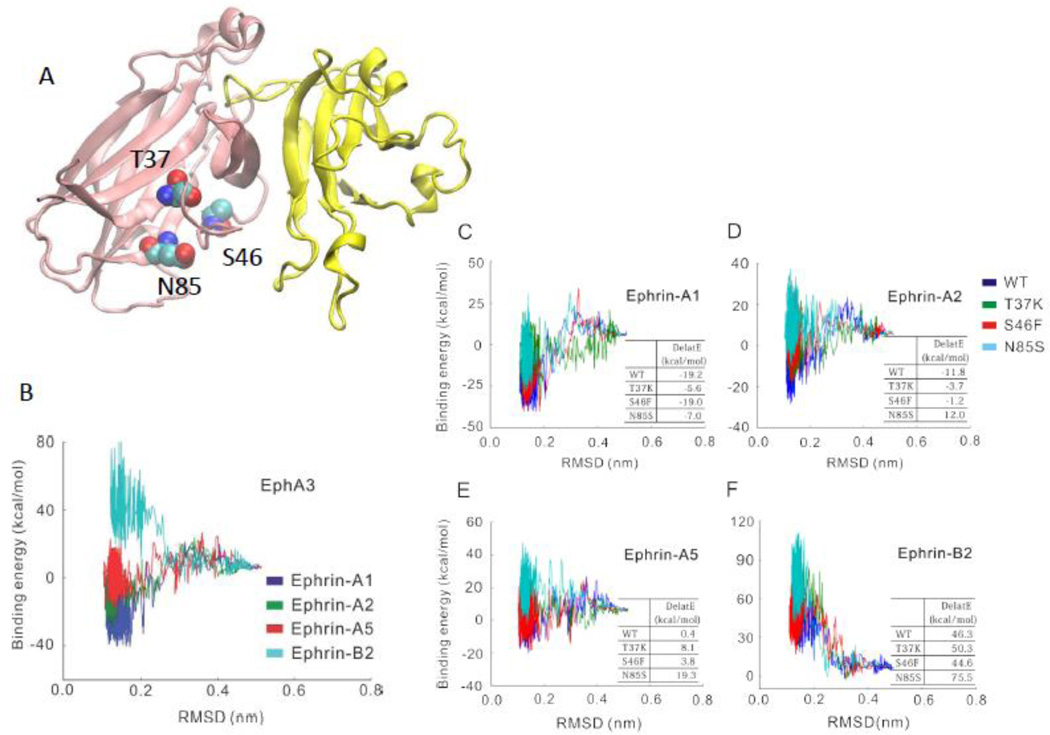
Eph/ephrin signaling plays an important role in tumor promotion and suppression. Point mutations T37K, S46F, and N85S in Eph receptor ephrin-binding domain were reported to relate to cancer [31] (Figure 7A). We simulated the recognition between the wild-type and three point mutants (T37K, S46F, and N85S) of EphA3 and the four ephrin ligands (ephrin-A1, ephrin-A2, ephrin-A5 and ephrin-B2). The energy landscapes (Figure 7B) correctly reflected the known selective recognition between EphA3 and these ligands. EphA3 can interact [43] with ephrin-A1, ephrin-A2, and ephrin-A5, but not with ephrin-B2 [52]. As can be seen in Figure 7B, EphA3 interaction with ephrin-A1, ephrin-A2, and ephrin-A5 are attractive, but repulsive with ephrin-B2.
Figure 7.
The energy landscapes of the wild type and three EphA3 point mutants interacting with four ephrins (A1, A2, A5, and B2) reflect the effect of point mutations on EphA3-ephrin interactions. (A) The location of the mutations in EphA3 based on the structure of EphA4 ephrin-B2 (pdb code: 3wo2). (B) The energy landscapes suggest that EphA3 can recognize the three A-type ephrins, but not ephrin-B2. (C)-(F) The influence of T37K, S46F, and N85S mutations on the energy landscapes and the binding energies of EphA3 the four ephrins.
Even though these mutation sites are not directly on the Eph-ephrin interface (Figure 7A), our simulation found that these mutations in the ephrin-binding domain differentially decrease the binding energies between EphA3 and the ephrin ligands (Figures 7C-7F). S46F point mutation mainly decreases the binding energy between EphA3 and ephrin-A2 and ephrin-A5. The T37K and N85S mutations decrease the binding energy by about 10 kcal /mol for all three A-type ephrins. All three mutations were identified in lung cancer and co-occurrence of T37K and N85S mutations was observed in colorectal cancer [53]. However, all these mutation do not change the repulsive interaction with Ephrin-B2. Our results suggest that Eph receptors cancer mutations disrupt binding selectivity of A-type ephrins, and each mutation differentially influences the Eph-ephrin interaction.
Discussion and Conclusions
The recognition of Eph receptors and ephrin ligands plays an important role in key biological processes. While several models have been proposed for describing dynamic mechanisms of molecular recognition, application of these models to biological problems has been difficult due to limitations imposed by both the microscopic atomic detail and the macroscopic energy landscape. Notwithstanding, accounting for protein flexibility is essential for understanding how a single protein can bind multiple ligands at the same or at different sites [54, 55]. Conventional molecular dynamics simulations aiming to efficiently reproduce protein recognition processes often fail to overcome local barriers. Steered molecular dynamics can simulate the disassociation of protein-protein complexes, but not the recognition steps. Even with flexible docking, it is challenging to explore the conformational space of protein recognition. In this study, we employed an efficient method to sample large and complex conformational changes in protein-protein recognition ensembles. Unlike conventional all atom force field simulation with explicit water solvation, the SBM model we used does not account for the contributions of water molecules in protein-protein interactions, which are known to be important[56, 57]. The steps used in the structure-based models are in reduced units, different from many other MD simulations. A 5 million steps simulation is far from being a true representation of an equilibrium representing whole protein-protein recognition process. However, the combination of the SBM model and MM-GBSA method allow us to provide a comprehensive description of the energy landscape as it relates to biological function. Using the energy landscape, we correctly predicted the promiscuity and specificity of the interaction among the four Eph receptors and four ephrin ligands and provided insights into their recognition.
4.1 Eph-ephrin recognition is a conformational selection process
Depending on the extent of the conformational changes, both ‘lock and key’ and ‘induced fit’ mechanisms have been used to describe the Eph-ephrin recognition. It was suggested that the A-class Eph receptor/ephrin interactions are better described by a 'lock-and-key'-type binding mechanism due to the smaller conformational rearrangements than the B-class molecules, which fit the 'induced fit' mechanism [58]. The alternative is a specific open conformation which is directly selected by an incoming ligand with subsequent minor induced fit optimizing it to reach the final bound state [10]. Structural plasticity in Eph-ephrin recognition has been documented experimentally [59–61]. Qin et al. [60] observed that heterogeneous free EphA4 conformations, including both the open and closed loop conformations, already exist before binding to the ephrin ligands, supporting the ‘conformational selection and population shift’ mechanism [1–7].
Here, however, we observed progressive conformational selection spanning the entire recognition process from the initial separated states to the final Eph-ephrin complex. When Eph receptors are in open conformations, Eph receptor and ephrin recognize each other quickly by directly conformational selection. However, it is not necessary for Eph receptor to have a fully open conformation to form complex with an ephrin molecule. Gradual conformational adjustments and energy changes are observed during the recognition process when binding starts with a closed Eph receptor conformation. The progressive conformational selection may help specific Eph-ephrin recognition. Many protein receptors also have similar mechanisms, including the kinases [62, 63], Hsp70s [64], caspases [65], and T-cell receptors (TCR) [66]. Hsp70s also have a conserved mechanism exploiting the pliable substrate binding groove to ensure substrate specificity [64]. The energy landscapes of the loops in TCR also allow cross-reactivity and specificity through both conformational selection and induced fit [66]. Conformational selection also operates in protein-nucleic acid (RNA and DNA) recognition, such as in initiation factor 3 ribosome interaction [67] and the PBX1 homeodomain DNA binding [68]. Induced fit, conformational selection and other mechanisms are often mixed. For example, a subtle interplay between flexibility and rigidity of two loops in snake venom metalloprotease seems to be the prerequisite for its multi-specificity to bind several partners with good affinity [66]. Small Angle X-ray Scattering has shown the progressive conformational change in the cGMP-dependent Protein Kinase[69]. Thrombin binding also involves progressive conformational changes which look like induced fit events [70, 71]. Still, trypsin-like proteases recognitions are conformational selection processes[72].
4.2 The energy landscapes control Eph-ephrin recognition and function
It is commonly believed that different binding affinities among receptors and their ligands relate to their distinct functions. For example, the binding affinities of EphA4 with ephrin-A1, ephrin-A2, ephrin-A4, ephrin-A5, and ephrin-B2 are 1.2 M, 2.3 µM, 36 nM, 360 nM, and 10.8 M, respectively [59], indicating varied ligand selectivity. However, the weak binding affinities of EphA4 across the ephrin-B ligands subclass do not imply functional deficiency, and the apparent paradox of decoupling of binding affinity from function may also be understood in terms of the entire energy landscape of the Eph-ephrin recognition, not only by the final binding energy.
Generally, the energy landscapes of the allowed Eph-ephrin interaction are attractive and those of the disallowed are repulsive. The patterns of ephrin-A5 interacting with Eph receptors differ (Figure 3), and the energy landscapes of some of the interactions are repulsive. While the repulsive energy landscape of EphB4 ephrin-A5 correctly reflects the class B-only selectivity of EphB4, it is counter intuitive. This might be due to ephrin-A5 being a specific ligand in the A-class ephrins that is able to interact with the B-class Eph receptors. Therefore, while ephrin-A5 is promiscuous and binds most Eph receptors, the energy landscape of ephrin-A5 needs to differentiate among the receptors to achieve functional selectivity. A recent study shows that while EphA2 ephrin-A5 and EphA4 ephrin-A5 have similar binding affinities, ephrin-A5 induces smaller EphA4 clusters than EphA2 and with different, circular versus array organizations, respectively [46]. This could reflect the repulsive energy landscape. Importantly, the crystal structures indicate that EphA4 dimerizes already prior to ephrin binding, with the conformational change likely also resulting in enhanced selectivity.
Ephs/ephrins cluster temporally, increasing their effective local concentration. This facilitates binding of their ligands and effectors, and encodes bursts of signaling. Among the ephrin ligand family, ephrin-A5 is most commonly involved in topographic map specification through graded expression in the target region [73–75]. Recently, it was found that ephrin-A5 EphA4 signaling controls the specific targeting to cochlear hair cells [76, 77]. The changes of the ephrin-A5 energy landscapes with different receptors may also increase its ability to assemble into different signaling complexes as a function of the concentration and affinities of ephrin-A5 and its Eph receptors [16].
4.3 Dynamic anchoring residues enable both repulsive and attractive selection
Protein sequence encodes its dynamics and the related distribution of the protein shapes; these combine with specific residues at the binding site that encode further specificity. These specific residues may recognize and help distinguish among ephrin ligands. During the recognition process, the number of interface residues gradually increases, and common anchoring residues shift from initial recognition contact to final binding pairs. Such a mechanism can only be possible by progressive conformational selection, with the open conformation embodying all the common anchoring residues available to interact with incoming ligands. A closed conformation may need only to open sufficiently to allow the early interaction of the common anchoring residues with the ligand, and may not require reaching the final open conformation as in the complex. In addition to these common anchoring residues, dynamic contacting residues appearing at an early recognition stage may also contribute to specific recognition. EphB4 and ephrin-B2 form two such dynamic salt bridges, which contribute to their specific recognition.
The dynamic anchoring residues, either common ones for promiscuous interaction or unique ones for specific recognition are subjected to allosteric perturbations away from binding sites. We found that oncogenic mutations of EphA3, which are not directly involved on the Eph-ephrin interface, decrease the binding energies between EphA3 and ephrin ligands (A1, A2, A5, and B2).
Supplementary Material
Highlights.
Promiscuous and specific Eph-ephrin binding between cells controls communication
Conformational ensembles and energies of sixteen Eph - ephrins complexes can illuminate the recognition mechanism
The process of recognition appears to involve progressive conformational selection events
Dynamics control specificity , anchoring residues may offer promiscuity
Conformational dynamics may simultaneously allow promiscuous and specific binding
Acknowledgements
The authors are funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Q. Huang was supported in part by the grants from the Hi-tech Research and Development Program of China (No. 2008AA02Z311), the Shanghai Natural Science Foundation (No. 13ZR1402400), and the Shanghai Leading Academic Discipline Project (B111).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ma B, Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr Opin Chem Biol. 2010;14:652–659. doi: 10.1016/j.cbpa.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein engineering. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birdsall B, Feeney J, Roberts GC, Burgen AS. The use of saturation transfer NMR experiments to monitor the conformational selection accompanying ligand-protein interactions. FEBS letters. 1980;120:107–109. doi: 10.1016/0014-5793(80)81057-x. [DOI] [PubMed] [Google Scholar]
- 5.Bruns RF. Conformational induction versus conformational selection: evidence from allosteric enhancers. Trends in pharmacological sciences. 1996;17:189. doi: 10.1016/0165-6147(96)20027-6. discussion 190–181. [DOI] [PubMed] [Google Scholar]
- 6.Berger C, Weber-Bornhauser S, Eggenberger J, Hanes J, Pluckthun A, Bosshard HR. Antigen recognition by conformational selection. FEBS letters. 1999;450:149–153. doi: 10.1016/s0014-5793(99)00458-5. [DOI] [PubMed] [Google Scholar]
- 7.Vértessy BG, Orosz F. From "fluctuation fit" to "conformational selection": evolution rediscovery and integration of a concept. Bioessays. 2011;33:30–34. doi: 10.1002/bies.201000068. [DOI] [PubMed] [Google Scholar]
- 8.Ma B, Nussinov R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J Mol Biol. 2012;421:172–184. doi: 10.1016/j.jmb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussinov R, Tsai CJ, Ma B. The underappreciated role of allostery in the cellular network. Annual review of biophysics. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csermely P, Palotai R, Nussinov R. Induced, fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Regenmortel MH. Molecular recognition in the post-reductionist era Journal of molecular recognition. JMR. 1999;12:1–2. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<1::AID-JMR449>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 12.Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. The Biochemical journal. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- 13.Hirai H, Maru Y, Hagiwara K, Nishida J, Takaku F. A novel putative tyrosine kinase receptor encoded by the eph gene. Science. 1987;238:1717–1720. doi: 10.1126/science.2825356. [DOI] [PubMed] [Google Scholar]
- 14.Pitulescu ME, Adams RH. Eph/ephrin molecules--a hub for signaling and endocytosis. Genes & development. 2010;24:2480–2492. doi: 10.1101/gad.1973910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Lackmann M, Boyd AW. Eph a protein family coming of age: more confusion insight or complexity? Sci Signal. 2008;1:re2. doi: 10.1126/stke.115re2. [DOI] [PubMed] [Google Scholar]
- 17.Frisen J, Holmberg J, Barbacid M. Ephrins and their Eph receptors: multitalented directors of embryonic development. The EMBO journal. 1999;18:5159–5165. doi: 10.1093/emboj/18.19.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- 19.Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17:230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Kandouz M. The Eph/Ephrin family in cancer metastasis: communication at the service of invasion. Cancer metastasis reviews. 2012;31:353–373. doi: 10.1007/s10555-012-9352-1. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Winterbottom E, Daar IO. Eph/ephrin signaling in cell-cell and cell-substrate adhesion. Front Biosci (Landmark Ed) 2012;17:473–497. doi: 10.2741/3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genander M, Frisen J. Ephrins and Eph receptors in stem cells and cancer. Current opinion in cell biology. 2010;22:611–616. doi: 10.1016/j.ceb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Beauchamp A, Debinski W. Ephs and ephrins in cancer: ephrin-A1 signalling. Seminars in cell & developmental biology. 2012;23:109–115. doi: 10.1016/j.semcdb.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvucci O, Tosato G. Essential roles of EphB receptors and EphrinB ligands in endothelial cell function and angiogenesis. Advances in cancer research. 2012;114:21–57. doi: 10.1016/B978-0-12-386503-8.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine & growth factor reviews. 2004;15:419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Noberini R, Mitra S, Salvucci O, Valencia F, Duggineni S, Prigozhina N, Wei K, Tosato G, Huang Z, Pasquale EB. PEGylation potentiates the effectiveness of an antagonistic peptide that targets the EphB4 receptor with nanomolar affinity. PloS one. 2011;6:e28611. doi: 10.1371/journal.pone.0028611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Zhuang G, Frieden L, Debinski W. Eph receptors and Ephrins in cancer: common themes and controversies. Cancer research. 2008;68:10031–10033. doi: 10.1158/0008-5472.CAN-08-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi HQ, Wu XS, Wei B, Chen L. Eph receptors and ephrins as targets for cancer therapy. Journal of cellular and molecular medicine. 2012;16:2894–2909. doi: 10.1111/j.1582-4934.2012.01612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquale EB. Eph receptors ephrins in cancer: bidirectional signalling beyond Nature reviews. Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber K. Of Ephs and ephrins: companies target guidance molecules in cancer. Journal of the National Cancer Institute. 2010;102:1692–1694. doi: 10.1093/jnci/djq479. [DOI] [PubMed] [Google Scholar]
- 31.Lisabeth EM, Falivelli G, Pasquale EB. Eph receptor signaling and ephrins. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funk SD, Orr AW. Ephs ephrins resurface in inflammation, immunity, and atherosclerosis. Pharmacological research : the official journal of the Italian Pharmacological Society. 2013;67:42–52. doi: 10.1016/j.phrs.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Marsh GA, Wang LF. Hendra and Nipah viruses: why are they so deadly? Current opinion in virology. 2012;2:242–247. doi: 10.1016/j.coviro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Nussinov R, Ma B, Tsai CJ. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophysical chemistry. 2014;186:22–30. doi: 10.1016/j.bpc.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN. SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 2010;38:W657–W661. doi: 10.1093/nar/gkq498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of chemical information and modeling. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 38.Fuchs JE, von Grafenstein S, Huber RG, Margreiter MA, Spitzer GM, Wallnoefer HG, Liedl KR. Cleavage entropy as quantitative measure of protease specificity. PLoS Comput Biol. 2013;9:e1003007. doi: 10.1371/journal.pcbi.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen H, Xu F, Hu H, Wang F, Wu Q, Huang Q, Wang H. Coevolving residues of (beta/alpha)(8)-barrel proteins play roles in stabilizing active site architecture and coordinating protein dynamics. Journal of structural biology. 2008;164:281–292. doi: 10.1016/j.jsb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Noel JK, Whitford PC, Onuchic JN. The shadow map: a general contact definition for capturing the dynamics of biomolecular folding and function. The journal of physical chemistry. B. 2012;116:8692–8702. doi: 10.1021/jp300852d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: what determines the structural details of the transition state ensemble and "en-route" intermediates for protein folding? An investigation for small globular proteins. Journal of molecular biology. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 42.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophysical journal. 2009;96:L7–L9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasquale EB. The Eph family of receptors. Current opinion in cell biology. 1997;9:608–615. doi: 10.1016/s0955-0674(97)80113-5. [DOI] [PubMed] [Google Scholar]
- 44.Irie N, Takada Y, Watanabe Y, Matsuzaki Y, Naruse C, Asano M, Iwakura Y, Suda T, Matsuo K. Bidirectional signaling through ephrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J Biol Chem. 2009;284:14637–14644. doi: 10.1074/jbc.M807598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuo K, Otaki N. Bone cell interactions through Eph/ephrin: bone modeling remodeling and associated diseases. Cell adhesion & migration. 2012;6:148–156. doi: 10.4161/cam.20888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiradake E, Schaupp A, Del Toro Ruiz D, Kaufmann R, Mitakidis N, Harlos K, Aricescu AR, Klein R, Jones EY. Structurally encoded intraclass differences in EphA clusters drive distinct cell responses. Nat Struct Mol Biol. 2013;20:958–964. doi: 10.1038/nsmb.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chrencik JE, Brooun A, Kraus ML, Recht MI, Kolatkar AR, Han GW, Seifert JM, Widmer H, Auer M, Kuhn P. Structural and biophysical characterization of the EphB4*ephrinB2 protein-protein interaction and receptor specificity. J Biol Chem. 2006;281:28185–28192. doi: 10.1074/jbc.M605766200. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen TM, Arthur A, Hayball JD, Gronthos S. EphB and Ephrin-B interactions mediate human mesenchymal stem cell suppression of activated T-cells. Stem cells and development. 2013;22:2751–2764. doi: 10.1089/scd.2012.0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawano H, Katayama Y, Minagawa K, Shimoyama M, Henkemeyer M, Matsui T. A novel feedback mechanism by Ephrin-B1/B2 in T-cell activation involves a concentration-dependent switch from costimulation to inhibition. European journal of immunology. 2012;42:1562–1572. doi: 10.1002/eji.201142175. [DOI] [PubMed] [Google Scholar]
- 50.Huan X, Shi J, Lim L, Mitra S, Zhu W, Qin H, Pasquale EB, Song J. Unique Structure and Dynamics of the EphA5 Ligand Binding Domain Mediate Its Binding Specificity as Revealed by X-ray Crystallography, NMR and MD Simulations. PloS one. 2013;8:e74040. doi: 10.1371/journal.pone.0074040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magliery TJ, Regan L. Sequence variation in ligand binding sites in proteins. BMC bioinformatics. 2005;6:240. doi: 10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falivelli G, Lisabeth EM, Rubio de la Torre E, Perez-Tenorio G, Tosato G, Salvucci O, Pasquale EB. Attenuation of eph receptor kinase activation in cancer cells by coexpressed ephrin ligands. PloS one. 2013;8:e81445. doi: 10.1371/journal.pone.0081445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuang G, Song W, Amato K, Hwang Y, Lee K, Boothby M, Ye F, Guo Y, Shyr Y, Lin L, Carbone DP, Brantley-Sieders DM, Chen J. Effects of cancer-associated EPHA3 mutations on lung cancer. J Natl Cancer Inst. 2012;104:1182–1197. doi: 10.1093/jnci/djs297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levy Y, Onuchic JN. Mechanisms of protein assembly: lessons from minimalist models. Accounts of chemical research. 2006;39:135–142. doi: 10.1021/ar040204a. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed MH, Habtemariam M, Safo MK, Scarsdale JN, Spyrakis F, Cozzini P, Mozzarelli A, Kellogg GE. Unintended consequences? Water molecules at biological and crystallographic protein-protein interfaces. Comput Biol Chem. 2013;47:126–141. doi: 10.1016/j.compbiolchem.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Ahmed MH, Spyrakis F, Cozzini P, Tripathi PK, Mozzarelli A, Scarsdale JN, Safo MA, Kellogg GE. Bound water at protein-protein interfaces: partners, roles and hydrophobic bubbles as a conserved motif. PloS one. 2011;6:e24712. doi: 10.1371/journal.pone.0024712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Himanen JP, Goldgur Y, Miao H, Myshkin E, Guo H, Buck M, Nguyen M, Rajashankar KR, Wang B, Nikolov DB. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO reports. 2009;10:722–728. doi: 10.1038/embor.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowden TA, Aricescu AR, Nettleship JE, Siebold C, Rahman-Huq N, Owens RJ, Stuart DI, Jones EY. Structural plasticity of eph receptor A4 facilitates cross-class ephrin signaling. Structure. 2009;17:1386–1397. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin H, Lim L, Song J. Protein dynamics at Eph receptor-ligand interfaces as revealed by crystallography, NMR and MD simulations. BMC Biophys. 2012;5:2. doi: 10.1186/2046-1682-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singla N, Goldgur Y, Xu K, Paavilainen S, Nikolov DB, Himanen JP. Crystal structure of the ligand-binding domain of the promiscuous EphA4 receptor reveals two distinct conformations. Biochemical and biophysical research communications. 2010;399:555–559. doi: 10.1016/j.bbrc.2010.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu M, Yu L, Wan B, Huang Q. Predicting inactive conformations of protein kinases using active structures: conformational selection of type-II inhibitors. PloS one. 2011;6:e22644. doi: 10.1371/journal.pone.0022644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu X, Bruschweiler-Li L, Davulcu O, Skalicky JJ, Bruschweiler R, Chapman MS. Arginine kinase: joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility. J Mol Biol. 2011;405:479–496. doi: 10.1016/j.jmb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcinowski M, Rosam M, Seitz C, Elferich J, Behnke J, Bello C, Feige MJ, Becker CF, Antes I, Buchner J. Conformational selection in substrate recognition by Hsp70 chaperones. J Mol Biol. 2013;425:466–474. doi: 10.1016/j.jmb.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 65.Fuchs JE, Von Grafenstein S, Huber RG, Wallnoefer HG, Liedl KR. Specificity of a protein-protein interface: Local dynamics direct substrate recognition of effector caspases. Proteins. 2013 doi: 10.1002/prot.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott DR, Borbulevych OY, Piepenbrink KH, Corcelli SA, Baker BM. Disparate degrees of hypervariable loop flexibility control T-cell receptor cross-reactivity specificity and binding mechanism. J Mol Biol. 2011;414:385–400. doi: 10.1016/j.jmb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elvekrog MM, Gonzalez RL., Jr Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat Struct Mol Biol. 2013;20:628–633. doi: 10.1038/nsmb.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farber PJ, Mittermaier A. Concerted dynamics link allosteric sites in the PBX homeodomain. J Mol Biol. 2011;405:819–830. doi: 10.1016/j.jmb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J, Trewhella J, Corbin J, Francis S, Mitchell R, Brushia R, Walsh D. Progressive cyclic nucleotide-induced conformational changes in the cGMP-dependent protein kinase studied by small angle X-ray scattering in solution. J Biol Chem. 1997;272:31929–31936. doi: 10.1074/jbc.272.50.31929. [DOI] [PubMed] [Google Scholar]
- 70.Lechtenberg BC, Johnson DJ, Freund SM, Huntington JA. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc Natl Acad Sci U S A. 2010;107:14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamath P, Huntington JA, Krishnaswamy S. Ligand binding shuttles thrombin along a continuum of zymogen- and proteinase-like states. J Biol Chem. 2010;285:28651–28658. doi: 10.1074/jbc.M110.154914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pozzi N, Vogt AD, Gohara DW, Di Cera E. Conformational selection in trypsin-like proteases. Curr Opin Struct Biol. 2012;22:421–431. doi: 10.1016/j.sbi.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frisen J, Yates PA, McLaughlin T, Friedman GC, O'Leary DD, Barbacid M. Ephrin-A5 (AL-1/RAGS) is essential for proper retinal axon guidance and topographic mapping in the mammalian visual system. Neuron. 1998;20:235–243. doi: 10.1016/s0896-6273(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 74.Dufour A, Seibt J, Passante L, Depaepe V, Ciossek T, Frisen J, Kullander K, Flanagan JG, Polleux F, Vanderhaeghen P. Area specificity and topography of thalamocortical projections are controlled by ephrin/Eph genes. Neuron. 2003;39:453–465. doi: 10.1016/s0896-6273(03)00440-9. [DOI] [PubMed] [Google Scholar]
- 75.Feldheim DA, Kim YI, Bergemann AD, Frisen J, Barbacid M, Flanagan JG. Genetic analysis of ephrin-A2 and ephrin-A5 shows their requirement in multiple aspects of retinocollicular mapping. Neuron. 2000;25:563–574. doi: 10.1016/s0896-6273(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 76.Defourny J, Poirrier AL, Lallemend F, Mateo Sanchez S, Neef J, Vanderhaeghen P, Soriano E, Peuckert C, Kullander K, Fritzsch B, Nguyen L, Moonen G, Moser T, Malgrange B. Ephrin-A5/EphA4 signalling controls specific afferent targeting to cochlear hair cells. Nature communications. 2013;4:1438. doi: 10.1038/ncomms2445. [DOI] [PubMed] [Google Scholar]
- 77.Kimura K, Hikida T, Yawata S, Yamaguchi T, Nakanishi S. Pathway-specific engagement of ephrinA5-EphA4/EphA5 system of the substantia nigra pars reticulata in cocaine-induced responses. Proc Natl Acad Sci U S A. 2011;108:9981–9986. doi: 10.1073/pnas.1107592108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.