Abstract
The transcription activator-like effector (TALE) nucleases, or TALENs, are customizable restriction enzymes that may be used to induce mutations at nearly any investigator-specified DNA sequence in zebrafish. The DNA-binding specificities of TALENs are determined by a protein array comprised of four types of TALE repeats, where each repeat recognizes a different DNA base. Here, we describe methods for constructing TALEN vectors that have been shown to achieve high success rates and mutation efficiencies in zebrafish. In addition, we discuss simple techniques and protocols that can be used to detect TALEN-induced mutations at almost any genomic locus. These methods should enable zebrafish researchers to quickly generate targeted mutations at their genes-of-interest.
Keywords: Site-specific nuclease, Transcription activator-like effector, nuclease, Knockout, Genome engineering, Gene targeting, Gene disruption, Zebrafish
1. Introduction
Engineered site-specific endonucleases are essential tools for targeted gene editing in zebrafish, human cells and many other important model organisms (reviewed in [1–4]). These enzymes can be used to create double-stranded DNA breaks at investigator- specified genomic loci, enabling sequence modifications and introduction of exogenous DNA sequences at their target sites [1–4]. To date, three major types of customizable endonucleases have been developed, namely zinc finger nucleases (ZFNs) [5–13], transcription activator-like effector nucleases (TALENs) [10,14–21] and clustered regularly interspaced short palindromic repeats (CRISPR) RNA-guided Cas9 (CRISPR/Cas) nucleases [22–32]. All three classes have been shown to cause efficient target gene disruption in zebrafish [5,6,8,9,14,17,19,27,33–39]. Nevertheless, each platform differs in the ease of their construction methods, potential off-target activities and the theoretical targeting range. In terms of nuclease construction, highly effective ZFNs are generally the most difficult to obtain, while CRISPR/Cas are the easiest. Both TALENs and CRISPR/Cas can reach very high on-target activities. However, in some cases, CRISPR/Cas nucleases have been shown to elicit very high off-target activities [40–43]. Among these approaches, TALENs exhibit the broadest targeting range, with almost no restrictions in target sequence [18]. Thus, TALENs may sometimes be the only possible choice for targeting a particular sequence of interest. Investigators can use the free online software ZiFiT Targeter (http://zifit.partners.org/ZiFiT/) to identify potential target sites for all of these approaches [6,9,19,27,44–46].
TALENs are chimeric proteins comprised of the endonuclease domain of FokI restriction enzyme and a site-specific DNA-binding domain derived from the transcription activator-like effectors (TALEs) of plant pathogens Xanthomonas species. The DNA-binding domain of TALEs is composed of a series of TALE repeats where each recognizes a specific DNA nucleotide [47,48]. These 32 to 35-amino acid repeats are almost identical except for two residues at positions 12 and 13 which govern DNA binding specificity [49,50]. We and others have demonstrated that customized TALENs based on this simple code can confer robust on-target activities in zebrafish [14,17,19,33,35,36]. Based on our experience, TALEN-induced mutation rates in somatic cells are found to be as high as 76% [33]. Moreover, TALENs induce heritable mutations in zebrafish efficiently [33]. Thus, TALENs are accessible and powerful research tools for zebrafish genome editing. Here, we discuss potential considerations and our preferred methods for generating zebrafish gene mutations using TALENs.
2. Considerations for choosing a TALEN framework
Currently, there are several public resources for TALEN construction (see review [2] or the website http://www.TALengineering.org). TALENs constructed on different platforms may have differences in their architectures, such as the sequences and the numbers of the TALE repeats, the DNA-binding domain outside of the TALE repeats, and modifications in the FokI nuclease domain. Some of these differences may affect the stability of the TALEN vectors, which can be subject to recombination due to the highly repetitive nature of the TALE repeats. Other differences may affect the efficiencies of the enzymes themselves. We have adopted the TALEN framework initially developed by Miller et al. based on the TALE13 of Xanthomonas axonopodis [21]. In this scaffold, the DNA-binding domain contains a truncated, 63-amino acid C-terminal segment, which has been found to increase TALEN activities in a serial deletion analysis. Moreover, slight sequence variations have been built into the TALE repeat modules to prevent recombination of TALEN vectors. To date, this TALEN platform has been successfully exploited in numerous model systems such as human cells, zebrafish, rats, and worms [10,16,18–20,33].
3. Considerations for choosing a target site
To construct customized TALENs for a gene of interest, the first step is to identify candidate target sites in its genomic sequence. Researchers can conduct this search using one of the two “TALE Nucleases” functions in ZiFiT Targeter (http://zifit.partners.org/ZiFiT/) depending on the researchers’ choice of TALEN construction methods (as discussed below). It is important to note that there are sometimes polymorphisms in a zebrafish colony [33]. Thus, investigators may wish to check their stock to confirm that there is no polymorphic sequence in the planned TALEN target sites. In addition, it has been shown that TALE DNA-binding domains are sensitive to DNA methylation [51]. Methylation on the cytosine of a CpG dinucleotide can be identified via bisulfite sequencing. Alternatively, researchers may want to avoid target sites containing CpG sequences if there are other options [36]. Beyond avoidance of polymorphisms and methylation sites, it is important to avoid highly repetitive sequences, which may increase risk of off-target cleavage. However, some earlier guidelines proposed by Cermak et al. based on a computational analysis of natural TAL effectors have been found to be non-critical [18,52].
4. Construction of customized TALEN vectors
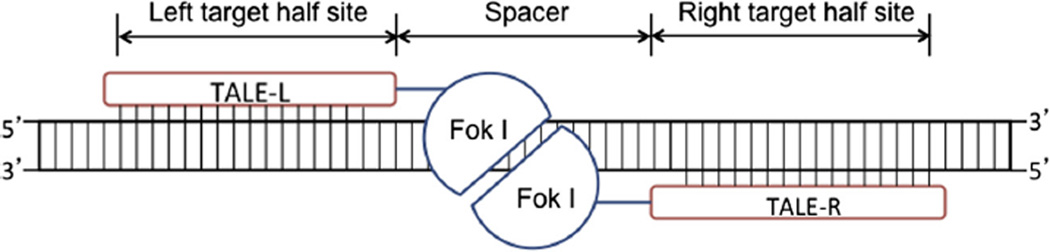
Since the FokI nuclease domain cleaves DNA in a dimeric form, two customized TALENs are needed for any given target locus. The DNA-binding domains of these two TALENs should bind to the left and the right target half sites, respectively. This will facilitate the dimerization of the nuclease domains in the spacer region between the two target half sites (Fig. 1). Thus, an optimal spacer length is important for efficient DNA cleavage activity. Previously Miller et al. have shown that, using their TALEN framework, the nucleases are the most active when the spacer lengths are between 14 and 19 basepairs (bps) [21]. We have found that spacer lengths ranging between 16 and 18 bps can be effective in zebrafish [19]. The same spacer range has also been validated in a large-scale test of TALENs that target endogenous human genes in cultured cells [18], and is used as the default setting in ZiFiT Targeter.
Fig. 1.
A schematic representation of TALENs binding to their cognate target sites. The DNA-binding domains of two TALENs (TALE-L and TALE-R) bind to the two target half sites, facilitating dimerization of their FokI nuclease domains. The dimerized nucleases can then create double-stranded DNA breaks in the spacer region.
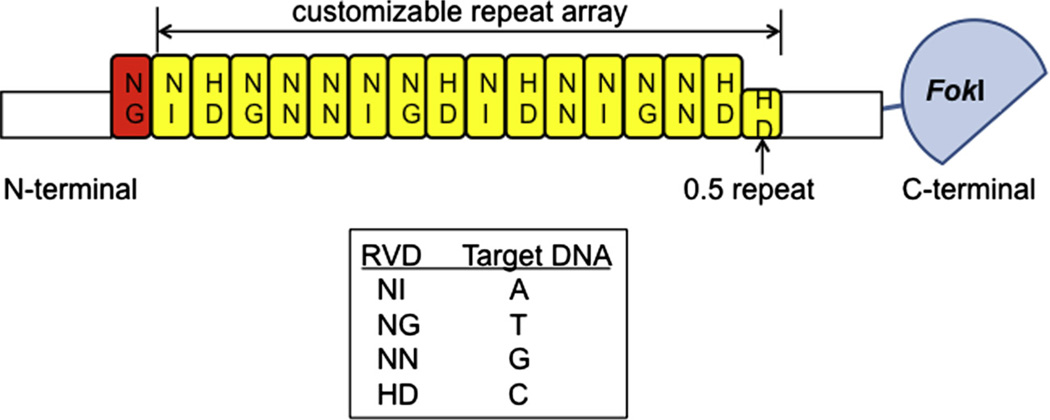
The number of TALE repeats in the DNA-binding domain can also influence the efficiency of the TALENs constructed [18]. Native TALEs often contain 17.5 TALE repeats where the final “0.5” repeat is only 20 amino acids long [21]. In addition, the first repeat typically binds to a thymidine nucleotide [21]. Thus, engineered TALENs normally have a fixed first repeat, which is followed by a customized TALE repeat array (Fig. 2). By evaluating the mutation efficiencies and cytotoxicities induced by TALENs harboring 8.5–19.5 TALE repeat arrays, Reyon et al. have proposed that TALENs composed of 14.5- to 16.5-repeat arrays (which will recognize a total of 16–18 DNA nucleotides including the 5′ T) are most likely to be effective [18]. Following this guideline, our overall success rate of customized TALEN pairs that can mutate their target loci in zebrafish is above 80% ([19,33] and unpublished results).
Fig. 2.
An engineered TALEN contains a customizable TALE repeat array that determines its DNA specificity. TALENs typically have a fixed first repeat that targets thymidine (red highlighted), followed by a customized array of TALE repeats (yellow highlighted). Each repeat domain is 32–35 amino acids except the final “0.5” repeat which is only 20 amino acids long. The identity of two amino acids located at positions 12 and 13 in a repeat domain (termed a repeat variable di-residue or RVD) forms a simple code that determines its nucleotide target (as shown in the box).
Construction of customized TALENs involves the generation of a TALE repeat array and the cloning of this array into a TALEN expression vector that has the coding sequences for the rest of the TALE DNA-binding domain and the FokI nuclease domain. We use TALE repeat arrays generated using three different methods developed by Joung and colleagues. The first method is a restriction enzyme and ligation-based protocol called REAL [19,44]. This method has the lowest throughput, but it does not require a large number of plasmid archives as the starting materials. Thus, it is suitable for researchers who are interested in producing only a few TALEN pairs. The second method, called REAL-Fast, is a modified version of the REAL method [44]. The major difference between these two methods is that REAL-Fast takes advantage of a larger number of plasmid archives as the starting materials (32 plasmids for REAL and 380 plasmids for REAL-Fast). Thus, REAL-Fast will not only increase the throughput but also shorten the construction time to 7 days as opposed to 9 days using REAL for arrays containing 12.5–16.5 TALE repeats. The third method is called Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) assembly [18,33,45]. The FLASH method can be conducted in a 96-well plate by automation or manually. Since the assembly is performed on solid phase, it does not call for cloning of the intermediate products as the other two methods do. Using FLASH, 96 arrays containing 16.5 TALE repeats or less can be built in one day. It is worth noting that these methods require either none or at most one PCR amplification throughout the whole process, thus lowering the chances of sequence errors introduced by PCR. Detailed protocols for constructing customized TALE repeat arrays and TALEN vectors using these methods have been described elsewhere [44,45]. The Joung lab REAL assembly TALEN kit is available from Addgene (http://www.addgene.org/talengineering/TALENkit/). Plasmid archives required for the REAL-Fast and the FLASH methods can be requested from the Joung lab (http://eGenome.org).
After assembly, a TALE repeat array can then be cloned into a TALEN expression vector. While most early studies used a wild-type homodimeric FokI nuclease domain in the TALEN framework, we have conducted a direct comparison between homodimeric TALENs and TALENs composed of obligate heterodimeric FokI nuclease domains in zebrafish. The results indicate that heterodimeric TALENs can in some cases significantly improve the efficiencies of their homodimeric counterparts [33]. Heterodimeric TALENs in theory should also be more specific than homodimeric TALENs, because they do not form unwanted homodimers that may have potential off-target effects. Thus, we have modified the previously published methods, and describe here the protocols for (1) constructing heterodimeric TALENs using TALE repeat arrays generated by REAL, REAL-Fast, or FLASH and (2) subcloning TALE repeat arrays from an existing homodimeric TALEN vector to a heterodimeric TALEN vector. The latter can be useful for researchers who have already made their customized homodimeric TALEN constructs but would like to test their TALENs in heterodimeric forms. Our protocol is designed to seamlessly integrate with the TALEN construction platforms developed by the Joung lab [44,45].
5. Protocols for constructing heterodimeric TALEN expression vectors
5.1. Cloning of TALE repeat arrays into heterodimeric TALEN vectors
5.1.1. Materials
-
–
Heterodimeric TALEN vectors: JDS80 (#40316), JDS82 (#40317), JDS84 (#40318), JDS88 (#40319), JDS90 (#40320), JDS92 (#40321), JDS94 (#40322) and JDS98 (#40323) from Addgene (https://www.addgene.org/).
-
–
BsmBI (#R0580S), BsaI (#R0535S) and BbsI (#R0539S) (New England Biolabs).
-
–
An agarose gel electrophoresis apparatus.
-
–
A spectrometer for measuring DNA concentration.
-
–
LigaFast™ Rapid DNA Ligation System (Promega).
-
–
Chemically competent or electrocompetent bacterial cells (recA1 strain for reducing nonspecific recombination in the cloned DNA, such as DH5α, TOP10 or XL1-Blue).
-
–
6-well LB/carbenicillinagarplates: Add7.5 g agar into500ml of LB. Autoclave. Add 0.5ml of 50mg/ml carbenicillin stock solution after it cools down but before it solidifies (~60 °C). Mix well. Pour 3ml into eachwell of 6-well plates. Store at 4 °C for up to 2months.
-
–
LB/carbenicillin medium: Add 0.5 ml of 50 mg/ml carbenicillin stock solution to 500 ml of autoclaved LB. Store at 4 °C for up to 2 months.
-
–
Sterile bacterial culture tubes.
-
–PCR primers:
- oSQT34, 5′-GACGGTGGCTGTCAAATACCAAGATATG-3′.
- oSQT35, 5′-TCTCCTCCAGTTCACTTTTGACTAGTTGGG-3′.
-
–Sequencing primers:
- oSQT1, 5′-AGTAACAGCGGTAGAGGCAG-3′.
- oSQT3, 5′-ATTGGGCTACGATGGACTCC-3′.
- JDS2980, 5′-TTAATTCAATATATTCATGAGGCAC-3′.
-
–
QIAquick PCR Purification Kit (Qiagen).
-
–
QIAprep Spin Miniprep Kit (Qiagen).
-
–
GoTaq® DNA Polymerase (Promega).
-
–
– 10 mM dNTPs.
5.1.2. Methods
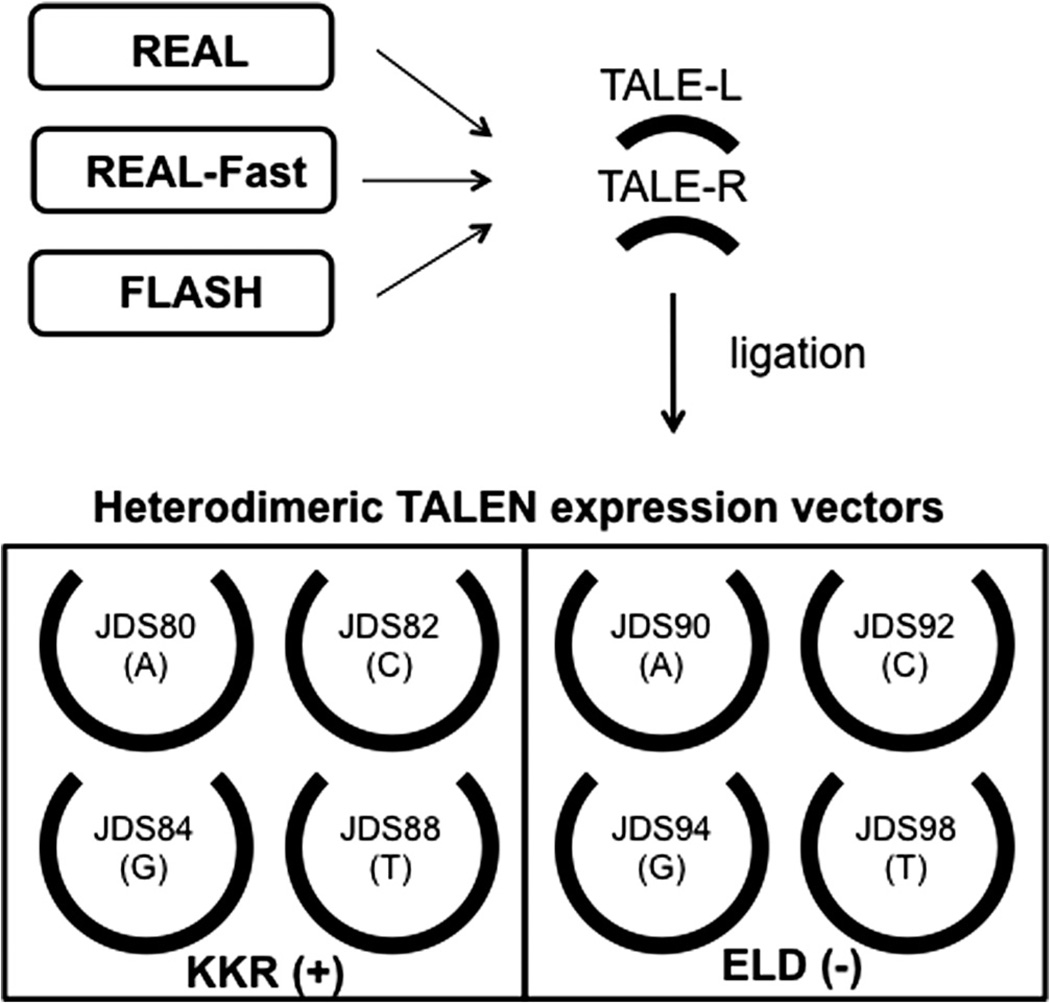
To construct obligate heterodimeric TALEN pairs, we use TALEN vectors that harbor either a KKR (+) or an ELD (−) FokI nuclease domain [53]. These nucleases are mutated such that neither the (+) nor the (−) nuclease can homodimerize effectively, but they retain the ability to heterodimerize. There is no restriction in terms of which of these two vectors should be used for the left or right target half site. However, the DNA fragments containing the TALE repeat arrays for two target half sites should be cloned into different expression vectors, (+) or (−). In addition, since TALEN vectors also carry the carboxyl-terminal 0.5 repeat domain, investigators should carefully choose the correct vector to use based on the nucleotide at the 3′ end of the TALEN binding site (Fig. 3). For example, to generate heterodimeric TALEN constructs for the following target sequences, 5′-TCCGGCAGACATCGTGAA (left site) and 5′-TTTGGGGAAGGCCCCAGG (right site), one can use JDS80 and JDS94 for the left and right TALEN arrays, respectively. Alternatively, the left and right TALEN arrays can be cloned into JDS90 and JDS84, respectively.
-
Linearize appropriate TALEN expression vectors with BsmBI by adding 5 µg of the vector DNA, 10 µl of 10× NEBuffer 3.1, 1 µl of BsmBI (10 units/µl) and sterile deionized water to a total volume of 100 µl in an eppendorf tube. BsmBI should be added last. Incubate the reaction at 55 °C for 2–3 h.
Note – The reaction temperature for BsmBI is 55 °C. Alternatively, Esp3I is an isoschizomer of BsmBI and can be used at 37 °C.
Run 50 ng of the uncut vector DNA and 5 µl of the reaction mixture on a 1% wt/vol agarose gel to verify that the digestion is complete. Purify the linearized DNA using the QIAquick PCR Purification Kit and elute the DNA with 50 µl of Buffer EB. Measure DNA concentration using a spectrometer. Adjust the final concentration to 10 ng/µl by adding the appropriate amount of Buffer EB.
Digest the TALE repeat arrays assembled using REAL, REAL-Fast or FLASH with BsaI and BbsI according to the instructions in the assembly protocols [44,45]. Continue to follow the protocols and purify the DNA fragments released by the digestion. The concentration of the purified TALE repeat arrays should be approximately 5–10 ng/µl.
-
Ligate the TALE repeat array into the appropriate TALEN expression vector backbone by mixing 1.5 µl of purified DNA fragment (Step 3), 0.5 µl of the linearized vector (Step 2), 2.5 µl of 2× Rapid Ligation Buffer and 0.5 µl of T4 DNA Ligase in an eppendorf tube. Set up a control ligation without the purified TALE repeat array. Incubate the reaction mixture at either room temperature for 1 h or 4 °C overnight. The ligated product can be used for transformation or stored at −20 °C.
Note – The optimal conditions for ligation may vary depending on the source of the ligase and the buffer system used. The conditions specified here are recommended if using the LigaFast™ Rapid DNA Ligation System from Promega. The molar ratio in this ligation is approximately 4:1–8:1 insert:vector.
-
Transform competent bacterial cells with 5 µl of the ligation product following the specific instructions for the cells that you are using. At the end of the transformation procedure, spread the cells on a LB/carbenicillin plate and incubate the plate at 37 °C overnight.
Note – The BsmBI digestion will create incompatible ends in the TALEN expression vectors. Thus, the control ligation should yield very few colonies if the digestion is complete. Meanwhile, the actual ligation should yield 10-fold more colonies than the control ligation.
Pick six colonies per transformation and resuspend each colony with 50 µl of sterile deionized water in a 96-well PCR plate. Store the plates at 4 °C.
Set up 20-µl colony PCR reactions to identify colonies containing a full-length TALE repeat array. Assemble 2 µl of the colony suspension (Step 6), 4 µl of 5× GoTaq Reaction Buffer, 0.4 µl of 10 mM dNTPs, 1 µl of the forward primer oSQT 34 (10 µM), 1 µl of the reverse primer oSQT 35 (10 µM), 0.1 µl of GoTaq DNA polymerase and sterile deionized water to a volume of 20 µl. Use the following cycling condition: Step 1 – 98 °C, 2 min; Step 2 – 98 °C, 10 s; Step 3 – 68 °C, 15 s; Step 4 – 72 °C, 75 s; Step 5 – go back to Step 2 for 25 times; Step 6 – 72 °C, 5 min; Step 7 – hold at 4 °C.
Run 2 µl of the completed reaction on a 1% wt/vol agarose gel to identify colonies that have the full-length array. The size of the PCR product should be (N × 102) + 538 bp, where N is the number of TALE repeats in the array.
-
Re-inoculate the colonies (Step 6) with the correct band size by transferring 20 µl of the colony suspension into a culture tube containing 3 ml of LB/carbenicillin medium. Incubate the culture tube at 37 °C overnight with agitation. Isolate plasmid DNA from the culture using the QIAprep Spin Miniprep Kit according to manufacturer’s instructions.
Note – We recommend not to incubate the culture for over 16 h at 37 °C to reduce the risk of plasmid deletion.
The plasmids should be verified by sequencing using a forward primer oSQT1, a reverse primer oSQT3 and a third primer JDS2980 for the reverse read that includes the 0.5 repeat domain. This should be done in a facility that can provide long sequencing reads (>750 bps). Expected DNA sequences can be acquired from the ZiFiT website (http://ZiFiT.partners.org) by clicking on “TALEs: Generate expected nucleotide sequence” under “Support Tools”
Fig. 3.
The construction scheme of the heterodimeric TALEN vectors. Customized TALE repeat arrays generated by REAL, REAL-Fast or FLASH can be cloned into the heterodimeric TALEN expression vectors. For each TALEN pair, researchers should use one (+) and one (−) backbone vector. In addition to a heterodimeric FokI nuclease domain, each expression backbone vector also contains a different 0.5 TALE repeat domain that specifies the last nucleotide of the target sequence (shown in the parentheses).
5.2. Transferring TALE repeat arrays from homodimeric TALEN vectors to heterodimeric FokI vectors
5.2.1. Materials
-
–
Heterodimeric FokI vectors: pCLR2068 (#28062) and pCLR2070 (#28063) from Addgene (https://www.addgene.org/) [33,35]; alternatively, heterodimeric TALEN expression vectors JDS80 (#40316), JDS82 (#40317), JDS84 (#40318), JDS88 (#40319), JDS90 (#40320), JDS92 (#40321), JDS94 (#40322) and JDS98 (#40323) from Addgene can also be used (see Section 5.2.2 Step 3 for details).
-
–
Homodimeric TALEN vectors: These are existing customized TALEN constructs generated by individual investigators in homodimeric TALEN expression vector JDS70, JDS71, JDS74 or JDS78 [18,19,44,45].
-
–
NheI (#R0131S) and BamHI (#R0136S) (New England Biolabs).
-
–
An agarose gel electrophoresis apparatus.
-
–
A spectrometer for measuring DNA concentration.
-
–
LigaFast™ Rapid DNA Ligation System (Promega).
-
–
Chemically competent or electrocompetent bacterial cells (recA1 strain for reducing nonspecific recombination in the cloned DNA, such as DH5α, TOP10 or XL1-Blue).
-
–
LB/carbenicillin agar plates (see Section 5.1.1).
-
–
LB/carbenicillin medium (see Section 5.1.1).
-
–
Sterile bacterial culture tube.
-
–
QIAquick Gel Extraction Kit (Qiagen).
-
–
QIAprep Spin Miniprep Kit (Qiagen).
5.2.2. Methods
It is possible to convert a pair of existing homodimeric TALEN constructs into a pair of heterodimeric TALEN constructs, which may increase their efficiencies in zebrafish. This can be done by cutting out the entire DNA-binding domains from the homodimeric TALEN constructs and inserting them into a pair of vectors that contain obligate heterodimeric FokI nuclease domains [33].
Digest a pair of homodimeric TALEN constructs with NheI and BamHI by adding 2 µg of the vector DNA, 10 µl of 10× NEBuffer 2.1, 1 µl of 100X BSA, 1 µl of NheI (10 units/µl), 0.5 µl of BamHI (20 units/µl) and sterile deionized water to a total volume of 100 µl in an eppendorf tube. Enzymes should be added last. Incubate the reaction at 37 °C for 2–3 h.
-
Run all of the reaction mixture on a 1% wt/vol agarose gel. There should be two digested products at 5.6 and 2.2–2.4 kilobase pairs (kb), respectively. Cut the 2.2–2.4 kb band out of the agarose gel and purify the DNA using the QIAquick Gel Extraction Kit. Elute the DNA with 30 µl of Buffer EB. Measure the DNA concentration using a spectrometer. Dilute to a final concentration of approximately 10 ng/µl with Buffer EB.
Note – In this reaction, the size of the smaller fragment, which contains the TALE DNA-binding domain, is determined by the number of TALE repeats in the homodimeric TALEN construct. A TALEN construct carrying a customized 14.5 or 16.5 TALE repeat array would yield the size of 2.2 or 2.4 kb, respectively.
-
Digest heterodimeric FokI vector pCLR2068 and pCLR2070 with NheI and BamHI as described in Step 1.
Note – Since double digestion of NheI and BamHI will release the entire DNA-binding domain from homodimeric TALEN constructs (Step 2), the fragments purified in Step 2 can be cloned into the vectors specified here that contain only KKR(+) or ELD(−) FokI nuclease domain but does not contain any portion of a TALE DNA-binding domain. However, investigators can also use heterodimeric TALEN expression vectors if they prefer (see Section 5.2.1). In this case, one KKR and one ELD vector should be used. It does not matter which 0.5 repeat domains are on those vectors because they will be removed by the NheI and BamHI double digestion.
Run all of the reaction mixture on a 1% wt/vol agarose gel. The reaction should yield 5.6 and 0.1-kb products. Cut out the band at 5.6 kb and purify the DNA using the QIAquick Gel Extraction Kit. Elute the DNA with 50 µl of Buffer EB. Measure DNA concentration using a spectrometer. Dilute to a final concentration of approximately 10 ng/µl with Buffer EB.
-
Ligate the TALE fragment into the heterodimeric FokI vector backbone by mixing 1 µl of the purified TALE fragment (Step 2), 1 µl of the linearized vector (Step 4), 2.5 µl of 2× Rapid Ligation Buffer and 0.5 µl of T4 DNA Ligase in an eppendorf tube. Incubate the reaction mixture at either room temperature for 1 h or 4 °C overnight. The ligated product can be used for transformation or stored at −20 °C.
Note – Please see the Note in Section 5.1.2 Step 4 about the ligation condition. The molar ratio in this ligation is approximately 2:1 insert:vector.
Transform competent bacterial cells with 5 µl of the ligation product following the specific instructions for the cells that you are using. At the end of the transformation procedure, spread the cells on a LB/carbenicillin plate and incubate the plate at 37 °C overnight.
-
Pick three colonies per transformation and inoculate individually in a culture tube containing 1.5 ml of LB/carbenicillin. Incubate the culture tubes at 37 °C overnight with agitation.
Note – We recommend not to incubate the culture for over 16 h at 37 °C to reduce the risk of plasmid deletion.
-
On the next day, extract plasmid DNA from the cultures using the QIAprep Spin Miniprep Kit. Perform a diagnostic digestion with NheI and BamHI. The correct clones should yield two products at 5.6 and 2.2–2.4 kb, respectively. These are the heterodimeric TALEN constructs.
Note – Please see the Note above in Step 2 about the size of the smaller fragment in this reaction. In addition, investigators may use the primers listed in Section 5.1.2 Step 10 for sequence verification.
6. Protocols for generation and detection of TALEN-induced mutations in zebrafish
To generate site-specific mutations in zebrafish, a pair of customized TALENs is in vitro transcribed into mRNAs, which can then be co-injected into one-cell stage zebrafish embryos. Once these TALENs successfully create a double-stranded DNA break (DSB) at the target genomic locus, this DNA damage is often repaired via an error-prone mechanism called non-homologous end joining (NHEJ). Consequently, extraneous insertions or deletions (indels) may be introduced at the target site, causing frameshift mutations [1–4]. A DSB can also be repaired via a homology-directed repair (HDR) mechanism in the presence of a homologous DNA donor template, although it usually occurs at a much lower rate compared to NHEJ [1–4]. We have found that engineered TALENs have high success rates for generating targeted mutations in zebrafish by NHEJ [19,33]. Moreover, TALENs have been successfully used to enhance HDR-mediated gene editing that would otherwise occur in almost undetectable frequencies [14,54].
Here, we focus on the method for generating targeted indel mutations using customized TALEN constructs, and we provide protocols for three different methods that can be used to detect indel mutations. All three methods can be used for almost any target sequence and without special optimization based on genomic target loci except for gene-specific PCR conditions. The T7 Endonuclease I (T7EI) assay, which we routinely use to assay for mutation generation, utilizes a mismatch-sensitive endonuclease that can discriminate heteroduplexes formed between mutant and wild-type PCR products [27]. This method is relatively fast and does not require any special equipment or service, but it may not be suitable for target loci containing polymorphisms. In addition, while the T7EI assay can be used to estimate mutation frequencies, it does not reveal the nature of the mutations as the “subcloning & sequencing” method would. We have also used PCR-fluorescent fragment length (PCR-FFL) analysis [34,55]. This method utilizes DNA analyzers that can determine PCR fragment lengths at single base pair resolution. It is simple, sensitive and fast. It can also easily distinguish different mutant alleles and reveal the sizes of insertions and deletions. Most DNA sequencing facilities have the DNA analyzers that can be used for this purpose. Researchers can choose between these methods based on their specific application and preference.
6.1. Generating site-specific mutations using TALENs
6.1.1. Materials
-
–
PmeI (#R0560S) (New England Biolabs).
-
–
QIAGEN Plasmid Mini Kit (Qiagen).
-
–
QIAquick PCR Purification Kit (Qiagen).
-
–
An agarose gel electrophoresis apparatus.
-
–
A spectrometer for measuring DNA and RNA concentration.
-
–
mMESSAGE mMACHINE® T7 Ultra Kit (Life Technologies).
-
–
Nuclease-Free Water (not DEPC-treated) (Life Technologies).
-
–
0.5% Phenol Red Solution (Sigma).
-
–
Danieau solution: 58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES, pH 7.6; filter-sterilized. Store at 25 °C for up to 1 year.
6.1.2. Methods
-
Isolate customized TALEN construct DNA (from Section 5.1.2 or 5.2.2) using a QIAGEN Plasmid Mini Kit. Linearize it with PmeI by mixing 5 µg of the vector DNA, 10 µl of 10× NEBuffer 4, 1 µl of 100× BSA, 1 µl of PmeI (10 units/µl) and sterile deionized water to a total volume of 100 µl.
Note – Plasmid DNAs isolated using QIAprep Spin Miniprep Kit (the final step of Section 5.1.2 or 5.2.2) can also be used here. However, we have found that plasmid DNAs purified using QIAGEN Plasmid Mini Kit often produce higher yields in the subsequent in vitro transcription reaction. Thus, we often reinoculate the bacteria containing sequence-confirmed constructs and prepare a new batch of plasmid DNAs using this kit.
Incubate the reaction at 37 °C for 2–3 h. Run 50 ng of uncut DNA and 2 µl of cut DNA on a 1% wt/vol agarose gel to verify that the digestion is complete. The size of the linearized construct will depend on the length of the TALEN repeat array in the construct but should be ~8 kb.
Purify the digested DNA using a QIAquick PCR Purification Kit and elute the DNA with 50 µl of Buffer EB. Measure DNA concentration using a spectrometer. Continue to the next step or store the purified DNA at −20 °C.
-
Synthesize TALEN-coding RNA using a mMESSAGE mMACHINE® T7 Ultra Kit following manufacturer’s instructions. We use approximately 0.3 µg of purified DNA template for a 10-µl reaction or 0.6 µg for a 20-µl reaction. Perform a poly(A) tailing reaction and precipitate RNAs using the Lithium Chloride Precipitation Solution as instructed in the manual.
Note – It is important to save a small aliquot of the sample before and after the poly(A) tailing reaction as instructed in the manual. Run these aliquots on a 1% agarose gel using the Formaldehyde Loading Dye provided in the kit. The RNA bands should not appear smeared which would indicate degradation. In addition, the size of the RNA should increase after the poly(A) tailing reaction. We have not encountered any problem of RNA degradation during electrophoresis using regular homemade solutions and gels. However, if this becomes a problem, one may want to use special nuclease-free solutions and gels for RNA electrophoresis.
-
Dissolve the RNA pellet with 20 µl of Nuclease-Free Water. Once it is dissolved, keep the tube on ice at all times. Measure RNA concentrations using a spectrometer. Make multiple aliquots in nuclease-free eppendorf tubes. In every tube, mix mRNAs for a pair of TALENs (750 ng each) and store at −80 °C.
Note – Do not use DEPC-treated RNase-free water as it could reduce the viability of zebrafish embryos. Storing RNA in aliquots can prevent RNA degradation during freeze–thaw cycles.
-
On the day of injection, prepare the injection solution by adding to one tube of TALEN mRNAs (1500 ng), 0.5 µl of 0.5% Phenol Red Solution and Danieau solution to a total volume of 5 µl. Thus, the final RNA concentration is 300 ng/µl. Keep the injection solution on ice to prevent RNA degradation. Carefully inject 2 nl of the RNA solution into single-cell stage zebrafish embryos. Injections should be made inside the cytoplasm. Save some uninjected embryos from each clutch as controls. Incubate both control and injected embryos at 28.5 °C overnight.
Note – Save some uninjected embryos from the same clutch as the wild-type control for the mutation analysis described below.
-
Examine the injected embryos at 24 h post-fertilization (hpf). Classify both the injected and uninjected embryos as normal, deformed, or dead by visual inspection. Repeat with the control embryos. Record these numbers. The phenotypically normal embryos may be used for determining the somatic mutation efficiency or raised to adulthood.
Note – The deformity of the injected embryos may have resulted from the injection itself, the toxicities or off-target effects caused by TALENs, or the loss of function of the target gene. We often check the somatic mutation efficiencies in phenotypically normal embryos, because we are interested in knowing the mutation rates in the potential founders. Thus, it is possible that we sometimes underestimate the actual mutation rates if the target gene plays a critical role in early embryonic development. Some researchers may want to use both normal and deformed embryos for the mutation analysis depending on the purpose of the analysis. In addition, sometimes researchers may want to adjust their injection concentrations based on the percentages of phenotypically normal embryos. If the purpose of the experiment is to create mutant founder lines, we suggest that injections should be made at a concentration where the percentage of phenotypically normal embryos is greater than 50% to avoid embryo losses. Reducing injection concentrations may also reduce the chances of off-target DNA cleavage [8].
6.2. Detecting TALEN-induced mutations
6.2.1. T7 Endonuclease I (T7EI) Assay
A. Materials
-
–
PCR primers to amplify target genomic locus. The primers should anneal to sequences approximately 150–200 bps upstream and downstream of the target site.
-
–
Taq DNA polymerase such as GoTaq® DNA Polymerase (Promega) or other PCR enzymes such as Phusion® High-Fidelity DNA Polymerase (New England Biolabs).
-
–
A thermocycler.
-
–
QIAquick PCR Purification Kit (Qiagen).
-
–
A spectrometer for measuring DNA concentration.
-
–
T7 Endonuclease I (T7EI) (#M0302) and NEBuffer 2 (#B7002S) (New England Biolabs).
-
–
0.25 M ethylenediaminetetraacetic acid (EDTA).
-
–
An agarose or polyacrylamide gel electrophoresis apparatus.
-
–
15% Tris/borate/EDTA (TBE) acrylamide gels (Bio-Rad) or 2% w/v agarose gels.
-
–
100 bp DNA ladder (New England Biolabs).
-
–
Ethidium bromide.
-
–
A gel documentation system.
B. Methods
T7EI from the New England Biolabs is similar to the SURVEYOR Nuclease from Transgenomics®. Both have been commonly used for mutation detection [18,27,56], but T7EI is less expensive. Our protocol is specifically optimized for T7EI. We use genomic DNA from single embryos for this assay. It is important to set up one or two control reactions using uninjected embryos from the same clutch as the injected embryos to verify that the T7EI-sensitive PCR products have not resulted from endogenous polymorphisms in the fish colony. The sensitivity of this assay depends on the sensitivity of the gel documentation system. If the lower limit of detecting a band on a gel is approximately 10 ng of DNA, the lowest mutation rate that can be detected using this assay will be 3% based on our protocol (200 ng of DNA input in Step 3) and the formula described in Step 6 below.
Set up 50-µl PCR reactions to amplify the genomic DNA encompassing the target site using gene-specific primers.
Purify the PCR products using a QIAquick PCR Purification Kit and elute with 30 µl of Buffer EB. Measure DNA concentration using a spectrometer.
- Set up the hybridization by mixing 200 ng of the purified PCR product, 2 µl of NEBuffer 2 (10×) and sterile deionized water to a total volume of 19 µl in a PCR plate. Put the plate in a thermocycler and run the following annealing sequence:
Step Temperature Time 1 95 °C 5 min 2 95–85 °C −2 °C/s 3 85–25 °C −0.1 °C/s 4 4 °C Hold At the end of the hybridization cycle, immediately add 1 µl of T7 Endonuclease I to the wells. Incubate the plate at 37 °C for exactly 15 min. Stop the reaction by adding 2 µl of 0.25 MEDTA.
Load all of the reaction products and analyze the sizes of the DNA fragments on a 15% TBE acrylamide gel or a 2% agarose gel. Stain the gel in water containing 0.5 µg/ml ethidium bromide at room temperature for 30 min. Inspect the gel using a UV light box. Compare the injected samples to the control samples and identify any additional restricted fragments of the expected sizes in the injected samples (Fig. 4).
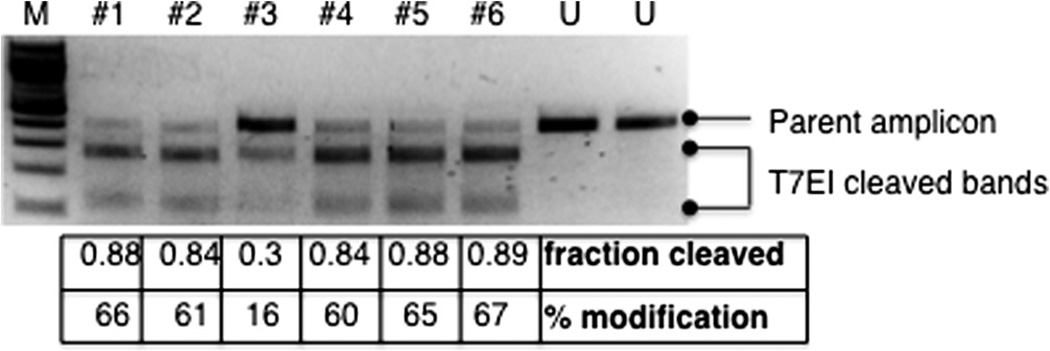
To calculate the somatic mutation rate, the following equation is used: % gene modification = 100 × (1−(1−fraction cleaved)½) [56]. The fraction cleaved can be quantified based on the intensities of the parent amplicon band and the T7EI cleavage bands as determined by a gel documentation system such as the Bio-Rad Gel Doc™ XR + System with the Quantity One® 1-D Analysis Software.
Fig. 4.
A representation of T7EI assay results. T7EI assay was performed as described in Section 6.2.1 using single embryo lysis from six nuclease-injected embryos (marked as #1–6) and two uninjected embryos (marked as “U”). An agarose gel image of the results is shown. While the parent amplicons from the uninjected embryos remained intact after the assay, some portions of the parent amplicons from the injected embryos were cleaved by T7EI into two bands of expected sizes. The “fraction cleaved” and the “% modification” calculated according to the protocol are shown at the bottom of each lane. M, 100 bp DNA ladder (New England Biolabs).
6.2.2. Subcloning & sequencing
A. Materials
-
–
PCR primers to amplify target genomic locus. The primers should anneal to sequences approximately 150–200 bps upstream and downstream of the target site.
-
–
Taq DNA polymerase such as GoTaq® DNA Polymerase (Pro-mega) (see Note in Step 3 below).
-
–
A thermocycler.
-
–
A spectrometer for measuring DNA concentration.
-
–
QIAquick PCR Purification Kit (Qiagen).
-
–
QIAprep Spin Miniprep Kit (Qiagen).
-
–
An agarose gel electrophoresis apparatus.
-
–
Chemically competent or electrocompetent bacterial cells (recA1 strain for reducing nonspecific recombination in the cloned DNA, such as DH5α, TOP10 or XL1-Blue).
-
–
LB/carbenicillin medium (see Section 5.1.1).
-
–
LB/carbenicillin agar plates (see Section 5.1.1).
-
–
X-gal solution, 20 mg/ml in dimethylformamide (DMF) or dimethylsulfoxide (DMSO).
-
–
Sterile bacterial culture tubes.
-
–
pGEM®-T system (Promega) (see Note in Step 3 below).
B. Methods
Genomic DNAs isolated from a pool of injected embryos (normally 5–10 embryos) are used for this assay. We normally pick 24 colonies for sequencing analysis, which would result in a false negative rate of 0.5%, 8% and 29% when the mutation efficiencies are approximately 20%, 10% and 5%, respectively. Thus, this method can fairly consistently identify TALENs that exhibit mutation rates >10%. The sensitivity of this method can be increased by analyzing more colonies.
In addition, after PCR subcloning, the colonies may also be used for direct PCR → sequencing [39] or PCR-FFL analysis (Section 6.2.3). Colonies picked from the transformation plates can be resuspended with 50 µl of sterile deionized water as described in Section 5.1.2 Step 6 and used for regular PCR or fluorescent PCR. These alternative approaches may save the time and cost associated with plasmid extraction. Moreover, PCR-FFL analysis is often cheaper and has a faster turnaround time compared to Sanger sequencing.
Set up 20-µl PCR reactions to amplify the genomic DNA encompassing the target site using gene-specific primers.
Examine the PCR products on an agarose gel. Purify the reaction products using a QIAquick PCR Purification Kit and elute with 50 µl of Buffer EB.
-
Subclone the PCR product into pGEM®-T vector, transform the ligated products and plate the transformants on a LB/carbenicillin agar plate with X-gal following the manufacturer’s instructions.
Note – Alternatively, we have also used Phusion High-Fidelity DNA Polymerase (New England Biolabs) for the PCR reaction in Step 1 and Zero Blunt TOPO PCR Cloning Kit (Life Technologies) here for cloning of blunt-ended PCR products.
Pick 24 white colonies from the transformation plates. Inoculate each single colony into 1.5 ml of LB/carbenecillin medium in a sterile culture tube. Incubate in a 37 °C shaker overnight.
Isolate plasmid DNA from the bacterial culture using a QIAprep Spin Miniprep Kit and submit for sequencing.
6.2.3. PCR- fluorescent fragment length (PCR-FFL) analysis
A. Materials
-
–
PCR primers to amplify target genomic locus. The primers should anneal to sequences approximately 150–200 bps upstream and downstream of the target site. One of the two PCR primers should be end-labeled with either NED, 6-FAM, PET, or VIC.
-
–
PCR lysis buffer with Proteinase K: Add 0.5 ml of 1 M Tris at pH 8, 0.2 ml of 0.5 M EDTA, 0.1 ml of Triton X-100 and water to 50 ml. Store at room temperature. Add Proteinase K to 100 µg/ml before use.
-
–
Taq DNA polymerase such as GoTaq® DNA Polymerase (Promega) or other PCR enzymes such as Phusion® High-Fidelity DNA Polymerase (New England Biolabs).
-
–
A thermocycler.
-
–
An agarose gel electrophoresis apparatus.
-
–
A DNA sequencing facility that offers fluorescent fragment length analysis. For example, Massachusetts General Hospital (MGH) DNA Core provides this service under the selection “Microsatellite analysis” (https://dnacore.mgh.harvard.edu/cgi-bin/jsp/sequencingFirst.action).
-
–
Peak Scanner™ or GeneMapper® software downloaded from the Applied Biosystems website (optional).
B. Methods
This is our preferred method for identifying founders and their offspring that carry target gene mutations. Single embryo lysis or fin biopsies from any generation of fish that may carry indel mutations are used in this protocol. However, genomic DNA isolated from pooled embryos (up to 6 embryos) may also be used [55].
Array embryos or fin biopsies individually into each well of a 96-well PCR plate. Add 30 µl of PCR lysis buffer with Proteinase K. Seal and incubate the plate at 50 °C overnight. On the next day, gently agitate the plate to make sure that the embryos have been lysed completely. Incubate the plate at 95 °C for 10 min to inactivate the Proteinase K. Spin the plate at 3000 rpm for 2 min at room temperature to bring down any debris. The supernatant contains genomic DNA, and it is now ready to be used for PCR.
-
Set up 20-µl PCR reactions encompassing the target site using gene-specific primers and a cycling condition that has been optimized for the primers, the buffer and the DNA polymerase that you are using. Be sure to set up a control PCR using genomic DNA isolated from a wild-type embryo or fin. Minimize sample exposure to light. Run 2 µl of PCR products on a 2–3% agarose gel to verify successful amplification.
Note – Sood et al. have described a modified protocol where, instead of a fluorescent conjugate, a universal linker is added onto the 5′ end of a gene-specific forward primer. Subsequently, PCR is conducted using three primers – a fluorescently labeled primer with the sequence of the universal linker, a gene-specific forward primer with the universal linker and a gene-specific reverse primer [39]. This setup will reduce the cost associated with individual gene-specific fluorescent primers.
-
Determine the sizes of the PCR products on an ABI 3730xl DNA analyzer or similar instrument. (We perform our analyses in a sequencing facility. Samples are processed with GeneScan™ 500 LIZ® Size Standard. Researchers can request data analysis, or choose to obtain raw data (“.ab1” files) and analyze fragment lengths using GeneMapper® by themselves). Identify the peaks of the PCR products and determine their lengths following the instructions of the software. Compare the chromatographs between the test and the control samples. Any additional size variants in the test samples are likely TALEN-induced indel mutations (Fig. 5).
Note – Usually an indel length less than 10 bp can be predicted accurately by calculating the difference in lengths between the mutant and the wild-type PCR products. However, whenever a new mutant allele is identified via this method, it is advised that a user should sequence the PCR product to determine the actual mutation.
Fig. 5.
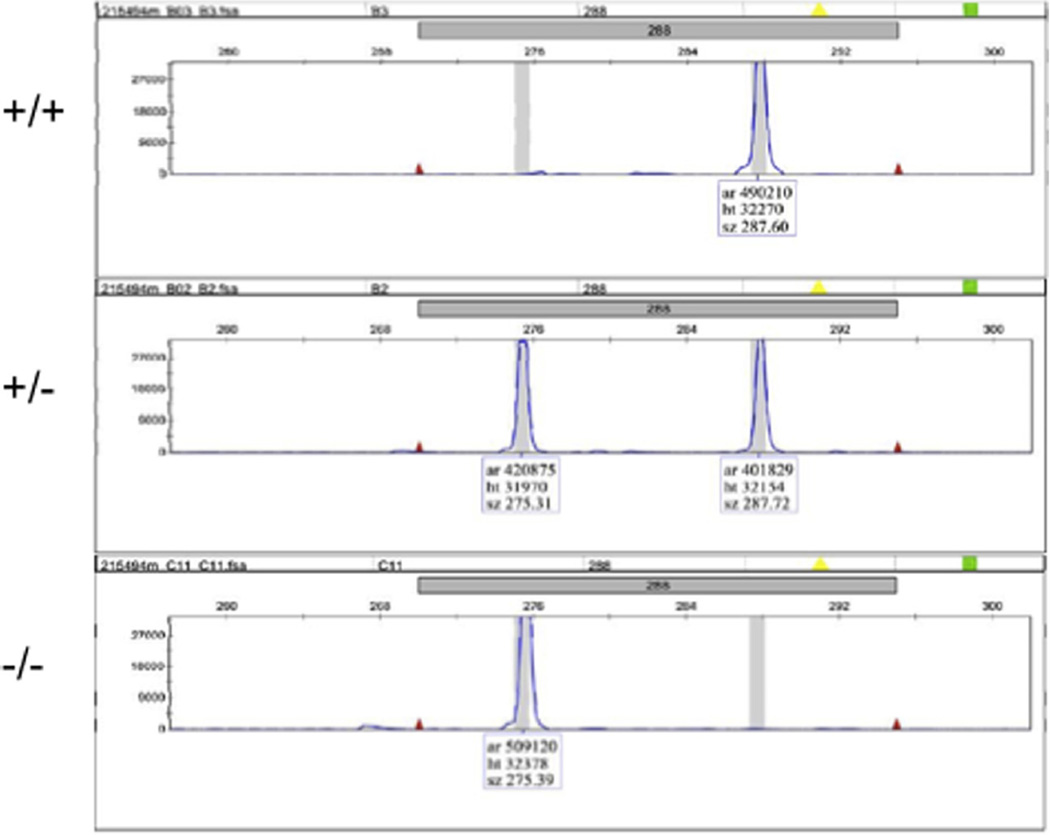
A representation of genotyping results by the PCR-fluorescent fragment length analysis. Fluorescent PCR products from the fin biopsies of a wild-type (+/+), a heterozygous (+/−) and a homozygous (−/−) fish were run on an ABI 3730xl DNA analyzer, and the fragment lengths were determined by GeneMapper®. The results show that while the wild-type sample contains a single fragment of 287 basepairs (bps), the homozygous mutant sample contains a single fragment of 275 bps. Moreover, the heterozygous sample contains two fragments at 287 and 275 bps.
7. Anticipated results
Depending on the method used, TALEN construction and sequence verification should take approximately one to three weeks. In our experience, creating zebrafish gene knockouts by TALENs is robust and efficient ([19,33] and our unpublished results). Over 80% of the TALEN pairs that we have made and tested were able to induce mutations at their target loci. The percentage of modification successfully induced by TALENs ranged between 1.5% and 76.3% based on subcloning and sequence analysis of pooled injected embryos. The distribution of somatic mutation efficiencies is approximately1/3 in all of the following three categories – below 10%, between 10% and 30% and above 30%. For the majority of the TALEN pairs exhibiting a somatic mutation rate at above 20%, all of the F0 fish that we screened were founders, indicating that TALEN-induced mutations can be efficiently transmitted through the germline. At present, among the three commonly used nuclease platforms including ZFN, TALEN and CRISPR/Cas9, TALEN is the only type that has an almost unlimited target range. Thus, customized TALENs should enable facile generation of zebrafish knockout lines for nearly all endogenous genes.
Acknowledgements
We thank Drs. J. Keith Joung, Deepak Reyon, Jeffry D. Sander, Ms. Lindsay Cade and Mr. Samir Patel for their contribution to the development of the methods described in this article. This work is supported by the National Institutes of Health (R01 GM088040 and R01 CA140188) and by the Charles and Ann Sanders MGH Scholar Award.
References
- 1.Gaj T, Gersbach CA, Barbas CF., 3rd Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joung JK, Sander JD. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Esvelt KM, Church GM. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 5.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. Plant J. Cell Mol. Biol. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 12.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christian M, Qi Y, Zhang Y, Voytas DF. G3 (Bethesda) 2013;3:1697–1705. doi: 10.1534/g3.113.007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 18.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Nat. Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. Nat. Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 21.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 22.Bassett AR, Tibbit C, Ponting CP, Liu JL. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho SW, Kim S, Kim JM, Kim JS. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 24.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Nat. Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. Nat. Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Teng F, Li T, Zhou Q. Nat. Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 31.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J, Lee DA, Antoshechkin I, Prober DA. Nucleic Acids Res. 2013;41:2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jao LE, Wente SR, Chen W. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sood R, Carrington B, Bishop K, Jones M, Rissone A, Candotti F, Chandrasekharappa SC, Liu P. PLoS One. 2013;8:e57239. doi: 10.1371/journal.pone.0057239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyon D, Khayter C, Regan MR, Joung JK, Sander JD. Frederick M, Ausubel, et al., editors. Current Protocols in Molecular Biology. 2012;Chapter 12(Unit 12):15. doi: 10.1002/0471142727.mb1215s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyon D, Maeder ML, Khayter C, Tsai SQ, Foley JE, Sander JD, Joung JK. Frederick M, Ausubel, et al., editors. Current Protocols in Molecular Biology. 2013;Chapter 12(Unit 12):16. doi: 10.1002/0471142727.mb1216s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 48.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 49.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valton J, Dupuy A, Daboussi F, Thomas S, Marechal A, Macmaster R, Melliand K, Juillerat A, Duchateau P, Biol J. Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 54.Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, et al. Nat. Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 55.Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Nat. Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]