Abstract
Integration of viral DNA into the host chromosome is an obligatory step in retroviral replication and is dependent on the activity of the viral enzyme integrase. To examine the influence of chromatin structure on retroviral DNA integration in vitro, we used a model target comprising a 13-nucleosome extended array that includes binding sites for specific transcription factors and can be compacted into a higher-ordered structure. We found that the efficiency of in vitro integration catalyzed by human immunodeficiency virus type 1 (HIV-1) integrase was decreased after compaction of this target with histone H1. In contrast, integration by avian sarcoma virus (ASV) integrase was more efficient after compaction by either histone H1 or a high salt concentration, suggesting that the compacted structure enhances this reaction. Furthermore, although site-specific binding of transcription factors HNF3 and GATA4 blocked ASV DNA integration in extended nucleosome arrays, local opening of H1-compacted chromatin by HNF3 had no detectable effect on integration, underscoring the preference of ASV for compacted chromatin. Our results indicate that chromatin structure affects integration site selection of the HIV-1 and ASV integrases in opposite ways. These distinct properties of integrases may also affect target site selection in vivo, resulting in an important bias against or in favor of integration into actively transcribed host DNA.
Stable integration of viral DNA into a host cell's genome is an essential step in the replication of retroviruses. The process is catalyzed by the viral enzyme integrase and establishes the integrated viral DNA (the provirus) as a genetic component that persists for the life of the cell. Because of this unique property, retroviruses have important potential value as vectors for gene therapy in the treatment of human disease (for a review, see reference 27). However, many sites in host chromatin can be targets for retroviral DNA integration, and insertion near or within a cellular gene may lead to inappropriate expression of that gene. The occasional integration in the vicinity of proto-oncogenes or tumor suppressor genes accounts for the oncogenicity of several retroviral genera (13, 32), and this possibility seriously limits the use of retroviral vectors for gene therapy (32). Recent cell culture experiments with human immunodeficiency virus type 1 (HIV-1) have revealed a bias for integration into highly expressed genes, especially genes activated in response to infection (34). Further characterization of a large number of murine leukemia virus (MLV) and HIV-1 integration sites have indicated that MLV is integrated preferentially near the start of actively transcribed genes, whereas HIV-1 shows a bias for integration in many places within such genes but not in the transcriptional start region (42). These data suggest that transcriptionally active chromatin is favored for integration of HIV-1 and MLV DNAs and, conversely, transcriptionally silent chromatin is disfavored; the basis for this distinction is unknown. On the other hand, cell culture experiments with avian sarcoma virus (ASV) have shown that transcriptionally active DNA is not a preferred target for integration, and transcriptional activity may be correlated with a decrease in integration for the DNA of this retrovirus (41). A better understanding of the factors that affect the integration target site selection of different retroviruses may make it possible to design safer and/or more appropriate retroviral vectors for gene therapy.
Studies with purified enzymes and DNA substrates have failed to show any sequence specificity for target site selection in vitro, but the integration site patterns exhibited by different retroviral integrases with the same naked DNA target are not identical (23, 35). Other experiments have shown that the binding of certain proteins can either prevent integration by blocking access to the target DNA or stimulate integration by distorting the target DNA structure (1, 14, 22) such as by bending (20, 26). In addition, experimental systems using mononucleosome particles (28) or minichromosome targets of core histone octamers and circular DNA (29, 30) have shown that retroviral DNA integration occurs preferentially into DNA that is wrapped around the nucleosome compared to nucleosome-free linker DNA. A 10-bp periodicity for integration has been observed at sites where the major groove of the target DNA is exposed on the face of the nucleosome. Integrase proteins from different retroviral species can respond differently to the changes in DNA structure caused by the presence of nucleosomes (30). However, all of these earlier studies have used open or uncompacted chromatin, leaving unexplored the role played by higher-order chromatin structure in the retroviral integration process.
To investigate the role of chromatin structure in retroviral DNA integration, we have asked how its compaction affects the efficiency of integrase-mediated joining of viral sequences to target DNA, with a well-characterized reconstituted chromatin substrate (Fig. 1A) (8, 37) and purified integrase proteins. Our defined-length oligonucleosome target comprises an array of 13 phased nucleosomes, which accurately models the features of native, extended chromatin, and contains binding sites for specific transcription factors (8). The array is also sufficiently long to be able to assume a compacted chromatin structure under appropriate salt conditions or upon addition of linker histone H1. With this system, we have uncovered unexpected differences in the target preferences of integrase proteins from HIV-1 and ASV.
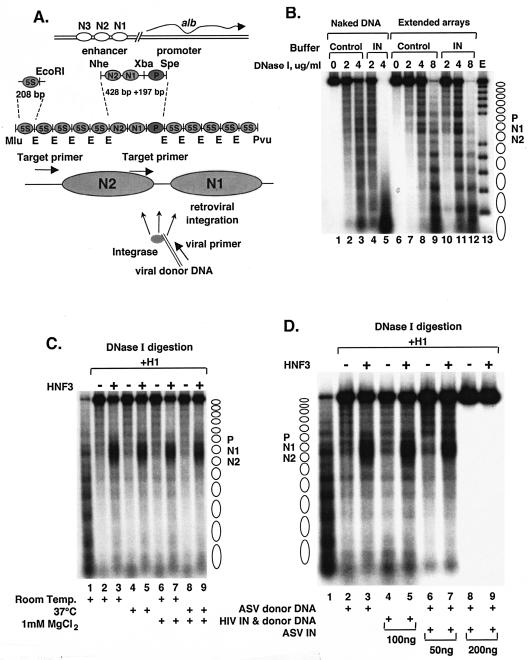
FIG. 1.
Control experiments to examine the influence of retroviral integration reaction conditions and components on nucleosome arrays. (A) Diagram of the MluI-PvuII DNA fragment used to reconstitute the albumin (alb)-5S nucleosome arrays. The locations of the PCR target and viral DNA primers are shown by arrows at the bottom. (B) Effects of integrase and integration reaction components of nucleosome arrays. DNase I digestion analysis of extended nucleosome arrays in the presence of array buffer with (IN lanes) or without (Control lanes) ASV integration reaction components. Lane E, partial EcoRI digest of a DNA end-labeled, extended nucleosome array. Positions of nucleosomes on reconstituted arrays are indicated to right. (C) Effect of temperature and MgCl2 on nucleosome compaction and accessibility of HNF3. Lanes: 1, extended nucleosome array; 2 to 9, array compacted with histone H1 in the absence (−) or presence (+) of HNF3; 2 and 3, incubation of the compacted nucleosome array for 1 h at room temperature; 4 and 5, incubation at 37°C; 6 and 7, incubation with 1 mM MgCl2 at room temperature; 8 and 9, incubation with 1 mM MgCl2 at 37°C. The locations of the nucleosomes corresponding to the albumin enhancer and promoter N1, N2, and P are indicated at the right. DNase I concentrations used: 4 μg/ml for lane 1 and 8 μg/ml for lanes 2 to 9. (D) Effects of viral DNA substrates and viral integrases on nucleosome compaction and accessibility of HNF3. Lanes: 1, extended nucleosome array; 2 to 9, array compacted with histone H1 in the absence (−) or presence (+) of HNF3 (the compacted nucleosome array was incubated for 1 h in lanes 2 and 3 with 1 mM MgCl2 and ASV donor DNA at 37°C); 4 and 5, array compacted with 100 ng of soluble HIV-1 (sIN) integrase protein and HIV-1 donor DNA; 6 and 7, array compacted with 50 ng of ASV integrase protein and ASV donor DNA; 8 and 9, array compacted with 200 ng of ASV integrase protein and ASV donor DNA amounts similar to those in lanes 6 and 7. DNase I concentrations used: 4 μg/ml for lane 1 and 8 μg/ml for lanes 2 to 11.
MATERIALS AND METHODS
Viral DNA substrates and PCR primers.
A synthetic oligodeoxyribonucleotide HIV-1 duplex substrate (21 bp) and an analogous ASV substrate (18 bp) were described previously (22). The single-stranded oligodeoxyribonucleotides were purified by 20% polyacrylamide gel electrophoresis, and complements were annealed at equimolar concentrations in TE buffer (10 mM Tris HCl, 1 mM EDTA, pH 8.0) by heating to 95°C for 5 min, followed by slow cooling to room temperature. Two fixed oligodeoxynucleotide primers complementary to target DNA sequences were used for PCR-based detection of joining events. The target primers were specific for regions upstream of the predicted positions of nucleosomes N1 and N2 in the albumin region, respectively (Fig. 1A) (25): 5′-GCC TAG AAA ATA ACC TGC GTT ACA-3′ and 5′-GGC AAC CCA CAC ATC CTT AGG CAT-3′. These target DNA primers were labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. The virus-specific primer for PCR-based detection of HIV-1-mediated joining events was 5′-GTG TGG AAA ATC TCT AGC A-3′, and that for detection of ASV-mediated joining was 5′-ATT GCA TAA GAC TA CA-3′.
Nucleosomal array reconstitution.
Core histones were purified from sheep liver and used in nucleosome array assembly reaction mixtures containing 2 μg of a 32P-end-labeled double-stranded DNA fragment and core histones at an approximately equal molar ratio of nucleosomal sites and octamers, as described previously (8). Several assembly reaction mixtures containing a range of DNA-histone concentrations were performed, and ideal array saturation (i.e., at a molar ratio of 1:1.2) was determined by the EcoRI digestion assay (8).
Binding reactions, chromatin compaction, and DNase I assays.
Binding reactions for the in vitro integration and DNase I assays were carried out with a nucleosome array concentration of 1 nM (1.5 ng/μl) as previously described (8). To prepare compacted nucleosome arrays, a mixture of 7.5 or 13 nM purified histone H1 (Boehringer) and an extended nucleosome array concentration of 1 nM was incubated at room temperature for 1 h to obtain different degrees of array compaction. For NaCl-mediated nucleosome array compaction, the final NaCl concentration was 100 or 120 mM and the mixtures were incubated at room temperature for 1 h. To achieve HNF3-mediated chromatin opening, the H1-compacted array was incubated with 20 nM mouse HNF3α transcription factor for 2 h at room temperature (8). The mouse HNF3α and GATA4 proteins were purified from Escherichia coli (9, 43).
DNase I digestion was carried out as previously described (8). Briefly, the reaction mixture included 2.5 to 10 μg of DNase I per ml diluted in 50 mM MgCl2, and incubation was at room temperature for 1 min. Digestions were terminated by addition of an equal volume of stop buffer (20 mM Tris-HCl [pH 7.5], 50 mM EDTA, 1% sodium dodecyl sulfate, 0.2 mg of proteinase K per ml), followed by incubation at 50°C for 30 min. Purified digestion products were subjected to electrophoresis in 1% agarose-0.5× Tris-borate-EDTA gels. Gels were fixed in 10% acetic acid-10% methanol and dried.
HIV-1 and ASV in vitro integration assays.
Following incubation with histone H1 or transcription factors as indicated, the volume of each mixture was increased from 10 to 20 μl by adding the integration reaction mixture. The final concentrations of integration reaction components were 0.25 mg of bovine serum albumin per ml, 6.4% glycerol, 5.7 mM dithiothreitol, 0.02% Nonidet P-40, 12 mM HEPES (pH 7.5), 45 mM KCl, and 0.1 mM MgCl2. Assays of ASV and HIV-1 integrase activities were performed under conditions that were optimal for each enzyme. ASV reaction mixtures included 50 ng (0.15 nM) of integrase, 1 pmol of ASV oligodeoxyribonucleotide duplex substrate (0.1 nM), and 1 mM MgCl2; HIV-1 reaction mixtures contained 100 ng (0.3 nM) of integrase, 1 pmol (0.1 nM) of HIV-1 oligodeoxyribonucleotide duplex substrate, and 1 mM MnCl2, as the activity of this protein in MgCl2 is too low to detect any joining in this assay. For HIV-1 integration assays, we used bacterially produced HIV-1sIN integrase (NY5 strain sequence) that included the following amino acid substitutions to improve solubility: C56S, C65S, W131D, F139D, F185H, and C280S (40). The activity of this enzyme was comparable to that of the His-tagged HIV-1 wild-type version of this IN protein (HIV-1wt/His). Both derivatives display wild-type processing and joining activities and can be stored and assayed at relatively high concentrations.
Following incubation for 1 h at 37°C with ASV or HIV-1 integration components, 10-μl samples were removed for DNase I analysis. The remainder of the reaction mixtures were deproteinized by increasing the volume to 130 μl by addition of EDTA (final concentration of 4.25 mM), sodium dodecyl sulfate (final concentration of 0.44%), and proteinase K (final concentration of 0.06 mg/ml). After digestion for 60 min at 37°C, the reaction mixtures were extracted with phenol, followed by phenol-chloroform-isoamyl alcohol (25:24:1 mixture), and the DNA was precipitated with ethanol. The DNA pellets were then dissolved in 10 μl of water. As the nucleosomal DNA was labeled with 32P at one end, concentrations of all samples were estimated by analysis in a scintillation counter.
For both ASV and HIV-1 reaction mixtures, joining of viral DNA to specific regions in the target was detected with a PCR-based assay. Equal amounts of the DNA from a joining reaction were incubated with [γ-32P]dATP-labeled target and viral DNA primers. PCR mixtures (50 μl) contained 50 mM KCl, 20 mM Tris-HCl buffer (pH 8.4), 5 mM MgCl2, 200 μM deoxynucleoside triphosphate, and 2 U of Taq polymerase (Gibco BRL). Reaction mixtures were cycled as follows: 92°C for 3 min and 30 cycles of 92°C for 40 s, 50°C for 40 s, and 72°C for 45 s. PCR products were analyzed on 6% sequencing gels that included a 10-bp ladder that was also 5′ end labeled.
HIV-1wt/His integrase purification.
Frozen resuspended cells expressing HIV-1wt/His integrase, harvested from 2.4 liters of Luria broth culture, were thawed and resuspended in lysis buffer (25 mM BisTris [pH 6.1], 1 M KCl, 1 M urea, 1% thiodiglycol, 5 mM imidazole) at 0.13 g of cells/ml. Lysis was performed by French press, followed by sonication for 30 s, and the preparation was then subjected to centrifugation for 30 min at 30,000 × g. The supernatant fraction was filtered through a 0.22-μm-pore-size filter and applied to a 5-ml HiTrap iminodiacetic acid column freshly charged with 50 mM NiSO4 and equilibrated with lysis buffer containing 10% glycerol. The column was washed with 20 column volumes of the equilibrating buffer, and bound protein was eluted with a gradient of 5 mM to 1 M imidazole containing 25 mM BisTris (pH 6.1), 1 M KCl, 1 M urea, 1% thiodiglycol, and 10% glycerol. Fractions containing integrase were pooled, EDTA was added to a final concentration of 0.5 mM, and the preparation was frozen in aliquots at −70°C.
Single aliquots of the IN preparation were thawed and diluted fourfold with 25 mM BIS-Tris (pH 6.1)-1 M urea-2 mM β-mercaptoethanol-0.1 mM EDTA-10% glycerol to reduce the KCl concentration to 0.25 M. This solution was immediately applied to a 5-ml HiTrap heparin column equilibrated with 25 mM BIS-Tris (pH 6.1)-0.25 M KCl-1 M urea-2 mM β-mercaptoethanol-0.1 mM EDTA-10% glycerol with a syringe. Following a wash step with the equilibrating buffer, bound protein was eluted with a gradient to 1 M KCl. Fractions containing IN were pooled, concentrated, and dialyzed against 25 mM BIS-Tris (pH 6.1)-0.5 M KCl-1% thiodiglycol-1 mM dithiothreitol-0.1 mM EDTA-40% glycerol. After dialysis, the IN preparation was subjected to centrifugation at 30,000 × g for 15 min to remove any precipitated protein, and aliquots were frozen in liquid nitrogen.
RESULTS
Model chromatin substrate.
To examine the influence of higher-order chromatin structure on retroviral DNA integration in vitro, we used a 2,705-bp target DNA that can be assembled into a phased array of 13 nucleosomes. This target includes a trinucleosome-sized DNA section corresponding to the albumin gene enhancer and promoter, flanked on either side by five copies of a sea urchin 5S rRNA gene nucleosome positioning sequence (Fig. 1A) (8). The presence of binding sites for specific transcription factors in the albumin enhancer region makes it possible to examine the effects of their binding on retroviral DNA integration in vitro. Extended nucleosome arrays were assembled from end-labeled DNA templates and purified core histones. The nucleosomal DNA migrated faster on an agarose gel than did the naked DNA, as expected, and digestion of the extended array with EcoRI restriction endonuclease yielded less than 5% free DNA in this preparation (data not shown; see also references 2, 16, and 39). Partial digestion of the arrays with DNase I revealed cleavages and protections indicative of 13 evenly spaced nucleosomes (Fig. 1B, lanes 7 to 9). As a control for stability under our assay conditions, the extended nucleosome arrays first were incubated with components of the integration reaction mixture (purified ASV integrase protein, viral donor DNA, 1 mM MgCl2) at 37°C for 1 h. Partial digestion with DNase I showed that none of these components or the incubation temperature caused any change in the nucleosomal digestion pattern (Fig. 1B, lanes 10 to 12). Naked target DNA was used as a control for DNase I activity in the presence of integration reaction mixture components (Fig. 1B, lanes 1 to 5). The greater extent of digestion of the extended nucleosome array in the presence of the integration reaction mixture components is the result of more favorable conditions for DNase I rather than an effect on the array (compare lanes 9 and 12 and lanes 3 and 5). Similar control experiments with HIV-1 integration reaction mixture components also failed to show any effect on DNase I digestion patterns of the extended nucleosome array (data not included).
The extended arrays were then compacted by addition of the linker histone H1 to nucleosomes at a 1:1 molar ratio. As illustrated in Fig. 1C (compare lanes 1 and 2), compaction with H1 rendered the arrays virtually resistant to the same concentration of DNase I (8 μg/ml) that is able to digest the extended array almost completely (Fig. 1B, lane 12). As expected from previous studies (8), addition of the transcription factor HNF3 increased DNase I sensitivity in the vicinity of its binding site in the DNA of nucleosome N1 (Fig. 1C and the diagram in Fig. 2B). Addition of the HIV-1 integration reaction mixture components at the standard incubation temperature (37°C) had no effect on nucleosome compaction or chromatin opening by HNF3 as measured by DNase I digestion (Fig. 1D, lanes 4 and 5). Similar results were observed upon addition of 50 ng of ASV integrase, our standard condition (Fig. 1D, lanes 6 and 7). However, upon addition of a larger amount of protein (200 ng), with the same donor DNA concentration, DNase I digestion was totally inhibited (Fig. 1D, lanes 8 and 9). It is possible that at this high protein concentration, the binding of ASV integrase to the chromatin target blocks access to the DNase I (6).
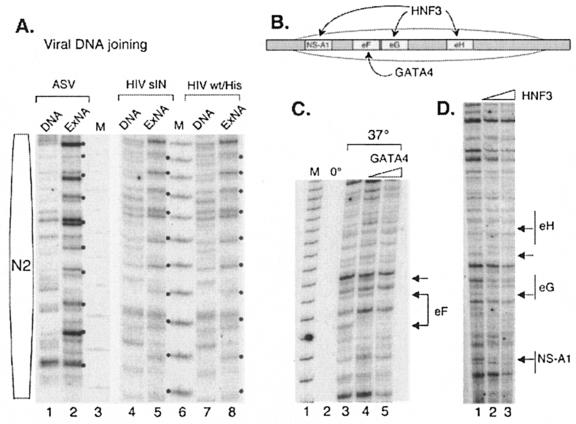
FIG. 2.
Retroviral integrase-mediated joining to the extended nucleosome array. (A) Controls for the retroviral integration reaction. Lanes: 1 and 2, ASV integrase-mediated joining of the cognate viral donor DNA to the naked target (DNA) and to the extended nucleosome array (ExNA) (dots mark the 10-bp periodicity for joining); 4 and 5, HIV-1sIN-mediated joining of the cognate viral donor DNA to the naked target and extended nucleosome array; 7 and 8, HIV-1wt/His integrase-mediated joining of the cognate viral donor DNA to the naked target and extended nucleosome array; 3 and 6, 10-bp DNA molecular weight marker (M). The position of the N2 nucleosome is shown at the left. (B) Sites of HNF3 and GATA4 binding to the nucleosome N1 region in the albumin enhancer region. The position of the nucleosome is indicated by the oval. Labeled boxes indicate transcription factor binding sites. (C) ASV DNA joining is blocked at sites at which transcription factor GATA4 is bound to the nucleosome array. Lanes: 1, 10-bp DNA molecular weight marker (M); 2, ASV DNA joining after incubation at 0°C; 3, ASV DNA joining after incubation at 37°C; 4, ASV DNA joining after incubation at 37°C after preincubation in 20 nM GATA4; 5, ASV DNA joining after incubation after preincubation in 40 nM GATA4. The GATA4 binding site is designated eF. Arrows show sites of target DNA protection (footprinting) from ASV DNA joining. (D) ASV DNA joining is blocked at sites at which transcription factor HNF3 is bound to the nucleosome array. Lanes: 1, ASV DNA joining to a nucleosome array at 37°C; 2, ASV DNA joining to a nucleosome array preincubated with 20 nM HNF3; 3, ASV DNA joining to a nucleosome array preincubated with 40 nM HNF3. HNF3 binding sites are designated NS-A1, eG, and eH. Arrows show sites of target DNA protection (footprinting) from ASV DNA joining.
Transcription factors HNF3 and GATA4 block the integration of viral DNA at their binding sites in the extended nucleosome array.
To detect joining of the viral DNA sequences to the target DNA by purified retroviral integrase, we used a PCR-based assay with a labeled primer similar to that described previously (7, 22, 30). The size of the PCR product reflects the distance between the joining site and a fixed point in the target DNA. We used two primers, complementary to different regions in the target DNA, to monitor viral donor DNA joining to DNA associated with nucleosome N2 or N1 and the upstream linker region (Fig. 1A, bottom section). Comparable viral DNA joining patterns were observed with the different primer sets (data not shown), confirming the validity of the assay. Several control experiments (data not shown) were carried out to verify that a PCR product was generated only when integrase and the cognate donor viral DNA were present. No PCR product was detected in the presence of EDTA or if the joining reaction mixtures were left on ice.
Patterns of integrase-mediated joining to naked DNA and the extended nucleosome array in the N2 region are shown in Fig. 2A. As expected (23, 35), different patterns were observed in the same naked DNA targets with ASV and HIV-1 integrases, with some sites preferred by one system and not the other. However, both enzymes produced a noticeable 10-bp periodicity for joining in the extended nucleosome array target (ExNA, Fig. 2A, lanes 2, 5, and 8), as has been reported previously for the MLV and HIV-1 integrases (30). We found that the joining patterns for the HIV-1sIN and HIV-1wt/His integrases are indistinguishable (compare lanes 4 and 7 and lanes 5 and 8), indicating that the solubility-improving substitutions do not affect the integration preferences. We next asked if the binding of transcription factors to the extended nucleosome array would affect the joining reaction. As illustrated in Fig. 2B, the HNF3 transcription factor is known to bind specifically to its sites, designated eG, eH, and NS, in the N1 nucleosome of the extended array in vitro and forms protected and hypersensitive sites, as detected by DNase I footprinting (8). The GATA4 transcription factor also binds at this location, at a site designated eF (Fig. 2B) (8). As illustrated in Fig. 2C, the presence of GATA4 inhibited ASV integrase-mediated joining of viral DNA to the binding site of this factor in the extended nucleosome array, as demonstrated by the loss of specific integration products in the N1 region (footprinting; see arrows). Similar results were observed for HNF3; ASV integrase-mediated joining of donor viral DNA was blocked at the eG, eH, and NS binding sites of N1 in the target DNA (Fig. 2D). No other changes in integration patterns were detected. Specifically, no preferred sites for joining were generated in the presence of these factors. From these data, we conclude that GATA4 and HNF3 transcription factor binding to the extended nucleosomal array limits access to their binding sites for the integration machinery, as expected from previous studies with naked DNA targets (1, 14, 22). This result is similar to the inhibition of DNase I cleavage in footprinting experiments (3, 36), with some pattern variations likely due to differences in the way IN and DNase I act on a nucleosomal target; integrase joins viral DNA to target DNA at exposed sites on the major groove (30), whereas DNase I cleaves the target DNA in the minor groove (38).
Integration of viral donor DNA into the H1-compacted nucleosome array.
To study the influence of higher-order chromatin structure on the efficiency of retroviral integration, the extended nucleosome array target was compacted with linker histone H1 (Fig. 3A). After the integration reactions were terminated, one-half of the mixture was analyzed by DNase I digestion, which served as a control to measure the reduced accessibility caused by histone H1-mediated compaction (e.g., Fig. 3B, left side). The other half of the mixture was incubated with proteinase K, followed by DNA purification, and viral DNA joining in the N1 region was detected by PCR (e.g., Fig. 3B, right side). Results obtained with the ASV integration system again showed that the pattern obtained with naked DNA was distinct from that observed with the extended nucleosome array (Fig. 3B, right side, compare lanes 1 and 2). Remarkably, the efficiency of joining increased substantially upon compaction of the array with histone H1 (compare lanes 2 and 3). Further compaction by histone H1 did not change the integration pattern but appeared to increase the efficiency of joining (compare lanes 2 and 4). In three independent experiments, we observed a more-than-sevenfold increase in ASV joining to targets compacted by 13 nM histone H1 compared to the extended array, on the basis of density scanning. This increase in efficiency with the compacted target was not uniform; some sites in the target were preferred over others for joining of viral DNA.
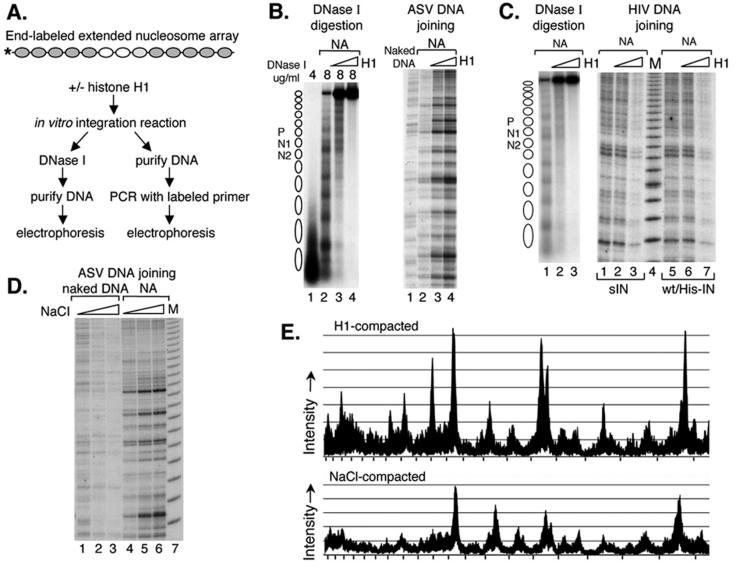
FIG. 3.
Influence of target nucleosome array compaction on integrase-mediated joining of viral DNAs. (A) Experimental design. The extended nucleosome array was incubated with increasing amounts of histone H1 for 1 h, followed by addition of the ASV or HIV-1 integration components. One hour later, the mixture was analyzed by DNase I assay and by PCR with the N2-specific target DNA primer and an HIV-1 or ASV donor DNA primer. (B) DNase I analysis of the target DNA after incubation with the ASV DNA joining components (on the left) and PCR detection of the ASV DNA joining pattern (on the right). NA, extended nucleosome array. On the left side, DNase I concentrations are shown above the gel. Right side: lane 1, naked DNA target; lane 2, extended nucleosome array without histone H1; lane 3, array compacted by addition of 7.5 nM histone H1; lane 4, extended array compacted by addition of 13 nM histone H1. (C) DNase I analysis of the DNA used as a target for HIV-1 DNA joining (on the left) and PCR detection of the HIV-1 DNA joining pattern (on the right). DNase I concentrations used (left side, 4 μg/ml for lane 1 and 8 μg/ml for lanes 2 and 3). Right side: lanes 1 to 3, HIV-1sIN-mediated DNA joining; lanes 5 to 7, HIV-1wt/His IN-mediated DNA joining; lanes 1 and 5, extended nucleosome array; lanes 2 and 6, extended array compacted by addition of 7.5 nM histone H1; lanes 3 and 7, extended array compacted by addition of 13 nM histone H1; lane 4, 10-bp molecular weight marker (M). (D) ASV integrase-mediated joining into NaCl-mediated compacted targets. Naked DNA or the extended nucleosome array was incubated with 100 mM NaCl (lanes 2 and 5, respectively) or 120 mM NaCl (lanes 3 and 6, respectively) and then incubated with the ASV integration components. Lanes 1 and 4, ASV DNA joining to naked DNA and the extended nucleosome array in the presence of control buffer; lane 7, 10-bp molecular weight marker (M). (E) Comparison of ASV integration patterns in the region of nucleosome N1 with extended and compacted nucleosomal arrays. The sequencing gels of panels B, right side, lanes 2 and 4 (top tracing), and D, lanes 4 and 6 (bottom tracing), were scanned with a Fuji BAS-250 Bio-imaging analyzer and analyzed with Image Gauge 4.0 software. Tick marks on the x axis correspond to the 10-bp DNA molecular weight marker.
As shown in Fig. 3C (left side), reduced target DNA accessibility due to histone H1-mediated compaction was again verified by DNase I digestion of the reaction mixture incubated with HIV-1 integration components. However, in sharp contrast to the results obtained with ASV, no increase in the efficiency of the donor viral DNA joining was observed with either the soluble derivative, sIN, or wt HIV-1 integrase in the presence of histone H1 (Fig. 3C, right side, compare lanes 1 and 2 and lanes 5 and 6). Furthermore, the efficiency of joining by the HIV-1 integrases actually decreased with increased concentration of histone H1 (compare lanes 2 and 3 and lanes 6 and 7). The similar behaviors of the two HIV-1 integrases indicate that differences observed in the integration preferences of the ASV and HIV-1 proteins are not due to the solubility-improving substitutions. The inhibition of HIV-1 integrase-mediated joining by chromatin compaction highlights the specificity of the enhanced joining observed with ASV integrase.
We considered the possibility that the observed ASV integrase-mediated increase in joining of viral donor DNA sequences to the compacted target DNA might be the result of interaction of this integrase protein with linker histone H1. Alternatively, this result could be due to an ASV integrase-specific preference for a compacted DNA target. To distinguish between these two possibilities, we used an H1-independent method of compaction; the extended nucleosome array was incubated in the presence of 100 or 120 mM NaCl for 1 h to produce salt-dependent compaction (15; also see Fig. 1F in reference 8). It was reported previously that variation in the monovalent-ion concentration within this range has no significant effect on the efficiency of the ASV integration reaction (12). As shown in Fig. 3D (lanes 1 and 2), under our conditions, 100 mM NaCl had a slight inhibitory effect on the efficiency of ASV integrase-mediated joining to naked target DNA. On the other hand, increased efficiency of joining to the nucleosome array was observed at this NaCl concentration, similar to that observed in the same region with histone H1 compaction (Fig. 3B, right side). This effect was even more striking in the presence of a higher salt concentration (120 mM NaCl; Fig. 3D, lane 6), even though these conditions were suboptimal for ASV integrase (Fig. 3D, compare lanes 1 and 3). We conclude that NaCl-compacted nucleosomal DNA is also a preferred target for ASV integrase-mediated joining. Figure 3E shows results from density scans of the data in panels B (lanes 2 and 4) and D (lanes 4 and 6). This comparison highlights the similarity in the patterns of ASV joining with extended arrays (open traces) and arrays compacted by either histone H1 or NaCl (shaded traces). Comparison of these traces provides a quantitative estimate of the increased efficiency of joining to the chromatin compacted by either method. It appears from these results that the compacted DNA structure, rather than an interaction with histone H1 per se, enhances this reaction.
Unfortunately, parallel experiments with the HIV-1 integration components were not possible, as the HIV-1 integration reaction was completely inhibited in the presence of 100 mM NaCl with both naked DNA and compacted nucleosome targets.
HNF3 binding to the compacted nucleosome array has no effect on ASV integrase-mediated joining.
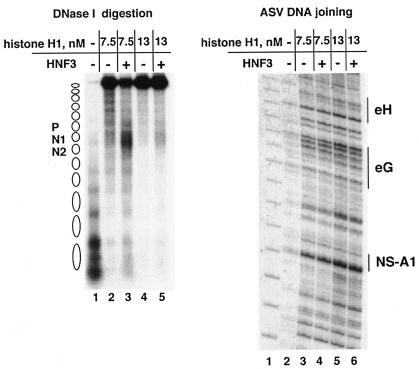
As illustrated in Fig. 1C and D, binding of transcription factor HNF3 to the histone H1-compacted array creates a DNase I-sensitive site in the vicinity of binding. To determine if this change creates a preferred or restricted site for integration, we added HNF3 to the H1-compacted nucleosome array and then incubated this substrate with ASV integration components. As a control, half of the incubation mixture was again analyzed by the DNase I digestion assay, as outlined in Fig. 3A, to verify that the target was compacted and HNF3 was bound at the expected site (Fig. 4, left side). As shown in Fig. 4 (right side), an increase in the efficiency of ASV-mediated joining was again observed with increasing nucleosome array compaction. However, the pattern of joining in the vicinity of the HNF3 binding site was unaffected by the presence of this transcription factor. Thus, locally open chromatin was not a preferred integration target in the compacted array.
FIG. 4.
ASV integrase-mediated joining of viral DNA to the compacted array opened locally by the HNF3 transcription factor. The histone H1-compacted nucleosome array was incubated for 2 h with transcription factor HNF3, and then the ASV integration components were added. One hour later, the mixture was analyzed by DNase I assay and by PCR with N1-specific target DNA and ASV donor DNA primers. DNase I analysis is shown in the left-side DNA. Lanes: 1, extended array; 2, extended array compacted by 7.5 nM histone H1; 3, 7.5 nM histone H1-compacted array opened locally by HNF3; 4, extended array compacted by 13 nM histone H1; 5, 13 nM histone H1-compacted array opened locally by HNF3. The DNase I concentrations used were 4 μg/ml for lane 1 and 8 μg/ml for lanes 2 to 5. Results from the joining assay are on the right side. Lanes:1, 10-bp marker; 2, viral DNA joining to the extended nucleosome array; 3 and 4, DNA joining to the extended nucleosome array compacted with 7.5 nM histone H1 in the absence (−) or presence (+) of HNF3; 5 and 6, with 13 nM histone H1 in the absence (−) or presence (+) of HNF3.
DISCUSSION
In these in vitro studies of retroviral DNA integration, we took advantage of a DNA target that has been used previously to reconstitute extended nucleosome arrays that can be compacted into higher-order structures comparable to chromatin in living cells (8). Our results showed that when an extended nucleosomal array is used as an integration target by ASV and HIV-1 integrases, the distribution of joining sites is distinct from that observed in the corresponding naked DNA, consistent with earlier findings (30). However, further compaction of the extended array by histone H1 or a high NaCl concentration did not cause major changes in the joining pattern but had dramatically opposite effects on the efficiency of the reaction with the ASV and HIV-1 integrases. We found that ASV integrase-mediated joining of the cognate viral DNA was substantially more efficient with a histone H1-compacted DNA chromatin target than with naked DNA or an extended nucleosome array. A similar increase was also observed with NaCl-compacted target chromatin, suggesting a critical role for the structure of the DNA in compacted chromatin, rather than interactions between the ASV integration machinery and histone H1. In contrast to the results obtained with ASV integrase, a decrease rather than enhancement of integration into the histone H1-compacted chromatin target was observed in reactions catalyzed by HIV-1 integrase. These observations indicate that the integration machineries of these two retroviruses may be markedly different in their site selection preferences.
Extended chromatin and compacted chromatin are characteristic of actively transcribed or silent regions of cellular chromosomes, respectively. Actively transcribed regions include a number of bound components that could potentially reduce the efficiency of retroviral DNA integration. Our data suggest that transcription factors bound to the chromatin may block ASV DNA integration in actively transcribed regions. The HNF3 transcription factor binds to three sites in the DNA associated with nucleosome N1 in the extended nucleosome array, and such binding results in DNA protection and exposure of hypersensitive sites, as detected by DNase I footprinting (8). In our ASV integrase-mediated joining reaction, accessibility was limited specifically at HNF3 binding sites in this target DNA but, interestingly, we did not observe any additional hot spots for viral donor DNA joining in the presence of this transcription factor. We also asked if HNF3 transcription factor binding to the histone H1-compacted target would affect ASV integrase-mediated joining in our system. Surprisingly, we observed no changes in the joining pattern when the compacted target was opened by HNF3 binding, even though our control experiment showed that sequences in the N1 nucleosome region became more accessible to DNase I digestion. These results underscore the preference of ASV for compacted chromatin, even when an open region is available on the same template.
The striking differences we observed between the target preferences of the ASV and HIV-1 integrases suggest that these proteins may interact differentially with extended and compacted chromatin in vivo. Although many factors may contribute to target site selection (11, 33), it is noteworthy that the joining preferences of the integrase proteins that we have uncovered in vitro are consistent with results reported for integration of ASV and HIV-1 DNAs in infected cells (34, 41). This observation underscores the importance of using more developed models of chromatin structure in vitro to replicate integration activities seen in vivo.
Retroviral integration site selection has become an important topic of study owing to the use of retroviral vectors in gene therapy for human disease. Several cases of insertional mutagenesis, believed to have been triggered by integration of the MLV vector DNA near the growth-promoting gene LMO2 were recently observed. In these clinical studies, 2 of 11 patients treated for severe combined immunodeficiency disease developed leukemias, with the malignant cells showing integrations at this locus (4, 5). A recent study with mice has also shown that integration of replication-deficient MLV vector DNA is associated with leukemia development (24). Our results suggest that an ASV vector might have some advantage over MLV or HIV-1 vectors if integration into actively transcribed regions is to be avoided. On the other hand, integration into regions of compacted chromatin might cause silencing of an integrated ASV vector. Additional engineering, for example, introduction of insulator sequences, might help to alleviate this problem (10, 18, 31). As ASV has in common with HIV-1 the ability to infect nondividing cells (17, 21), an ASV vector may be especially useful for particular therapeutic applications.
Integrases from different retroviruses produce unique and reproducible patterns of integration into naked DNA in vitro. The determinants for these patterns appear to be located within the core domain of the protein (19, 23, 35). Here we show that the HIV-1 and ASV integrases also have different integration preferences with respect to compacted chromatin targets. The molecular determinants for these properties of integrases remain to be determined. Detailed knowledge about the mechanism of integration site selection in chromatin may contribute to the development of safer gene therapeutic tools.
Acknowledgments
We thank G. Merkel and D. Colleluori for providing purified ASV and HIV-1 integrases.
This work was supported by National Institutes of Health grants AI40385, CA71515, CA06927, and GM47903; by the Mathers Charitable Foundation; and also by an appropriation from the Commonwealth of Pennsylvania. The following Fox Chase Cancer Center Shared Facilities were used in the course of this work: Cell Culture Facility, Biochemistry and Biotechnology Facility (DNA Synthesis), and Research Secretarial Services.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
REFERENCES
- 1.Bor, Y. C., F. D. Bushman, and L. E. Orgel. 1995. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc. Natl. Acad. Sci. USA 92:10334-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37:14776-14787. [DOI] [PubMed] [Google Scholar]
- 3.Chaya, D., T. Hayamizu, M. Bustin, and K. S. Zaret. 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 276:44385-44389. [DOI] [PubMed] [Google Scholar]
- 4.Check, E. 2003. Second cancer case halts gene-therapy trials. Nature 421:305. [DOI] [PubMed] [Google Scholar]
- 5.Check, E. 2002. A tragic setback. Nature 420:116-118. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P., W. Pluymers, A. Claeys, P. Proost, E. De Clercq, and Z. Debyser. 2002. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 14:1389-1399. [DOI] [PubMed] [Google Scholar]
- 7.Chow, S. A. 1997. In vitro assays for activities of retroviral integrase. Methods 12:306-317. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo, L. A., F. R. Lin, I. Cuesta, D. Friedman, M. Jarnik, and K. S. Zaret. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9:279-289. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, L. A., and K. S. Zaret. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4:961-969. [DOI] [PubMed] [Google Scholar]
- 10.Emery, D. W., E. Yannaki, J. Tubb, and G. Stamatoyannopoulos. 2000. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc. Natl. Acad. Sci. USA 97:9150-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman, A. 2003. The role of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 281:209-238. [DOI] [PubMed]
- 12.Fitzgerald, M. L., A. C. Vora, W. G. Zeh, and D. P. Grandgenett. 1992. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J. Virol. 66:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka (ed.). 2000. Principles of virology: molecular biology, pathogenesis, and control, p. 552-593. ASM Press, Washington, D.C.
- 14.Goulaouic, H., and S. A. Chow. 1996. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 70:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, J. C., J. Ausio, V. H. Stanik, and K. E. van Holde. 1989. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry 28:9129-9136. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, J. C., and D. Lohr. 1993. Assembly and structural properties of subsaturated chromatin arrays. J. Biol. Chem. 268:5840-5848. [PubMed] [Google Scholar]
- 17.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hejnar, J., P. Hajkova, J. Plachy, D. Elleder, V. Stepanets, and J. Svoboda. 2001. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. USA 98:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes-Son, M. L., R. S. Appa, and S. A. Chow. 2001. Molecular genetics and target site specificity of retroviral integration. Adv. Genet. 43:33-69. [DOI] [PubMed] [Google Scholar]
- 20.Katz, R. A., K. Gravuer, and A. M. Skalka. 1998. A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem. 273:24190-24195. [DOI] [PubMed] [Google Scholar]
- 21.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz, R. A., G. Merkel, and A. M. Skalka. 1996. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology 217:178-190. [DOI] [PubMed] [Google Scholar]
- 23.Katzman, M., and M. Sudol. 1995. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 69:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Z., J. Dullmann, B. Schiedlmeier, M. Schmidt, C. von Kalle, J. Meyer, M. Forster, C. Stocking, A. Wahlers, O. Frank, W. Ostertag, K. Kuhlcke, H. G. Eckert, B. Fehse, and C. Baum. 2002. Murine leukemia induced by retroviral gene marking. Science 297:497. [DOI] [PubMed] [Google Scholar]
- 25.McPherson, C. E., E. Y. Shim, D. S. Friedman, and K. S. Zaret. 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387-398. [DOI] [PubMed] [Google Scholar]
- 26.Müller, H.-P., and H. E. Varmus. 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13:4704-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeifer, A., and I. M. Verma. 2001. Gene therapy: promises and problems. Annu. Rev. Genomics Hum. Genet. 2:177-211. [DOI] [PubMed] [Google Scholar]
- 28.Pruss, D., F. D. Bushman, and A. P. Wolffe. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91:5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pryciak, P. M., A. Sil, and H. E. Varmus. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11:291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769-780. [DOI] [PubMed] [Google Scholar]
- 31.Rivella, S., J. A. Callegari, C. May, C. W. Tan, and M. Sadelain. 2000. The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J. Virol. 74:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-585. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 33.Sandmeyer, S. B. 2003. Integration by design. Proc. Natl. Acad. Sci. USA 100:5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder, A. R., P. Shinn, H. T. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 35.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 272:8361-8369. [DOI] [PubMed] [Google Scholar]
- 36.Shim, E. Y., C. Woodcock, and K. S. Zaret. 1998. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 12:5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suck, D., and C. Oefner. 1986. Structure of DNase I at 2.0A resolution suggests a mechanism for binding to and cutting DNA. Nature 321:620-625. [DOI] [PubMed] [Google Scholar]
- 39.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. J. Biochem. 36:11381-11388. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 43.Zaret, K. S., and K. Stevens. 1995. Expression of a highly unstable and insoluble transcription factor in Escherichia coli: purification and characterization of the fork head homolog HNF3α. Protein Expr. Purif. 6:821-825. [DOI] [PubMed] [Google Scholar]