Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that upon activation by the toxicant 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) stimulates gene expression and toxicity. AHR is also important for normal mouse physiology and may play a role in cancer progression in the absence of environmental toxicants. The objective of this report was to identify AHR-dependent genes (ADGs) whose expression is regulated by AHR in the absence of toxicants. RNA-Seq analysis revealed that AHR regulated the expression of over 600 genes at an FDR < 10% in MCF-7 breast cancer cells upon knockdown with short interfering RNA. Pathway analysis revealed that a significant number of ADGs were components of TCDD and tumor necrosis factor (TNF) pathways. We also demonstrated that siRNA knockdown of AHR modulated TNF induction of MNSOD and cytotoxicity in MCF-7 cells. Collectively, the major new findings of this report are: 1) endogenous AHR promotes the expression of xenobiotic metabolizing enzymes even in the absence of toxicants and drugs, 2) AHR by modulating the basal expression of a large fraction of TNF target genes may prime them for TNF stimulation and 3) AHR is required for TNF induction of MNSOD and the cellular response to cytotoxicity in MCF-7 cells. This latter result provides a potentially new role for AHR in MCF-7 cancer progression as a mediator of TNF and antioxidant responses.
Keywords: Aryl Hydrocarbon Receptor (AHR), gene expression, breast cancer, xenobiotics, tumor necrosis factor
1. Introduction
The environmental toxicant TCDD acts through a ligand-activated transcription factor, the aryl hydrocarbon receptor (AHR), to regulate gene expression and induce toxicity [1]. In the absence of TCDD, AHR localizes to the cytoplasm and is physically associated with heat shock protein 90 (HSP90), AHR interacting protein (AIP) and protein p23 in a protein complex [1]. TCDD stimulates AHR to undergo a conformational change that stimulates its translocation to the nucleus and dissociation away from HSP90, AIP and p23 [1]. Upon entering the nucleus, AHR physically interacts with AHR nuclear translocator (ARNT) to activate canonical TCDD target genes containing dioxin response elements (DREs), including CYP1A1, CYP1B1, NRF2 and AHR Repressor (AHRR) [1]. Prior pathway analyses have shown that TCDD regulated gene sets that are associated with metabolism of xenobiotics by cytochrome P450’s, xenobiotic metabolism signaling, and fatty acid and lipid metabolism pathways; these findings are consistent with induction of phase I and phase II drug metabolizing enzymes [2,3].
Several studies have shown that AHR inhibits and stimulates gene expression in the absence of TCDD [4,5,6,7]. For instance, Boutros et al. reported that knockdown of AHR in liver and kidney of mice disrupted the expression of 417 and 379 genes, respectively [4]. Adenoviral-mediated knockdown of AHR in primary mouse hepatocytes in vitro induced significant changes in the expression of 97 genes at 12 hours and 246 genes at 24 hr [5]. Chang et al reported that AHR knockdown altered the expression of 1133 genes in mouse embryonic fibroblasts [6]. Mouse hepatoma cells (Hepa-1) express an AHR that binds DREs, while a variant line, Hepa-1 C35, harbors a dysfunctional mutant AHR that fails to bind DREs [7]. Consistent with AHR being an endogenous regulator of gene expression, the Hepa-C35 transcriptome is dramatically disrupted compared to parent Hepa-1 cells [8]. The findings that AHR knockout mice are less fertile, exhibit higher rates of intestinal cancers, and have developmental and vascular defects suggests that AHR regulation of gene expression in rodent models is physiologically important [9,10,11,12,13].
AHR has been reported to play roles in breast tumorigenesis. Knockdown of AHR in breast cancer cells (BCCs) inhibits mitogen-induced proliferation (MCF-7 cell line), invasion/migration (MDA-MB-231 cell line) and xenograft tumorigenicity (rodent mammary fibroblasts) [14,15,16,17]. Further, rat mammary tumors have been shown to express higher levels of AHR than normal mammary tissue [18]. The mechanism(s) of AHR action in breast tumorigenesis is not clear. We reasoned that defining AHR-dependent genes (ADGs) in MCF-7 BCCs would identify pathways downstream of AHR that are important in cancer. To this end, we performed expression profiling via RNA-Seq on control and AHR knockdown MCF-7 cells in the absence of external stimuli. Pathway analysis of ADGs revealed new roles for AHR. First, MCF-7 cells maintain expression of xenobiotic metabolizing enzymes in the absence of toxicants. Second, AHR promotes basal expression of a large fraction of TNF target genes in MCF-7 cells. Finally, knockdown of AHR inhibited TNF-induced increases in MNSOD and promoted the cytotoxic response in MCF-7 cells. This latter result provides a potential new role for AHR in cancer as a mediator of MNSOD induction and the antioxidant cytoprotective response to TNF.
2. Methods
2.1. Materials and MCF-7 cell culture
Dulbecco's Modified Eagle Medium/High glucose (DMEM) with L-glutamine and sodium pyruvate, phenol red-free DMEM, phosphate buffered saline (PBS), fetal bovine serum (FBS), charcoal-treated FBS, penicillin, and streptomycin were purchased from Thermo Fisher Scientific (Pittsburgh, PA). Sodium dodecyl sulfate (SDS), 30 % acrylamide/bis solution, ammonium persulfate, Tween-20, and 2-mercaptoethanol was obtained from Bio-RAD (Hercules, CA). Non-specific control RNA (cRNAi) (cat # D-001810-01-20), short interfering RNA (siRNA) against AHR (AHR-siRNA, cat # J-004990-08-0010), RELA (RELA-siRNA, cat # J-003533-06-0010) and DharmaFECT 1 Transfection Reagent (#1) were purchased from GE Healthcare Life Sciences (Pittsburgh, PA). 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) was obtained from Cambridge Isotopes Laboratory (Andover, MA). MCF-7 human breast cancer cells were purchased from ATCC (Manassas, VA) and maintained in DMEM, 10% FBS, with penicillin (100 IU/mL) and streptomycin 100 (µg/mL).
2.2. AHR knockdown for RNA-Seq
To knockdown AHR for RNA-Seq analysis, 200,000 MCF-7 cells in 6-well tissue culture plates were transfected with 50 nM AHR-siRNA in phenol red-free DMEM, 10% charcoal-treated FBS and DharmaFECT 1 Transfection Reagent following the manufacturer’s protocols. After 36h, cells were serum starved overnight in phenol-red free DMEM. Control cells in 6-well tissue culture plates were transfected with 50 nM control-siRNA using the same methods used to knockdown AHR.
2.3. Whole transcriptome expression profiling via RNA-Seq
Total RNA was isolated from overnight serum starved control (5 replicates) and AHR knockdown MCF-7 (6 replicates) using RNA purification columns (Qiagen, Valencia, CA) with DNase treatment. DNase was purchased from Qiagen. RNA sample quality was assessed using Bioanalyzer RNA Nano chips (Agilent); all RNA samples had an RNA Integrity Number greater than or equal to 8. RNA-Seq libraries were prepared from 1 µg of total RNA using a TruSeq RNA Prep Kit (Illumina Inc., San Diego, CA).
2.4. RNA-Seq Analysis
RNA-Seq on AHR knockdown and control MCF-7 cells was performed using an Illumina HiSeq1000 in a 2 × 100 base paired end design yielding a minimum of 50 million reads per sample. Demultiplexing of samples was performed using CASAVA 1.8.2 (Illumina). Reads were aligned to the human reference genome (hg19/GRCh37) using TopHat 2.0.6 [19]. TopHat was configured to use BowTie 0.12.8 [20] and SAMtools 0.1.18 [21]. Aligned reads were then mapped to genes from the ensembl database using Bioconductor [22] packages Rsamtools and biomaRt [23]. Data were then analyzed using the DESeq Bioconductor package [24] as follows. Counts were normalized to account for differences in sequencing depth between samples. Samples were clustered using the top 30 expressing genes. One control sample, which did not cluster with the remaining control samples, was removed from further analysis. In order to mitigate the loss of statistical power from multiple hypothesis correction, we removed the lowest 40% of genes by total read count across all samples and performed differential expression analysis on the remaining 60%. Following standard practice (for example, [24]), genes statistically significant at a false discovery rate of 10% were reported, irrespective of fold change. To validate the low-expression filtering step, we repeated the analysis without removing the 40% of genes that were low expressers (data not shown). None of the filtered genes were identified as statistically significant in this analysis, while the loss of statistical power resulted in 126 of the unfiltered genes losing significance. Sequencing data were deposited in the Gene Expression Omnibus (GEO) database maintained by the National Center for Biotechnology Information (NCBI) and are accessible with accession number GSE52036.
2.5. Ingenuity pathway analysis (IPA)
Differentially expressed genes (FDR < 10%) were expressed as a ratio of AHR knockdown/control level and loaded into Ingenuity Pathway Analysis software (IPA; Ingenuity Systems, Redwood City, CA) in order to perform an IPA Core Analysis under default settings. Of the 634 RNAs, 496 were mapped to known functions and pathways by IPA. In IPA, a biological function is a process or disease with a pre-defined set of molecules (genes). IPA was used to compute significant associations between biological functions and our ADG set. Specifically, we ran a Core Analysis in IPA which used Fisher’s Exact Test to assign levels of statistical significance to associations between biological functions and our gene set. We configured the core analysis to report Benjamini-Hochberg corrected p-values. We also used the Upstream Regulator Analysis function to identify candidate regulators of ADG pathways.
2.6. Validation of RNA-Seq by qRT-PCR
Real-time reverse-transcription PCR (qRT-PCR) analysis from control and AHR knockdown MCF-7 cells (5 replicates) was carried out to validate RNA-Seq (AHR knockdown detailed in 2.2.). Total RNA was isolated using Qiagen RNA purification columns and DNase treated. Reverse transcription was performed with 100 ng of total RNA using Verso cDNA kit (Thermo Fisher Scientific; cat # AB-1453/B). PCR of cDNA was conducted with SYBER GREEN and ROX qPCR mix (Qiagen) with a 5 min denaturing step at 95°C, followed by 40 cycles of 15 s at 95°C, 30s at 60°C, 30s at 72°C. Relative gene expression was calculated using the formula 2−ΔΔCT, as described by Livak and Schmittgen [25]. Glyceraldehyde-3-phosphate (GAPDH) mRNA levels served as the internal control. Primer sequences GAPDH [forward 5'-catgagaagtatgacaacagcct 3' and reverse 5'-agtccttccacgataccaaagt-3'], OAS1 [forward 5'-cagacgatgagaccgacgat-3' and reverse 5'-cctggagtgtgctgggtcta-3'], PKD1L1 [forward 5'-cgcctctggattgtgataacag-3' and reverse 5'-cggtcccagtagcacacag-3'], PLA2G2 [forward 5'-accagacgtaccgagaggag-3' and reverse 5'-cgctggggattggtgactg-3'], SERPIN5A [forward 5'-atgcccttttcaccgacctg-3' and reverse 5'-tgcagagtccctaaagttggtag-3'], PYDC1 [forward 5'-cacacgtatagctaccggcg-3' and reverse 5'-cgcgtaagacaacagcagtg-3'], HMGCS2 [forward 5'-caatgcctgctacggtggta-3' and reverse 5'-gacggcaatgtctccacaga-3'], SERPIN3A [forward 5'-tgccagcgcactcttcatc and reverse 5'-tgtcgttcaggttatagtccctc-3'], CYP1A1 [forward 5'-cttcaccctcatcagtaatggtc-3' and reverse 5'-aggctgggtcagaggcaat-3'], CYP1B1 [forward 5'-ctgcactcgagtctgcacat-3' and reverse 5'-tatcactgacatcttcggcg-3'], NRF2 [forward 5'-tccagtcagaaaccagtggat-3' and reverse 5'-gaatgtctgcgccaaaagctg-3'], PGR [forward 5'-ttatggtgtccttacctgtggg-3' and reverse 5'-gcggattttatcaacgatgcag-3'], MGP [forward 5'-tccgagaacgctctaagcct-3' and reverse 5'-gcaaagtctgtagtcatcacagg-3'], ADORA [forward 5'-ccacagacctacttccacacc-3' and reverse 5'-taccggagagggatcttgacc-3'], CREB3L [forward 5'-cctcccgaagcctcctattct-3' and reverse 5'-ggggttgatttcccagcca-3'], AHR [forward 5'-acatcacctacgccagtgg-3' and reverse 5'-ctctatgccgcttggaaggat-3'], ALOX5 [forward 5'-ctcaagcaacaccgacgtaaa-3' and reverse 5'-ccttgtggcatttggcatcg-3'], ALDH3A1 [forward 5'-tgttctccagcaacgacaagg-3' and reverse 5'-agggcagagagtgcaaggt-3'], RELA [forward 5'-tccagaccaacaacaacccc-3' and reverse 5'- gatcttgagctcggcagtgt] and ABCG2 [forward 5'-acgaacggattaacagggtca-3' and reverse 5'-ctccagacacaccacggat-3']. The Harvard Primer Bank http://pga.mgh.harvard.edu/primerbank/ was used to design primers above. The primer sequences for the UGTA isoforms have been published [26]. Primers were purchased from Sigma (St. Louis, MO). Primer specificity was verified with melt curve analysis and NIH primer blast search engines located at http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome. Two-tailed, paired t tests with confidence intervals of 95% were used to determine statistically significant differences between controls and AHR knockdown cells.
2.7. Western blot analysis determination of MNSOD
AHR knockdown prior to western blot analysis was carried as detailed in Tomblin and Salisbury [15]. Briefly, MCF-7 cells (200,000) were mixed directly with siRNA (50 nM control or AHR-siRNA) and DharmaFECT 1 Transfection reagent (2 µL-per well), added to phenol red-free DMEM, 10% charcoal treated FBS in 6-well tissue culture plates and cultured for 24h. Following serum starvation in phenol red-free DMEM for 16h, cells were treated with either H2O vehicle or human recombinant TNF (10 ng/mL) (R & D Systems) for 12h. Treatments were removed, adherent and detached cells were collected and total cellular extract was isolated in 250 µL of 2× sample lysis buffer (Bio-RAD; cat #161-0737) and approximately 10 µg of protein was subjected to SDS PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked in PBS, .01% Tween 20 (PBS-T), 5% (wt/vol) low fat powdered milk for 1 h and incubated overnight with primary antibody at 4 °C with gentle mixing. Membranes were rinsed five times (five minutes each wash) with PBS-T and then incubated with an appropriate HRP-labeled secondary antibody (Thermo Fisher Scientific) (diluted 1:10,000 in PBS-T, 5% milk) for 1 h, followed with rinsing five times (five minutes each wash) in PBS-T. Membranes were developed with enhanced chemiluminescent substrate (Millipore Corporation, Billerica, MA) and exposure to X-ray film (MidSci, St. Louis, MO). Antibodies were purchased from the following vendors: (1) Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody from Millipore (cat # MAB374), (2) AHR antibody from Santa Cruz (Santa Cruz, CA, Cat # H-211) and (3) MNSOD antibody from Abcam (Cambridge, MA, cat #: ab13533). GADPH was diluted 1:10,000, while AHR and MNSOD were diluted 1:2000 in PBS, .01% Tween-20, 5% powdered milk. Densitometry was calculated with ImageJ PC-based software (National Institute of Health). The Student-Newman–Keuls (SNK) post-hoc test was used to determine statistically significant differences among groups following one-way analysis of variance (ANOVA).
2.8. qRT-PCR analysis TNF induction of SOD2
MCF-7 cells were reverse transfected in 6-well tissue culture plates as detailed in section 2.6 and then treated with H2O vehicle or TNF (10 ng/mL) (R & D Systems) for 12hr. Treatments were removed, adherent and detached cells were collected and total RNA was isolated in TRI-Reagent (Sigma-Aldrich., St. Louis, MO) and quantitated by NanoDrop spectrophotometry. RNA was reverse transcribed to cDNA (Verso cDNA kit; Thermo Fisher Scientific; cat # AB-1453/B). Resulting cDNAs were subjected to qRT-PCR with SYBR Green Rox Mix (Qiagen) using PCR reaction conditions detailed in 2.6. Relative gene expression among groups was calculated using the formula 2−ΔΔCT, as described by Livak and Schmittgen [25]. Primer sequences for SOD2 mRNA were (forward, 5'-GGAAGCCATCAAACGTGACTT-3'; reverse, 5'-CCCGTTCCTTATTGAAACCAAGC-3'). The SNK post-hoc test was used to determine statistically significant differences among groups following one-way analysis of variance (ANOVA).
2.9. qRT-PCR analysis of TCDD treated cells
MCF-7 cells plated in 35 mm tissue culture plates (200, 000 cells/mL) were serum starved overnight in phenol red-free DMEM. For TCDD stimulation, either .1 % (v/v) Dimethyl sulfoxide (DMSO) (Sigma-Aldrich) or TCDD (10 nM at the final concentration) was added directly to media along with either H2O vehicle or TNF (10 ng/mL) (R & D Systems) for 12 hr. Treatments were stopped and cells were rinsed once with PBS. Total RNA was isolated using TRI reagent (Sigma-Aldrich) and SOD2 mRNA was measured using real time RT-qPCR analysis.
2.10. Chromatin immunoprecipitation followed by qPCR (ChiP-qPCR)
For ChIP, one 80% confluent 150mm plate of MCF-7 cells was serum starved in phenol red-free DMEM and then treated with H2O vehicle or TNF (10 ng/mL) for 1h or 12h. Post treatment, cells were cross-linked with formaldehyde (Sigma-Aldrich) (.75% v/v) for 10min at room temp, followed by the application of glycine (.125M) (Sigma-Aldrich) for 5min. Cells were rinsed with cold PBS, pelleted by centrifugation, and cell pellets were lysed in 1 mL Lysis Buffer (50 mM Tris-HCl pH 7.5, 140mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Sodium Deoxycholate, 0.1% SDS plus protease inhibitors (Thermo Scientific #78410). After 15 min, extracts were sonicated (5 times, each time 10 s) and diluted 1:10 in dilution buffer (1% Triton X-100, 2mM EDTA pH8, 20mM Tris-HCl pH 8, 150 mM NaCl plus protease inhibitors), rotated overnight at 4 °C with 5 µg of non-specific IgG (Santa Cruz; cat# sc-2027), 5 µg of anti-AHR antibody (Santa Cruz; cat # H-211) or 5 µg of anti-p65 NFKB antibody (Santa Cruz; cat # sc-372). Antibody-chromatin complexes were collected using 10 µL of magnetic protein A beads (Invitrogen; cat # 100.01D) with rotation at 4 °C for 90 min. Using magnetic separation (Life-Technologies; part # 49-2025), beads were washed three times (10 min each wash) with wash buffer (20 mM Tris-HCl, pH 8; 150 mM NaCl; 2.0 mM EDTA; 0.1% SDS) and once with final wash buffer (.1% SDS, 1% Triton X-100, 2mM EDTA pH 8, 500 mM NaCl) and incubated at 65 °C for 4–6 h in elution buffer (1% SDS, 0.1 M NaHCO3) with proteinase K (20 mg/mL) (Invitrogen Life-Technologies., Carlsbad, CA). DNA was purified with phenol-chloroform extraction followed by isopropanol precipitation and analyzed using real time PCR. Phenol, chloroform and isopropanol were purchased from Sigma-Aldrich. Primers spanning NFKB response elements in intron 2 of SOD2 were: [forward 5'-GGAAAAGGCCCCGTGATTT-3' and reverse 5-TCCTGGTGTCAGATGTTGCC-3'] [27]. ChiP data was expressed as % input, in which signals obtained from the ChIP are divided by signals obtained from an input sample. Statistical differences among groups were determined by the SNK post-hoc test following oneway analysis of variance (ANOVA)
2.11. Cell viability
MCF-7 cells (200,000/mL) were mixed directly with 50 nM siRNA (either control or AHR-siRNA) and Dharmafect #1 transfection reagent (2 µL/mL), added to phenol red-free DMEM, 10% charcoal treated FBS and plated into 60 mm tissue culture plates (3 mL per plate) and cultured for 24h. Following serum starvation in phenol red-free DMEM for 16h, cells were treated with either H2O vehicle or human recombinant TNF (10 ng/mL) (R & D Systems) for 12h. Cell viability was measured with trypan blue stain (Thermo Fisher Scientific). The percentage of non-viable cells were calculated as: non-viable cell (%) = (total number of non-viable cells/total number of cells) multiplied by 100. The SNK post-hoc test was used to determine statistically significant differences among groups following one-way analysis of variance (ANOVA)
3. Results
3.1 Effect of AHR knockdown on MCF-7 gene expression
Expression profiling on control and AHR knockdown MCF-7 cells was conducted to identify a set of ADGs in the absence of stimuli. AHR knockdown inhibited the expression of 380 genes and promoted the expression of 254 genes at FDR < 10%, with all reported fold changes being at least 1.2 fold; we refer to the combined group of 634 genes as the ADG set. A full list of these genes is included as a supplemental file with NCBI GEO data deposit (accession number GSE52036). Real-Time qRT-PCR was used to validate RNA-Seq expression in a set of 30 genes. The rational for selecting validation genes listed in Table 1 is that they were among either the top downregulated (CYP1A1, HMGCS2, OAS1, PLA2G2, ALDH3A1, PKD1L1), the top upregulated (CREB3L1, PYDC1, MGP, ADORA1, PGR, SERPIN3A, and SERPIN5A) ADGs or known TCDD gene targets (CYP1A1, CYP1B1, ABCG2, ALDH3A1, NRF2 and UDP-glucuronosyltransferases (UGTAs). ALOX5 was selected for validation because it is the rate limiting gene in leukotriene synthesis [28].
Table 1.
Fold change expression value from AHR knockdown MCF-7 compared to controls.
| GENE ID | AHR-RNA-Seq | qRT-PCR |
|---|---|---|
| AHR | 0.24585 | 0.14400 ± .040 |
| CYP1A1 | 0.04738 | 0.43253 ± .215 |
| CYP1B1 | 0.44897 | 0.36685 ± .137 |
| HMGCS2 | 0.12976 | 0.53902 ± .114 |
| OAS1 | 0.13104 | 0.62920 ± .054 |
| PLA2G2 | 0.11346 | 0.42180 ± .073 |
| ABCG2 | 0.35987 | 0.51580 ± .146 |
| NRF2 | no difference | 0.55050 ± .003 |
| ALOX5 | 0.49086 | 0.47342 ± .024 |
| ALDH3A1 | 0.05942 | 0.17310 ± .067 |
| UGT1A1 | 0.31011 | 0.50581 ± .069 |
| UGT1A3 | 0.30741 | 0.43706 ± .037 |
| UGT1A4 | 0.32062 | 0.60735 ± .066 |
| UGT1A5 | 0.32055 | 0.60620 ± .079 |
| UGT1A6 | 0.32404 | 0.61148 ± .085 |
| UGT1A7 | 0.33345 | 0.53582 ± .058 |
| UGT1A8 | 0.33597 | no difference |
| UGT1A9 | 0.33357 | no difference |
| UGT1A10 | 0.33502 | no difference |
| PKD1L1 | 0.16430 | 0.29732 ± .031 |
| PYDC1 | 2.50200 | no difference |
| PGR | 1.87529 | 1.40625 ± .103 |
| MGP | 2.39768 | 1.2750 ± .034 |
| SERPIN3A | 1.92517 | 1.52785 ± .031 |
| CREB3L | 2.67290 | 1.40500 ± .018 |
| SERPIN5A | 1.88800 | 1.7890 ± .233 |
| ADORA | 2.03342 | 1.61912 ± .209 |
Column 2 is expressed as RNA-Seq ratio of AHR knockdown/control level (FDR < 10%). Column 3 is expressed as real-time qRT-PCR ratio of AHR knockdown/control normalized to GAPDH expression (P < .05).
In general, there is a good concordance between the RNA-Seq and qRT-PCR measurements. Levels of AHR mRNA were lower in knockdown MCF-7 cells than controls as measured by RNA-Seq (~4-fold) and qRT-PCR (~7-fold) from independent experiments (Table 1). Expression of known TCDD-target genes (CYP1A1 [29], CYP1B1 [29] and ALDH3A1 [30]) was lower in AHR knockdown MCF-7 cells by both RNA-Seq and qRT-PCR measurements (Table 1). Prior reports have shown that TCDD stimulates increased expression of UGTAs in mouse liver [31]. RNA-Seq and qRT-PCR assays revealed that UGT1A1, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7 mRNAs were lower in AHR knockdown cells than controls (Table 1). UGT1A8, UGT1A9, and UGT1A10 were not differently regulated by qRT-PCR, but their levels were lower (~3-fold) in AHR knockdown cells compared with controls based on RNA-Seq measurements (Table 1). The drug transporter, ABCG2, has been reported to be induced by TCDD in human cells (breast, colon and liver), but not in rodent cells [32]. ABCG2 mRNA was ~3 fold lower in AHR knockdown MCF-7 cells than controls in both RNA-Seq and qRT-PCR data sets (Table 1). NRF2 is a transcription factor that stimulates the expression of anti-oxidant enzymes [31]. Prior reports have shown that NRF2 is a TCDD gene target [33,34]. NRF2 expression was not differentially expressed by RNA-Seq, but its levels were lower (~50%) in AHR knockdown cells compared with controls when assayed by qRT-PCR (Table 1). The levels of the PLA2G2 and ALOX5 were lower in AHR knockdown MCF-7 cells than controls by RNA-Seq and qRT-PCR (Table 1).
AHR knockdown had modest stimulatory effects on the expression of several genes. As measured by RNA-Seq, CREB3L was the most upregulated gene (by 2.67290) in AHR knockdown MCF-7 cells compared with controls (Table 1). The expression of PGR, MGP, SERPIN3A, CREB3L, SERPIN5A, and ADORA were increased in AHR knockdown cells compared with controls by RNA-Seq and qRT-PCR (Table 1). Observed expression levels of IGHG2, IGHA1 and RNF128 were reduced by AHR knockdown by RNA-Seq analysis (GEO submission GSE52036), but not by qRT-PCR (data not shown). This discrepancy could be attributed to IGHG2, IGHA1 and RNF128 transcript levels that were below qRT-PCR detection limits (Ct values higher than 35; data not shown). We note that RNA-Seq fold changes were greater than qRT-PCR fold changes for several genes including: CYP1A1, HMGCS2, OAS1, PLA2G2, ALDH3A1, MGP, CREB3L, UGTAs and ADORA; however the direction of expression changes were the same (Table 1).
3.2 Pathway analysis of AHR-dependent genes
In order to determine functions and pathways regulated by ADGs, we analyzed the ADG set using the Ingenuity Pathway Analysis (IPA) core analysis tool which finds gene sets that are over-represented in defined, canonical cellular pathways and molecular functions. Of the 634 genes, 496 were mapped to known functions and pathways by IPA. These ADGs were significantly associated with cancer-related pathways including: cellular movement, cell cycle, cellular growth and proliferation, cell death and survival, cellular development and cellular morphology (Table 2). In addition, significant numbers of ADGs were over-represented in pathways involved in post-translational modification and in the metabolism of drugs, amino acids and small molecules (Table 2).
Table 2.
IPA cellular and molecular functions associated with RNA-Seq ADGs.
| Category | *B-H p-value | Target molecules in dataset |
|---|---|---|
| Cellular Movement | 1.37E-06-4.32E-02 | 101 |
| Cell Cycle | 1.96E-06-5.67E-02 | 90 |
| Cellular Growth and Proliferation | 2.38E-06-5.67E-02 | 149 |
| Cell Death and Survival | 4.46E-06-5.67E-02 | 150 |
| Amino Acid Metabolism | 2.24E-05-4.32E-02 | 18 |
| Drug Metabolism | 2.24E-05-4.32E-02 | 12 |
| Post-Translational Modification | 2.24E-05-3.04E-02 | 19 |
| Small Molecule Biochemistry | 2.24E-05-5.67E-02 | 73 |
| Cell Morphology | 2.53E-04-5.13E-02 | 85 |
| Cellular Development | 3.74E-04-5.67E-02 | 138 |
p-values are calculated by Fishers exact test and corrected for multiple testing by the Benjamini-Hochberger p-values (B–H) method (B–H p-value). Column 2 shows the range of B-H corrected p-values for the biofunctions in a given category. Target molecules in dataset are the number of RNA-Seq ADGs in a given biofunction.
We refined the pathway analysis by applying the IPA Upstream Regulator Analysis tool to determine if the ADGs are connected through a common upstream regulator. This analysis revealed that ADGs were enriched among the following IPA canonical regulatory pathways: beta-estradiol (endogenous hormone), tumor necrosis factor (TNF) (cytokine), tumor protein 53 (TP53) (transcriptional regulator), lipopolysaccharide (chemical drug), decitabine (chemical drug), calcitriol (chemical ligand), dexamethasone (glucocorticoid receptor), v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2) (kinase), cyclin-dependent kinase inhibitor 1A (CDKN1A) (kinase), TGFβ (growth factor) and TCDD (toxicant) (Table 3). Specifically, IPA reveals that 74 of 171 TNF pathway target genes are ADGs (Table 3). Of the 74 ADGs in the TNF pathway, 44 exhibited patterns of expression consistent with inhibition of TNF activity (Table 3). The finding that IPA revealed 87 of 197 beta-estradiol target genes are ADGs is not surprising, considering that AHR and the estrogen receptor (ER) have been reported to interact extensively [35,36] (Table 3). Finally, ADGs were found to be significantly enriched within the TCDD pathway (23 of 125 TCDD pathway genes were ADGs) (Table 3). The IPA-predicted inhibition of TCDD activity (Table 3) was based in part on the observed inhibition of conical TCDD target genes including: CYP1A1, CYP1B1 and ALDH3A1 in AHR knockdown cells compared with controls.
Table 3.
IPA upstream regulators associated with RNA-Seq ADGs
| IPA- upstream regulator |
IPA-upstream regulator activity prediction |
*p-value of overlap |
Target molecules in dataset |
Target molecules in IPA-upstream regulator pathway |
|---|---|---|---|---|
| beta-estradiol | 5.64E-19 | 87 | 197 | |
| TNF | Inhibited | 1.79E-13 | 74 | 171 |
| TP53 | Inhibited | 4.15E-13 | 66 | 137 |
| lipopolysaccharide | 6.29E-12 | 73 | 187 | |
| decitabine | 1.46E-11 | 34 | 160 | |
| calcitriol | Inhibited | 3.16E-11 | 33 | 159 |
| dexamethasone | 4.26E-11 | 70 | 170 | |
| ERBB2 | 2.66E-10 | 38 | 170 | |
| CDKN1A | 3.07E-10 | 22 | 123 | |
| TGFβ | 4.11E-09 | 66 | 198 | |
| TCDD | Inhibited | 5.13E-07 | 23 | 125 |
An IPA-upstream regulator regulates a IPA-defined set of target molecules (genes). Column 2 reveals IPA upstream regulator activity determined by comparing reported gene responses to a given upstream regulator to observed expression changes in AHR knockdown cells compared to controls. Column 4 shows the number of target molecules in the RNA-seq AHR knockdown dataset that are in a given IPA-upstream regulator pathway. Column 5 indicates that total number of target molecules that are in a given IPA-upstream regulator pathway.
p-value of overlap are calculated by Fisher exact test.
3.3 Comparison of AHR-dependent gene set with known TCDD and AHR effects
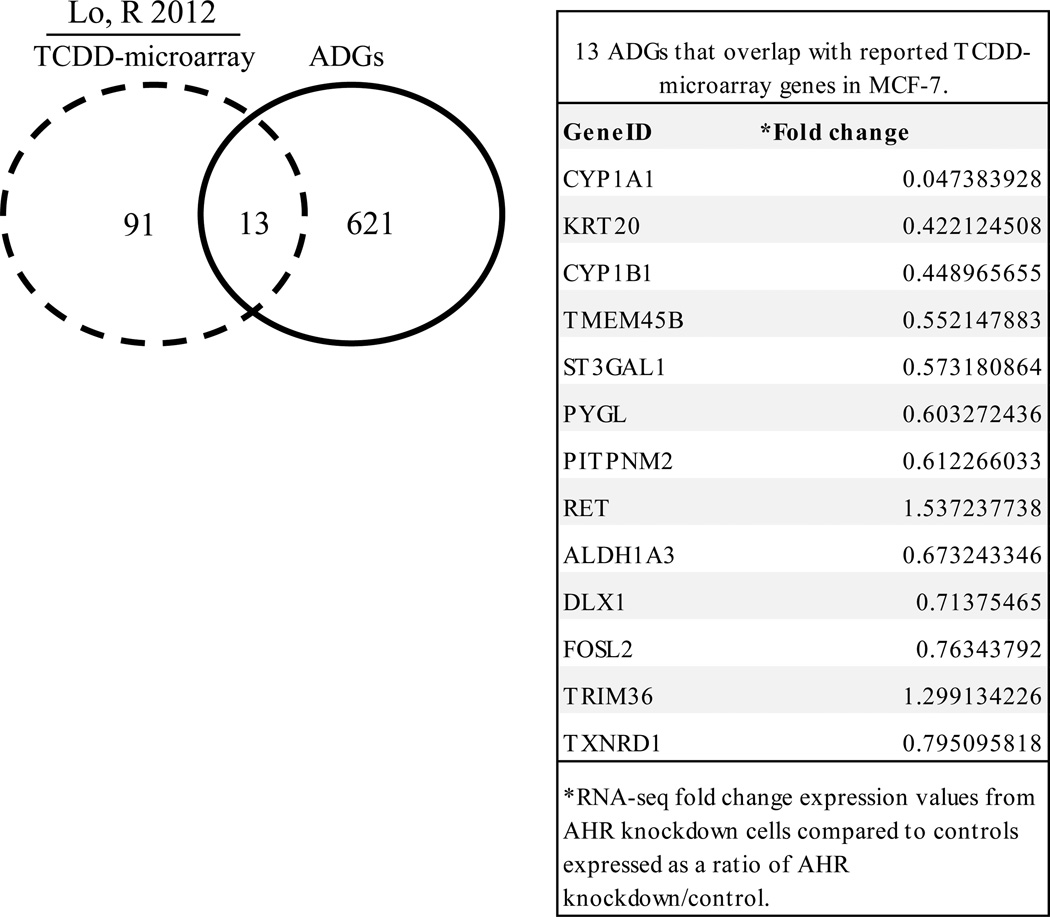
TCDD is a strong exogenous AHR ligand that is resistant to degradation [1]. TCDD has been reported to regulate the expression of 104 genes in MCF-7 cells [3]. To identify ADG genes that are induced by TCDD in MCF-7 cells, we overlapped published TCDD microarray data [3] and AHR knockdown RNA-Seq expression profiles. While the majority of ADGs (621) did not overlap with reported TCDD-regulated genes, there were 13 genes in both sets (Fig. 1). Common genes included CYP1A1, CYP1B1 and ALDH3A1, which are important in lipid metabolism, small molecule biochemistry, and drug metabolism (Fig. 1).
Fig. 1. Genes in common between AHR knockdown RNA-Seq expression profiles and TCDD-microarray data.
Analysis of reported TCDD-microarray data [3] demonstrated that 13 of 634 ADGs were TCDD-regulated genes. The specific 13 ADGs that overlapped with TCDD-microarray genes in MCF-7 cells are shown in the table.
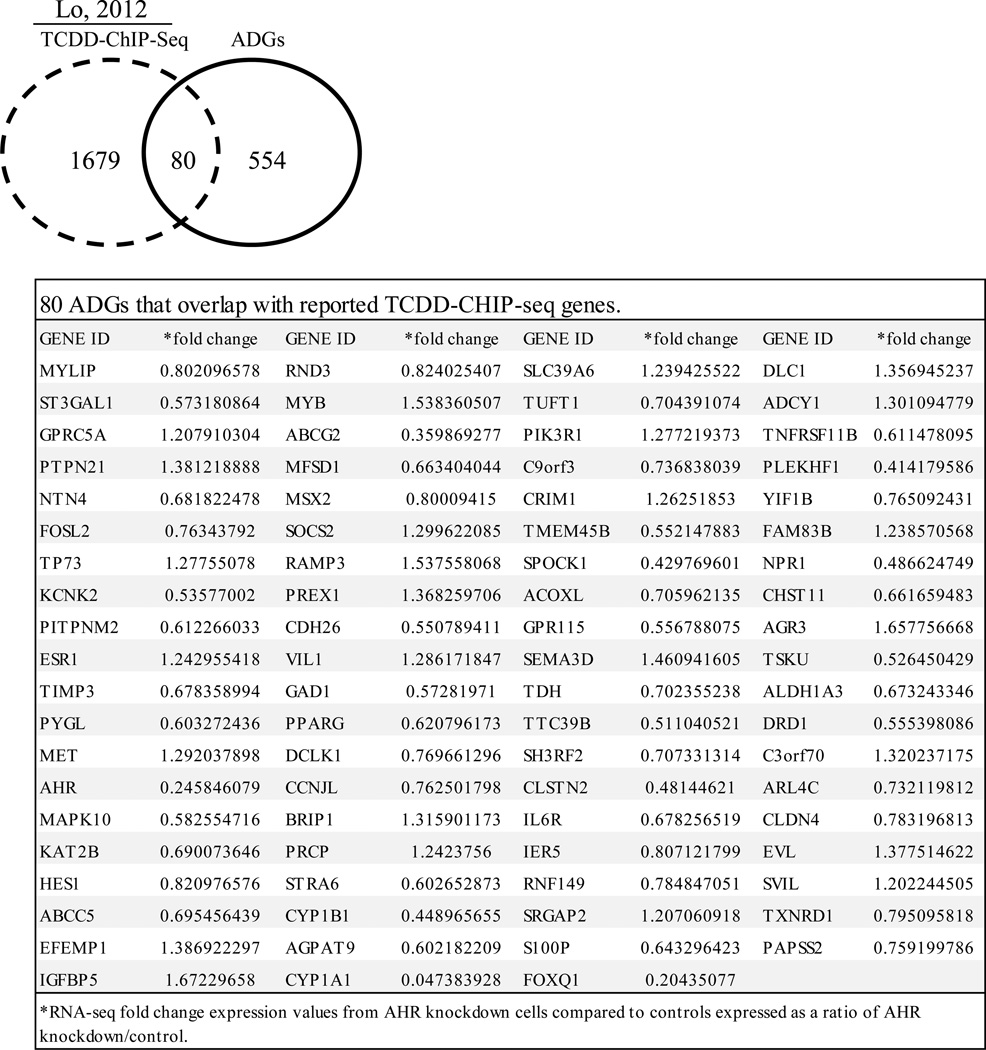
Lo and Matthews identified TCDD-induced binding sites in MCF-7 cells using ChIP-Seq technology [3]. Since these should represent AHR binding sites, we compared the TCDD-ChIP-Seq gene set with ADG set and found that approximately 15% of ADGs have a TCDD-AHR binding site. This finding suggests that the remaining 85% could be indirect AHR gene targets. The 80 specific TCDD-ChIP-Seq genes that overlap with the ADG set are shown in Fig. 2. Common target genes included ABCG2, CYP1A1 and CYP1B1 which are known TCDD-AHR target genes [3,32].
Fig. 2. Genes in common between AHR knockdown RNA-Seq expression profiles and TCDD-ChIP-Seq data.
Analysis of reported MCF-7 TCDD-ChIP-Seq data [3] revealed that 80 of the 634 AHR-RNA-Seq genes were TCDD-AHR bound genes. The specific 80 ADGs that overlapped with TCDD-ChIP-Seq genes in MCF-7 cells are shown in the table.
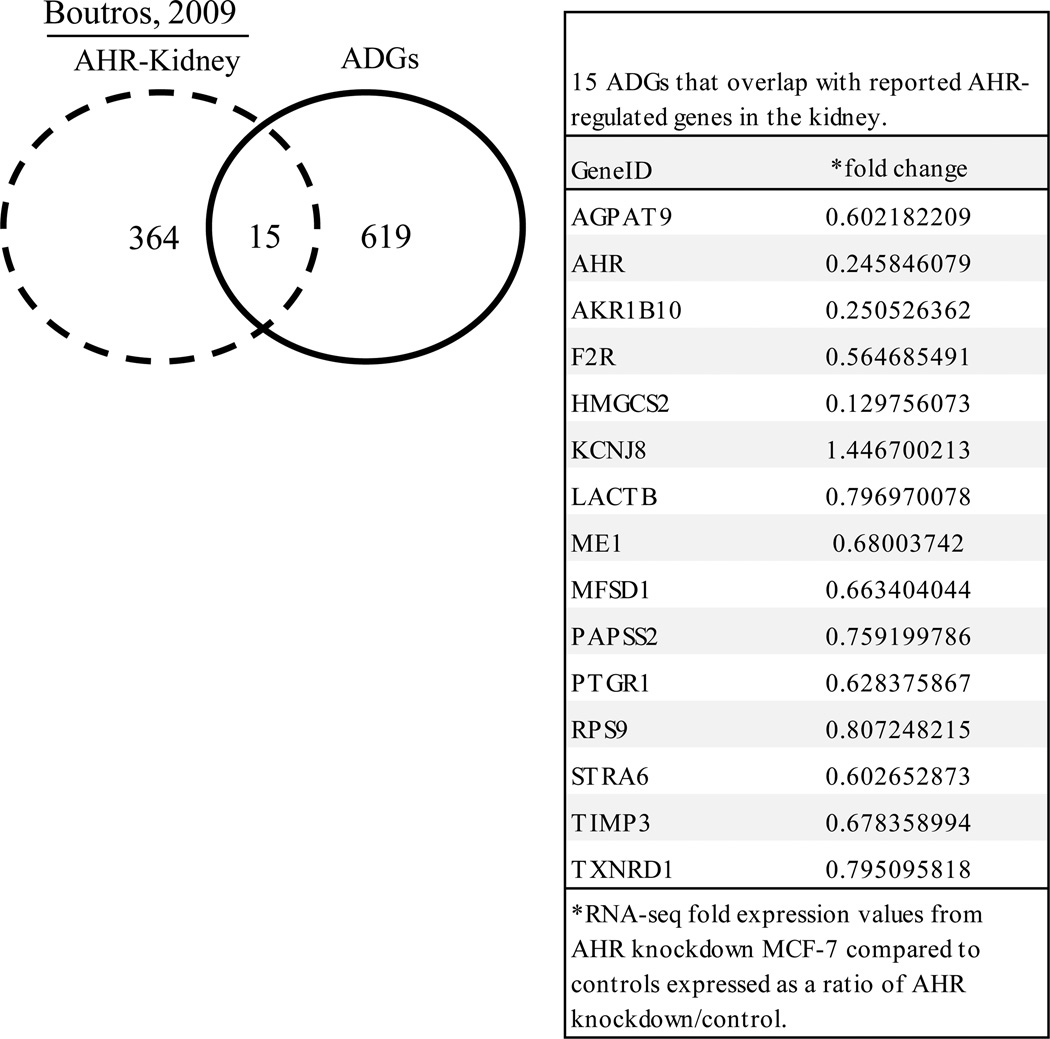
Microarray based expression profiles on liver and kidney from AHR null mice has been reported [4]. Twenty eight genes were shared between the mouse liver gene set and ADG set (Fig. 3). A small number of mouse kidney genes (15) overlapped with ADG set (Fig. 4). The specific ADGs that overlapped with AHR-liver and AHR-kidney are shown in Fig. 3 and Fig. 4, respectively. Differences in tissue- and species-specific expression may explain the limited overlap in these gene sets.
Fig. 3. Genes in common between AHR knockdown RNA-Seq expression profiles and AHR-regulated genes in mouse liver.
Analysis of reported AHR gene targets in mouse liver [4] revealed that 28 of the 634 ADGs were AHR targets in mouse liver. The specific 28 ADGs that overlapped with AHR-regulated genes in liver are shown in the table.
Fig. 4. Genes in common between AHR knockdown RNA-Seq expression profiles and AHR-regulated genes in mouse kidney.
Analysis of reported AHR gene targets in mouse kidney [4] revealed that 15 of the 634 ADGs were AHR targets in mouse kidney. The specific 15 ADGs that overlapped with AHR-regulated genes in the kidney are shown in the table.
3.4 AHR modulates TNF induction of MNSOD and cytotoxicity response
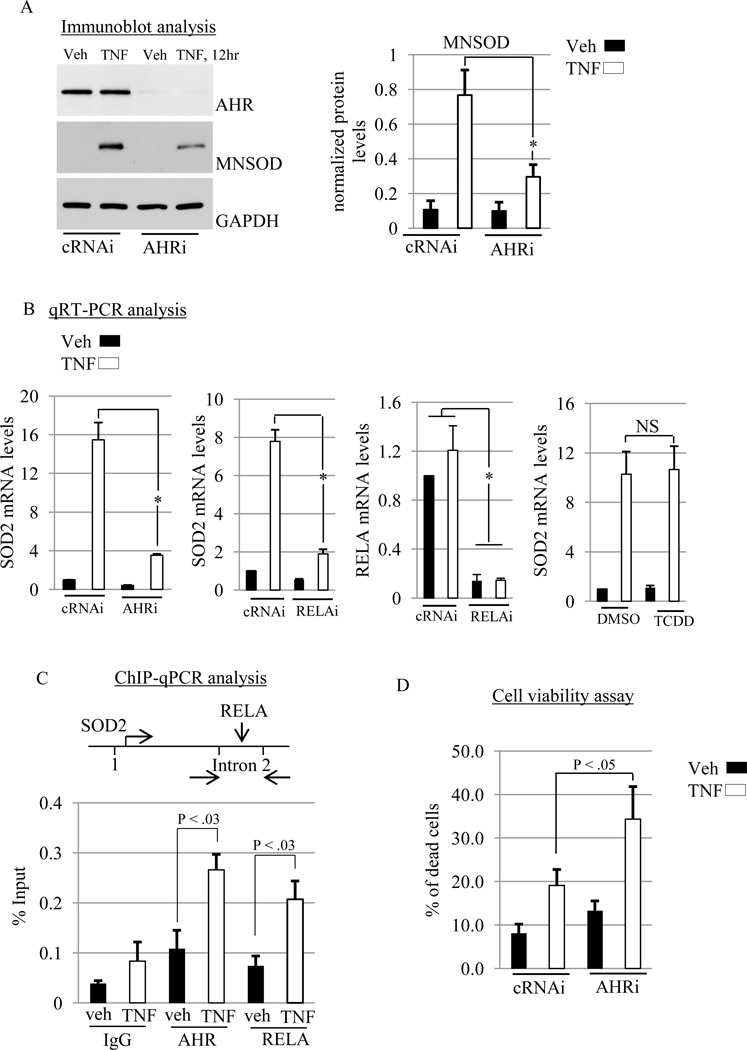
Based on the finding that the ADG set is significantly associated with the TNF pathway, we sought to determine if TNF induction of SOD2 requires AHR expression. SOD2 is a nuclear gene that encodes the mitochondrial superoxide dismutase (MNSOD). We focused on MNSOD regulation because it is inducible [37] and Rico de Souza et al [38] have reported that MNSOD levels are lower in AHR knockdown primary mouse lung fibroblasts than control cells [38]. Serum-starved control and AHR knockdown MCF-7 cells were treated with vehicle or TNF (10 ng/mL) for 12h. As expected, AHR protein levels were lower in knockdown cells than control cells (Fig. 5A). While TNF stimulated MNSOD protein levels ~8-fold in control cells, this induction was significantly abrogated in AHR knockdown cells by 60% (Fig. 5A). We then asked whether siRNA knockdown of AHR and NF-κB subunit RELA (also known as p65) inhibited TNF-stimulated induction of MNSOD mRNA expression. The impetus for including RELA is based on its requirement for TNF induction of the SOD2 gene [39,40]. RELA mRNA was reduced ~90% by siRNA treatment (Fig. 5B). Knockdown of AHR and RELA suppressed TNF induction of MNSOD mRNA levels (Fig 5B). We also asked whether TCDD would modulate TNF regulation of MNSOD. The level of MNSOD induction by TNF was not affected by TCDD (Fig. 5B). Collectively, these data indicate that endogenous AHR and RELA promote TNF induction of MNSOD in MCF-7 cells through a mechanism that is independent of TCDD effects.
Fig. 5. AHR promotes TNF induction of MNSOD.
(A) MCF-7 cells were transfected with control (cRNAi) or AHR (AHRi) siRNAs (please see methods for details regarding transfection) prior to treatment with vehicle (Veh) or TNF (10 ng/mL) for 12hr. Total cellular protein was then isolated and subjected to western blot analysis. The blot was then probed with the indicated antibodies. Relative level of MNSOD protein was expressed as a ratio of MNSOD/GAPDH. A significant decrease in MNSOD protein by AHRi is indicated by (*, P < .001). (B) RT-qPCR analyses of SOD2 and RELA mRNA levels in MCF-7 cells transfected with control (cRNAi), AHR (AHRi) or RELA (RELAi) siRNAs prior to vehicle (Veh) or TNF (10 ng/mL) treatment for 12 hr. Significant decreases in SOD2 or RELA mRNAs is indicated by (*, P < .001). For TCDD stimulation, MCF-7 cells were stimulated with either dimethyl sulfoxide vehicle (DMSO-veh) or TCDD (10 nM) in the presence of vehicle or TNF (10 ng/mL) for 12 hr, followed by measurement of SOD2 mRNA with RT-qPCR. No significant difference is indicated by (NS). Gene expression was normalized against GAPDH. (C) MCF-7 cells were treated with vehicle (Veh) or TNF (10 ng/mL) for 12 hr and chromatin immunoprecipitation (ChIP) experiments were conducted, followed by real-time Q-PCR (ChIP-qPCR). A significant increase in AHR and RELA binding on an intronic NFKB response element in the SOD2 gene induced by TNF is indicated (*, P < 0.03). (D) MCF-7 cells were transfected with cRNAi or AHRi prior to treatment with vehicle (Veh) or TNF (10 ng/mL) for 12 hr and cell viability was determined as outlined in the Materials and Methods. Significant increases is indicated (*, P < 0.05) (A–D) Data shown are the means ± S.E. of at least three independent experiments.
TNF-induced RELA stimulates SOD2 expression by binding to NF-κB response elements (κB-RE) in intron 2 [39,40]. Physical interactions between AHR and RELA have been reported [41]. We therefore tested whether TNF signaling results in recruitment of AHR and RELA to the SOD2 κB-RE. ChIP-qPCR experiments revealed that treatment with TNF (12 hr) increased the binding of AHR and RELA on the SOD2 κB-RE by ~2.5 fold in each case (Fig. 5C). AHR and RELA association with κB-RE in vehicle treated cells was not greater than non-specific IgG (Fig. 5C). These results indicate that TNF signaling recruits AHR and RELA to an active κB-RE in the SOD2 gene [38,39].
The finding that AHR modulates TNF induction of MNSOD prompted us to investigate whether AHR is required in the response to TNF-induced cytotoxicity. To this end, MCF-7 cells were transiently transfected with non-targeting control or AHR siRNAs prior to treatment with vehicle or TNF for 12 hr, followed by determination of the percentage of non-viable cells. As shown in figure 5D, TNF-induced cytotoxicity was significantly higher in AHR knockdown MCF-7 cells compared with controls (Fig. 5D). This result suggests that AHR suppresses TNF-induced cytotoxicity.
4. Discussion
In this report, RNA-Seq analysis revealed that the expression of over 600 genes in MCF-7 cells is dependent on AHR based on our knockdown experiments. Pathway analysis revealed that a significant number of ADGs were present in toxicant and TNF pathways (Table 3). TNF induction of MNSOD required AHR and RELA expression, and this process involved recruitment of RELA and AHR to a TNF-responsive NF-κB element in the SOD2 gene (Fig. 5). Consistent with AHR/RELA recruitment to MNSOD, the cellular response to TNF was dependent on AHR expression as demonstrated in knockdown experiments (Fig. 5).
There is little current evidence that demonstrates that cancer progression requires the expression of AHR; however, it is clear that AHR responds to and modulates cancer signals. From our prior report, we know that insulin like growth factor 2 (IGF-2) signaling rapidly increases AHR mRNA and protein levels in MCF-7 cells and that upregulated AHR promoted the activation of the CCND1 gene upon binding to the CCND1 gene promoter [15]. In this report we demonstrate that AHR modulates MCF-7 responsiveness to TNF. Together these findings indicate that AHR can modulate MCF-7 cancer progression by interacting with two major cancer signaling pathways, specifically IGF-2 and TNF.
Even though AHR expression has not been directly associated with cancer, AHR activity may be aberrant in cancer cells. AHRR is a putative tumor suppressor whose expression is downregulated in multiple cancers including breast tumors due to hypermethylation of its promoter [42]. AHRR inhibits AHR activity through a mechanism that could be mediated by AHRR binding with AHR [43]. Thus, AHR activity could be higher because AHRR expression is downregulated in human cancers [42].
There are several lines of evidence that AHR through interactions with RELA regulates proinflammatory genes; our data suggests this interaction is also important for regulating MNSOD, a major antioxidant enzyme. DiNatale and colleagues demonstrated that TCDD and interleukin 1 (IL-1) synergistically induce IL-6 transcription [44]. This was mediated through DREs in the IL-6 gene promoter [44]. Recently, AHR itself, in the absence of TCDD, has been reported to activate the IL-6 gene by pairing with RELA at κB-RE in the IL-6 gene [45]. AHR interaction with NF-κB is not restricted to RELA, considering that AHR binding with RELB activates the IL-8 gene [46,47]. TCDD inhibits NF-κB activity when measured with EMSA and a κB-RE-luciferase reporter construct [41]. We found that TNF induction of MNSOD is refractory to TCDD (Fig. 5). So in some cases TCDD interactions with NF-κB therefore could be gene specific.
AHR-deficient MCF-7 cells were more sensitive to TNF-induced cytotoxicity than controls (Fig. 5D). TNF signaling stimulates opposing cell survival and death pathways [48]. TNF-induced NF-κB protects cells from TNF-induced cell death by upregulating the expression of antioxidant and antiapoptotic genes [48]. Upregulation of MNSOD by NF-κB inhibits TNF-induced ROS accumulation and cell death [49,50]. The levels of MNSOD were lower in AHR knockdown MCF-7 cells in response to TNF compared with controls (Fig. 5). Thus, AHR could in part protect MCF-7 cells from TNF-induced cytotoxicity by promoting upregulation of MNSOD (Fig. 5).
Our RNA-Seq data and IPA analyses are consistent with many reports showing that AHR regulates gene expression in the absence of TCDD. There are potential mechanisms to explain AHR activity in MCF-7 cells in the absence of TCDD. Chiaro et al. 2008 discovered that the 5-lipoxygenase (5-LOX) pathway generates 5,6-dihydroxyeicosatetraenoic acid isomers (5,6- DiHETEs) that induce expression of a DRE-promoter reporter construct, the formation of AHR-DNA binding complexes in EMSA assays, and increases in CYP1A1 mRNA in hepatocytes [51]. DiNatale and colleagues reported that the tryptophan metabolite kynurenic acid induced CYP1A1 mRNA, DRE-promoter reporter activity and the formation of an AHR-DNA complex, and competitively displaced labeled AHR ligand from AHR in hepatocytes [52]. Kynurenine has been reported to be secreted at µM levels from glioma cells and to induce DRE-promoter reporter activity, CYP1A1 mRNA levels and to competitively displace labeled AHR ligand from AHR in glioma cells [53]. 5,6-DiHETEs, kynurenic acid and kynurenine therefore may serve as endogenous AHR ligands that stimulate AHR activity and expression of AHR target genes in MCF-7 cells in the absence of TCDD. Considering our data showing that TNF-induced AHR binding at an active NFKB-RE, we postulate that AHR may be recruited to gene promoters by activated RELA (perhaps in an AHR ligand independent mechanism).
In conclusion, our RNA-Seq data suggest a role for AHR in toxicant and TNF pathways. Further, AHR and RELA are clearly required for induction of MNSOD and the cytoprotective response to TNF. In a similar vein, AHR protects lung cells from cigarette induced cytotoxicity by maintaining MNSOD expression [38]. As a whole, our findings implicate unliganded AHR expression in a new aspect of cancer progression.
Acknowledgements
This work was supported in part by NIH grants P20RR016477 and P20GM103434 to the WV-INBRE program and Research Starter Grant from the PhRMA Foundation, Washington DC to Travis Salisbury. RNA-Seq and related data analyses were performed by the Marshall University School of Medicine Genomics Core Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare that they have no competing interests
Contributor Information
Travis B Salisbury, Email: salisburyt@marshall.edu.
Justin K. Tomblin, Email: tomblin36@marshall.edu.
Donald A. Primerano, Email: primeran@marshall.edu.
Jun Fan, Email: fanj@marshall.edu.
Jackie Fletcher, Email: fletcherjackie@gmail.com.
Nalini Santanam, Email: santanam@marshall.edu.
Estil Hurn, Email: hurn@marshall.edu.
Gary Z. Morris, Email: Gary.Morris@glenville.edu.
James Denvir, Email: denvir@marshall.edu.
Literature Cited
- 1.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo R, Matthews J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol Sci. 2012;130:349–361. doi: 10.1093/toxsci/kfs253. [DOI] [PubMed] [Google Scholar]
- 4.Boutros PC, Bielefeld KA, Pohjanvirta R, Harper PA. Dioxin-dependent and dioxin-independent gene batteries: comparison of liver and kidney in AHR-null mice. Toxicol Sci. 2009;112:245–256. doi: 10.1093/toxsci/kfp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper TA, Jr, Joshi AD, Elferink CJ. Identification of stanniocalcin 2 as a novel aryl hydrocarbon receptor target gene. J Pharmacol Exp Ther. 2013;344:579–588. doi: 10.1124/jpet.112.201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang X, Fan Y, Karyala S, Schwemberger S, Tomlinson CR, Sartor MA, Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, Zhang J, Hankinson O. A mutation in the aryl hydrocarbon receptor (AHR) in a cultured mammalian cell line identifies a novel region of AHR that affects DNA binding. J Biol Chem. 1997;272:31845–31854. doi: 10.1074/jbc.272.50.31845. [DOI] [PubMed] [Google Scholar]
- 8.Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, Medvedovic M, Puga A. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harstad EB, Guite CA, Thomae TL, Bradfield CA. Liver deformation in Ahr-null mice: evidence for aberrant hepatic perfusion in early development. Mol Pharmacol. 2006;69:1534–1541. doi: 10.1124/mol.105.020107. [DOI] [PubMed] [Google Scholar]
- 10.Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci U S A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Ochoa I, Karman BN, Flaws JA. The role of the aryl hydrocarbon receptor in the female reproductive system. Biochem Pharmacol. 2009;77:547–559. doi: 10.1016/j.bcp.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, Akiyama T, Kurosumi M, Poellinger L, Kato S, Fujii-Kuriyama Y. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci U S A. 2009;106:13481–13486. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salisbury TB, Morris GZ, Tomblin JK, Chaudhry AR, Cook CR, Santanam N. Aryl hydrocarbon receptor ligands inhibit igf-ii and adipokine stimulated breast cancer cell proliferation. ISRN Endocrinol. 2013;2013:104850. doi: 10.1155/2013/104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomblin JK, Salisbury TB. Insulin like growth factor 2 regulation of aryl hydrocarbon receptor in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2014;443:1092–1096. doi: 10.1016/j.bbrc.2013.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode G, Ballard BR, Manning HC, Freeman ML, Kang Y, Eltom SE. Knockdown of aberrantly upregulated aryl hydrocarbon receptor reduces tumor growth and metastasis of MDA-MB-231 human breast cancer cell line. Int J Cancer. 2013 doi: 10.1002/ijc.28297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulero-Navarro S, Pozo-Guisado E, Perez-Mancera PA, Alvarez-Barrientos A, Catalina-Fernandez I, Hernandez-Nieto E, Saenz-Santamaria J, Martinez N, Rojas JM, Sanchez-Garcia I, Fernandez-Salguero PM. Immortalized mouse mammary fibroblasts lacking dioxin receptor have impaired tumorigenicity in a subcutaneous mouse xenograft model. J Biol Chem. 2005;280:28731–28741. doi: 10.1074/jbc.M504538200. [DOI] [PubMed] [Google Scholar]
- 18.Trombino AF, Near RI, Matulka RA, Yang S, Hafer LJ, Toselli PA, Kim DW, Rogers AE, Sonenshein GE, Sherr DH. Expression of the aryl hydrocarbon receptor/transcription factor (AhR) and AhR-regulated CYP1 gene transcripts in a rat model of mammary tumorigenesis. Breast Cancer Res Treat. 2000;63:117–131. doi: 10.1023/a:1006443104670. [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- 27.Ennen M, Minig V, Grandemange S, Touche N, Merlin JL, Besancenot V, Brunner E, Domenjoud L, Becuwe P. Regulation of the high basal expression of the manganese superoxide dismutase gene in aggressive breast cancer cells. Free Radic Biol Med. 2011;50:1771–1779. doi: 10.1016/j.freeradbiomed.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Pidgeon GP, Lysaght J, Krishnamoorthy S, Reynolds JV, O'Byrne K, Nie D, Honn KV. Lipoxygenase metabolism: roles in tumor progression and survival. Cancer Metastasis Rev. 2007;26:503–524. doi: 10.1007/s10555-007-9098-3. [DOI] [PubMed] [Google Scholar]
- 29.Walker NJ, Portier CJ, Lax SF, Crofts FG, Li Y, Lucier GW, Sutter TR. Characterization of the dose-response of CYP1B1, CYP1A1, and CYP1A2 in the liver of female Sprague-Dawley rats following chronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 1999;154:279–286. doi: 10.1006/taap.1998.8595. [DOI] [PubMed] [Google Scholar]
- 30.Vasiliou V, Reuter SF, Williams S, Puga A, Nebert DW. Mouse cytosolic class 3 aldehyde dehydrogenase (Aldh3a1): gene structure and regulation of constitutive and dioxin-inducible expression. Pharmacogenetics. 1999;9:569–580. [PubMed] [Google Scholar]
- 31.Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the "TCDD-inducible AhR-Nrf2 gene battery". Toxicol Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan KP, Wang B, Yang M, Boutros PC, Macaulay J, Xu H, Chuang AI, Kosuge K, Yamamoto M, Takahashi S, Wu AM, Ross DD, Harper PA, Ito S. Aryl hydrocarbon receptor is a transcriptional activator of the human breast cancer resistance protein (BCRP/ABCG2) Mol Pharmacol. 2010;78:175–185. doi: 10.1124/mol.110.065078. [DOI] [PubMed] [Google Scholar]
- 33.Lo R, Matthews J. The aryl hydrocarbon receptor and estrogen receptor alpha differentially modulate nuclear factor erythroid-2-related factor 2 transactivation in MCF-7 breast cancer cells. Toxicol Appl Pharmacol. 2013;270:139–148. doi: 10.1016/j.taap.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Miao W, Hu L, Scrivens PJ, Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 35.Safe S, Wormke M. Inhibitory aryl hydrocarbon receptor-estrogen receptor alpha cross-talk and mechanisms of action. Chem Res Toxicol. 2003;16:807–816. doi: 10.1021/tx034036r. [DOI] [PubMed] [Google Scholar]
- 36.Safe S, Wormke M, Samudio I. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J Mammary Gland Biol Neoplasia. 2000;5:295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- 37.Wong GH, Goeddel DV. Induction of manganous superoxide dismutase by tumor necrosis factor: possible protective mechanism. Science. 1988;242:941–944. doi: 10.1126/science.3263703. [DOI] [PubMed] [Google Scholar]
- 38.Rico de Souza A, Zago M, Pollock SJ, Sime PJ, Phipps RP, Baglole CJ. Genetic ablation of the aryl hydrocarbon receptor causes cigarette smoke-induced mitochondrial dysfunction and apoptosis. J Biol Chem. 2011;286:43214–43228. doi: 10.1074/jbc.M111.258764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Z, Boekhoudt GH, Boss JM. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J Biol Chem. 2003;278:23570–23578. doi: 10.1074/jbc.M303431200. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, St Clair DK. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 41.Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- 42.Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martinez A, Narayan G, Kirsch I, Franklin W, Hirsch F, Birrer M, Cuttitta F. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, Sherr DH, Hahn ME. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol Cell Biol. 2009;29:3465–3477. doi: 10.1128/MCB.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiNatale BC, Schroeder JC, Francey LJ, Kusnadi A, Perdew GH. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J Biol Chem. 2010;285:24388–24397. doi: 10.1074/jbc.M110.118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen PH, Chang H, Chang JT, Lin P. Aryl hydrocarbon receptor in association with RelA modulates IL-6 expression in non-smoking lung cancer. Oncogene. 2012;31:2555–2565. doi: 10.1038/onc.2011.438. [DOI] [PubMed] [Google Scholar]
- 46.Vogel CF, Li W, Wu D, Miller JK, Sweeney C, Lazennec G, Fujisawa Y, Matsumura F. Interaction of aryl hydrocarbon receptor and NF-kappaB subunit RelB in breast cancer is associated with interleukin-8 overexpression. Arch Biochem Biophys. 2011;512:78–86. doi: 10.1016/j.abb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 49.Wong GH, Elwell JH, Oberley LW, Goeddel DV. Manganous superoxide dismutase is essential for cellular resistance to cytotoxicity of tumor necrosis factor. Cell. 1989;58:923–931. doi: 10.1016/0092-8674(89)90944-6. [DOI] [PubMed] [Google Scholar]
- 50.Delhalle S, Deregowski V, Benoit V, Merville MP, Bours V. NF-kappaB-dependent MnSOD expression protects adenocarcinoma cells from TNF-alpha-induced apoptosis. Oncogene. 2002;21:3917–3924. doi: 10.1038/sj.onc.1205489. [DOI] [PubMed] [Google Scholar]
- 51.Chiaro CR, Morales JL, Prabhu KS, Perdew GH. Leukotriene A4 metabolites are endogenous ligands for the Ah receptor. Biochemistry. 2008;47:8445–8455. doi: 10.1021/bi800712f. [DOI] [PubMed] [Google Scholar]
- 52.DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci. 2010;115:89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]