Abstract
The p28 and p65 proteins of mouse hepatitis virus (MHV) are the most amino-terminal protein domains of the replicase polyprotein. Cleavage between p28 and p65 has been shown to occur in vitro at cleavage site 1 (CS1), 247Gly↓Val248, in the polyprotein. Although critical residues for CS1 cleavage have been mapped in vitro, the requirements for cleavage have not been studied in infected cells. To define the determinants of CS1 cleavage and the role of processing at this site during MHV replication, mutations and deletions were engineered in the replicase polyprotein at CS1. Mutations predicted to allow cleavage at CS1 yielded viable virus that grew to wild-type MHV titers and showed normal expression and processing of p28 and p65. Mutant viruses containing predicted noncleaving mutations or a CS1 deletion were also viable but demonstrated delayed growth kinetics, reduced peak titers, decreased RNA synthesis, and small plaques compared to wild-type controls. No p28 or p65 was detected in cells infected with predicted noncleaving CS1 mutants or the CS1 deletion mutant; however, a new protein of 93 kDa was detected. All introduced mutations and the deletion were retained during repeated virus passages in culture, and no phenotypic reversion was observed. The results of this study demonstrate that cleavage between p28 and p65 at CS1 is not required for MHV replication. However, proteolytic separation of p28 from p65 is necessary for optimal RNA synthesis and virus growth, suggesting important roles for these proteins in the formation or function of viral replication complexes.
Two families of large positive-strand RNA viruses, the arteriviruses and the coronaviruses, belong to the order Nidovirales. Despite differences in the genome sizes of the arteriviruses (13 to 16 kb) and coronaviruses (27 to 32 kb), these virus families have several important features in common, including a polycistronic genome, an array of conserved protein domains, and a discontinuous RNA transcription strategy. Another important conserved feature of nidovirus replication is the expression of viral replicase proteins from large polyprotein precursors. These replicase polyproteins are cleaved co- and posttranslationally by virus-encoded proteinases to yield up to 15 mature replicase protein products. Along with putative cellular factors, the mature replicase proteins and precursors are thought to mediate all stages of viral RNA synthesis on membrane-bound viral replication complexes in the cytoplasm. Previous studies have demonstrated roles for arterivirus replicase proteins in the formation and function of viral replication complexes (17, 21, 22, 26). However, the functions of the majority of coronavirus replicase proteins during viral replication have not been determined.
The coronavirus replicase gene (gene 1) is composed of two overlapping open reading frames (ORF1a and ORF1b) that are translated by host cell ribosomes to produce the replicase polyproteins. Translation of ORF1a yields a polyprotein of nearly 490 kDa, while a −1 ribosomal frameshift allows translation of an ORF1a-ORF1b fusion polyprotein that is approximately 800 kDa. Cleavage of the coronavirus replicase polyproteins is mediated by one or two papain-like cysteine proteinases (PLP1 and PLP2) and a picornavirus 3C-like cysteine proteinase (3CLpro) to yield 12 to 16 mature replicase protein products. 3CLpro cleavage sites have been identified at conserved regions in all coronaviruses, including the newly discovered coronavirus associated with severe acute respiratory syndrome (SARS-CoV) (23, 26). In contrast, the conservation and activity of PLP1 and PLP2 domains vary among the three known coronavirus serogroups (groups 1, 2, and 3). The group 1 and 2 coronaviruses express both PLP1 and PLP2 activities and have in common the feature that PLP1 cleaves the first two amino-terminal products from the polyproteins (26). Specifically, viruses in group 1, such as human coronavirus 229E, liberate an amino-terminal 9-kDa protein and an 87-kDa protein after PLP1-mediated processing (12, 27). For group 2 coronaviruses, including bovine coronavirus and mouse hepatitis virus (MHV), PLP1 has been shown in vitro to cleave an amino-terminal 28-kDa protein and a 65-kDa protein, referred to as p28 and p65, respectively (Fig. 1) (1, 2, 7, 8, 10, 14). In contrast, SARS-CoV, as well as the avian coronaviruses (group 3), such as infectious bronchitis virus (IBV), only express active PLP2, which is responsible for cleavage of amino-terminal proteins (23, 26). These observations suggest that PLP1-mediated cleavages may not be essential for coronavirus replication or that the amino-terminal proteins liberated by PLP1s have group-specific functions during replication.
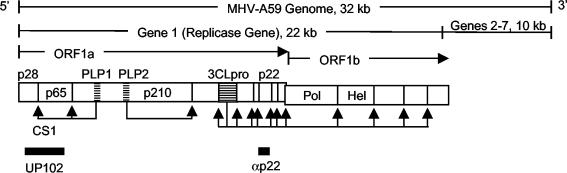
FIG. 1.
A59 genome organization and replicase antibodies. The A59 genome is 32 kb long. The replicase gene (gene 1, 22 kb) is shown, with replicase protein domains in overlapping ORF1a and ORF1b. Hatched boxes indicate proteinases: PLP1 domain (papain-like proteinase 1), PLP2 domain (papain-like proteinase 2), and 3CLpro (3C-like proteinase). The protein domains for mature proteins p28, p65, p210, p22, Pol, and Hel are indicated. Arrows beneath proteins indicate cleavage by the relevant proteinases. The first cleavage site between p28 and p65 (CS1) is shown. Black rectangles beneath the schematic indicate proteins generated to induce the rabbit polyclonal antibodies used in this study.
MHV has been extensively used as a model for studies of coronavirus replicase protein expression and processing. The determinants of MHV PLP1-mediated cleavages have been studied in detail by using in vitro expression and processing assays (2, 3, 10, 14). PLP1 has been shown to proteolytically process the first two cleavage sites in the MHV replicase polyprotein: between p28 and p65 at 247G↓V248 (referred to as cleavage site 1, or CS1, in this report) and between p65 and p210 at 832A↓G833 (CS2) (3, 10, 14). Until recently, the lack of a reverse genetics system has hindered the characterization of these cleavage sites in the context of MHV infection. Several lines of evidence indicate that continuous translation and processing of the MHV replicase polyprotein are required for virus replication (15, 18, 19). However, the requirements for processing at specific cleavage sites in the polyprotein, such as between p28 and p65, have not been shown. Biochemical experiments determined that both p28 and p65 are associated with intracellular membranes (4, 11, 20), and confocal immunofluorescence microscopy data have shown that both p28 and p65 localize to viral replication complexes in punctate perinuclear foci (20; S. M. Brockway, X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison, submitted for publication). These results suggest that p28 and p65 may play a role in replication complex formation or RNA synthesis.
In the present study, we used a reverse genetics approach to verify the critical CS1 residues necessary for cleavage and to determine if cleavage of p28 from p65 is required for MHV replication. By using the MHV strain A59 infectious clone, viruses were generated that had either predicted cleaving or noncleaving mutations in CS1. Viruses with predicted noncleaving CS1 mutations were viable but were unable to process p28 and p65, instead expressing an uncleaved p93 precursor protein. Although noncleaving CS1 mutants were able to replicate, these viruses had diminished peak titers, smaller plaques, and decreased RNA synthesis compared with wild-type virus and cleaving CS1 mutant viruses. Despite the defect in viral replication, the mutant viruses did not demonstrate phenotypic reversion or changes at introduced mutations after repeated passage in culture. This study demonstrates that while cleavage of p28 from p65 is not required for viral replication, alterations at the first cleavage site alter virus growth, protein processing, and RNA synthesis.
MATERIALS AND METHODS
Wild-type virus, cells, and antibodies.
MHV strain A59 was used as the wild-type control in all experiments. Delayed brain tumor (DBT) cells (13) and baby hamster kidney cells expressing the MHV receptor (BHK-MHVR) (5, 6) were grown in Dulbecco's modified Eagle medium that contained 10% fetal calf serum for all experiments. Media of BHK-MHVR cells were supplemented with G418 (0.8 mg/ml) for selection of cells expressing the receptor. Polyclonal antisera UP102 (anti-p28 and -p65) and α-p22, used for biochemical experiments, have been previously described (8, 16) (Fig. 1).
Construction of mutagenized A59 infectious-clone fragment A plasmids.
Site-directed mutations were made in the p28-p65 cleavage site (CS1) by PCR with the primers shown in Table 1. For all reactions, A59 infectious-clone cDNA fragment A construct pCR-XL-TopoA, which consists of nucleotides (nt) 1 to 4882, was used as template DNA (25). Primer-generated restriction sites (5′ AflII and 3′ BstZ17I) were used to clone PCR products into pCR-XL-TopoA in place of wild-type CS1.
TABLE 1.
Primers used for mutagenesis of CS1 in fragment A
| Primer | Sequencea |
|---|---|
| Common right | 5′-GCTCTT1298GTATACAGCATAGTCTCCACCAAACGG1285-3′ |
| Left (sense) | |
| mut3 | 5′-927GCTCTTCTTAAGGGCTATCACGGTGTTAAGCCCATC962-3′ |
| mut4 | 5′-927GCTCTTCTTAAGGGCTATGCCGGTGTTAAGCCCATC962-3′ |
| mut5 | 5′-927GCTCTTCTTAAGGGCTATCACGTTGTTAAGCCCATCCTG965-3′ |
| mut8 | 5′-927GCTCTTCTTAAGGGCTATCGCGGTGCTAAGCCCATCCTG965-3′ |
| mut9 | 5′-927GCTCTTCTTAAGGGCTATCGCGGTCATAAGCCCATCCTGTTT968-3′ |
| mutΔCS1 | 5′-927GCTCTTCTTAAGGGCTATAAGCCCATCCTGTTTGTG971-3′ |
Underlining indicates mutated or deleted nucleotides.
Generation of CS1 mutant viruses.
Viruses containing the PCR-generated mutations at CS1 were produced by the infectious-clone strategy for A59 described by Yount et al. (25). Briefly, plasmids containing the cDNA cassettes of the MHV genome were digested with MluI and BsmBI for fragment A; BglI and BsmBI for fragments B and C; BsmBI for fragments D, E, and F; and SfiI and BsmBI for fragment G. Digested, gel-purified fragments were ligated together in a total reaction volume of 200 μl overnight at 16°C. Following chloroform extraction and isopropanol precipitation of ligated DNA, full-length transcripts of MHV infectious-clone cDNA were generated in vitro with the mMessage mMachine T7 Transcription Kit (Ambion) in accordance with the manufacturer's protocol with modifications. Fifty-microliter reaction mixtures were supplemented with 7.5 μl of 30 mM GTP, and transcription was performed at 40.5°C for 25 min, 37.5°C for 50 min, and 40.5°C for 25 min. In parallel, transcripts encoding the MHV nucleocapsid protein (N) were generated in vitro with N cDNA generated by PCR (25). N transcripts and MHV infectious-clone transcripts were then mixed and electroporated into BHK-MHVR cells. Briefly, BHK-MHVR cells were grown to subconfluence, trypsinized, washed twice with phosphate-buffered saline (PBS), and then resuspended in PBS at a concentration of 107/ml. Eight hundred microliters of cells was then added to RNA transcripts in an electroporation cuvette with a 4-mm gap, and three electrical pulses of 850 V at 25 μF were delivered with a Bio-Rad Gene Pulser II electroporator. Transfected cells were then laid over a layer of 106 uninfected DBT cells in a 75-cm2 flask and incubated at 37°C for 30 h. Virus viability was determined by syncytium formation, and progeny were passaged and purified by plaque assay.
Viral passages, RT-PCR, and sequencing of viral RNA.
Viable viruses were plaque purified and then successively passaged 10 times at either 8- or 18-h intervals on DBT cells. After 10 passages, cell supernatant was harvested and centrifuged for 10 min at 700 × g at 4°C to clear cell debris. Clarified supernatant was then centrifuged at 150,000 × g for 90 min at 4°C to pellet virus. The virus pellet was then lysed with Tri-Reagent (Sigma), and viral RNA was isolated in accordance with the manufacturer's protocol. To generate viral cDNA corresponding to the first ∼5 kb of the genome, reverse transcription (RT)-PCR was performed. An antisense primer complementary to nt 5500 through 5531 of the MHV genome was used for reverse transcription. The RT product was then amplified by PCR with primers corresponding to nt 200 through 230 (sense) and 5500 through 5531 (antisense). Initial isolates at passage 2 (P2) were sequenced across 600 nt spanning the cleavage site coding region with an ABI Prism automated sequencer to confirm the introduced mutations. Passage 10 (P10) viruses were sequenced from nt 230 through nt 5500 to determine maintenance of mutations and any additional changes during passage in p28, p65, and PLP1.
Protein expression and immunoprecipitations.
Immunoprecipitations, pulse-labeling, and pulse-chase translation experiments were performed as previously described (7, 8).
Viral growth assays and metabolic labeling of viral RNA.
Mutant viruses were purified and analyzed by viral growth and RNA synthesis assays. For viral growth determination, DBT cells were infected at a multiplicity of infection (MOI) of either 0.1 or 5 PFU/cell. Aliquots of medium were collected from 2 h to 48 h postinfection (hpi), and titers were determined by plaque assay as described previously (15). For metabolic labeling of viral RNA, DBT cell monolayers (approximately 3 × 106 cells) were either mock infected or infected at an MOI of 5 PFU/ml. At 4.5 hpi, actinomycin D (Sigma) was added to the cells at a final concentration of 20 μg/ml. Cells were labeled with 100 μCi of [3H]uridine/ml in the presence of actinomycin D from 5 to 7 hpi. For harvesting of viral RNA, cells were washed twice with PBS and then lysed with 500 μl of cell lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 50 mM Tris [pH 8.0]). Lysates were centrifuged at 3,500 × g to remove nuclei, and then RNA in 200 μl of postnuclear supernatant was precipitated with trichloroacetic acid (TCA). Precipitated RNA was dried onto glass microfiber filters (Whatman) by vacuum filtration, and radioactivity was measured in a liquid scintillation counter (Beckman).
Northern blot analysis.
DBT cells were mock infected or infected at an MOI of 5 PFU/cell with wild-type or CS1 mutant virus. Cells were lysed at 10 hpi with Trizol (Invitrogen), and RNA was isolated in accordance with the manufacturer's protocol. RNA from approximately 4 × 105 cells was electrophoresed in a 0.8% agarose-formaldehyde gel at 144 V for 4 h. For Northern blot analysis, RNA was transferred overnight onto a nylon membrane (HyBond N+; Amersham Biosciences) with a wick transfer system. RNA was UV cross-linked to the nylon membrane and probed with a 32P-labeled probe complementary to the 3′ untranslated region (UTR) as described previously (24). RNA was visualized by autoradiography, and images were prepared with Adobe Photoshop 7.0 (Adobe).
RESULTS
Recovery, sequencing, and stability of A59 CS1 mutants.
To determine if cleavage between p28 and p65 at CS1 is required for virus growth, mutations were introduced at P2, P1, and P1′ of CS1, which were shown to either allow or abolish cleavage in vitro (10, 14) (Fig. 2). Mutated fragment A cDNAs were used to assemble full-length MHV genomic cDNA, which was transcribed in vitro to yield mutant MHV genomic RNA. BHK-MHVR cells were electroporated with in vitro-transcribed genome RNA and monitored for MHV-induced syncytium formation. Viral syncytia were detected within 24 h in cells electroporated with full-length wild-type genome RNA (icwt) and with genome RNA containing mutations that were predicted to allow cleavage at CS1 (mut8 and mut9) (Fig. 2). Unexpectedly, viral syncytia were also detected in cells electroporated with RNA containing predicted noncleaving CS1 mutations (mut3, mut4, and mut5). However, a delay in the timing and extent of the viral cytopathic effect (CPE) was noted during initial recovery of virus from electroporated cells producing predicted CS1 noncleaving mutant viruses. To verify the retention of the introduced mutations, both supernatant virus populations of P2 and plaque-purified virus clones from P2 of each mutant (P2 clone 1 [P2C1]) were sequenced across nucleotides encoding CS1. All viruses, including those predicted to be noncleaving, were found to have the correct CS1 nucleotide sequence, and no additional nucleotide changes were noted in the 300 nt flanking each side of CS1. Comparison of the sequence of the P2 population virus with the P2C1 cloned viruses demonstrated no heterogeneity or changes at any of the sequenced nucleotides (data not shown). These results demonstrated that mutations at CS1 predicted to either retain or abolish cleavage at CS1 allowed recovery of viable virus.
FIG. 2.
Mutagenesis of CS1. (Top) Organization and relative sizes of cDNA fragments A to G used for assembly of full-length MHV cDNA. (Middle) Expansion of genome fragment A showing coding domains for p28, p65, and the N-terminal 33% of p210, with the PLP1 domain intact. The amino acid (aa) and nucleotide termini of fragment A are indicated. (Bottom) P5 through P5′ of CS1 indicated by position and nucleotide and amino acid sequences, with amino acid numbering for P1 and P1′. The vertical arrow shows the site of cleavage. Nucleotide and amino acid mutations are shown for each mutant, and the deletion is indicated by the caret. Cleavage in vitro is based on previously published studies (10, 14), and virus viability is based on data from this study. N/T, not tested in vitro.
To determine if the CS1 mutations were stable over multiple passages, stocks from cloned mutant viruses (P2C1) were passaged 10 times in DBT cells at 8- to 18-h intervals, depending on the earliest appearance of CPE, to attempt to drive a reversion event. During passage in culture, the CS1 predicted noncleaving mutants demonstrated no change in the timing or extent of CPE in comparison with that originally observed in the electroporated culture. mut3, mut4, and mut5 demonstrated a visibly reduced plaque size, an extended time to the first detection of plaques (30 versus 24 hpi), and a reduction in the plaque area of 53 to 77% at 30 h, with the plaques never achieving the area of the wild-type virus (Fig. 3A). Even after 10 passages in cell culture, CS1 predicted noncleaving mutants retained their small plaque size, with no heterogeneity of plaque size observed. At the conclusion of passage 10 (P2C1-10) for each mutant, RNA was isolated from supernatant virus and sequenced across the first ∼5 kb of gene 1 (nt 230 through 5500). Sequence analysis of the P2C1-10 populations for each mutant demonstrated that all CS1 mutations were retained (Fig. 3B), and no additional changes were noted within p28, p65, or PLP1. In the present study, we did not sequence the entire 32-kb genomes of the original eight input viruses and the P2C1-10 viruses. Therefore, we cannot entirely exclude the possibility that compensatory second-site mutations occurred during virus passage. However, the lack of mutations in the first ∼5 kb of gene 1 (p28, p65, and PLP1 coding regions), as well as the complete retention of the wild-type and mutant virus phenotypes, supports the conclusion that CS1 mutant viruses were genetically stable following multiple passages in cell culture.
FIG. 3.
Plaque morphology and sequences of CS1 mutants. (A) Plaque morphology of CS1 mutants. All images were obtained at the same resolution (magnification, 10×) with a Zeiss Axiovert microscope. Black circles were visually drawn at limits of plaques. Percentages represent total plaque areas of black circles, with A59 arbitrarily set at 100%. icwt is assembled wild-type strain A59; the other viruses are mutants as in Fig. 2. (B) Sequence data from RT-PCR of virion RNA showing retained mutations following passage of cloned wild-type and mutant viruses in culture as described in the text. The location of CS1 is indicated by RGV and a vertical dotted line. Mutations are indicated by horizontal bars with dots at mutated nucleotides and the resulting amino acid change underneath the bar. Δ indicates a nucleotide deleted in mutΔCS1.
Reduced peak titers of predicted noncleaving CS1 mutants.
Viral growth experiments were performed to determine if CS1 predicted noncleaving mutants had defects in virus replication compared to predicted cleaving and wild-type controls (Fig. 4). DBT cell monolayers were infected at MOIs of 0.001, 0.1, and 5 PFU/cell; supernatant virus was harvested at various times from 2 to 24 hpi; and viral titers were determined by plaque assay. Infections at MOIs of 0.1 and 5 PFU/cell both demonstrated consistent single-cycle growth kinetics (Fig. 4B). Because of this consistency and the difficulty of producing higher-titer stocks of the predicted noncleaving mutants, an MOI of 0.1 PFU/cell was chosen for subsequent experiments.
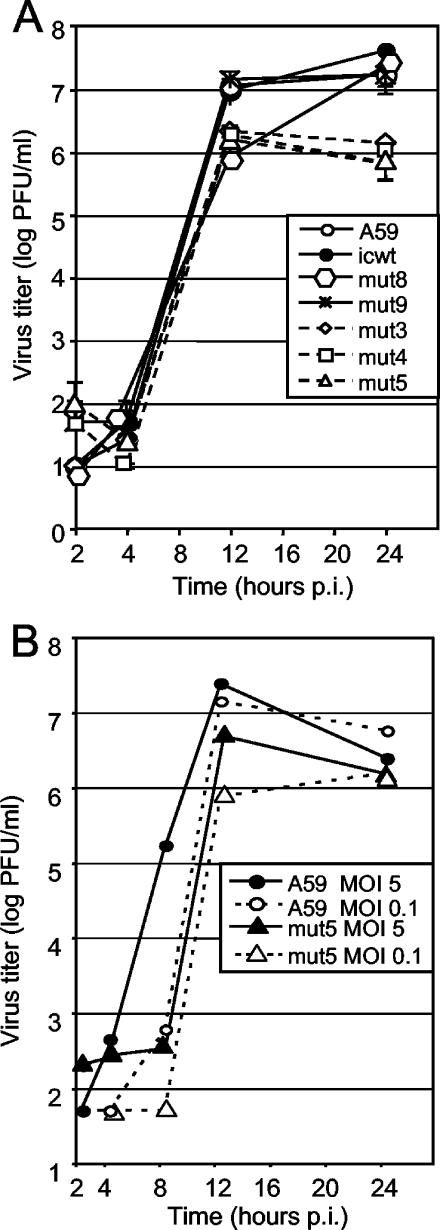
FIG. 4.
Growth of CS1 mutant viruses in DBT cells. (A) Growth of CS1 mutants at an MOI of 0.1 PFU/cell. A59, icwt, and mutant viruses mut8, mut9, mut3, mut4, and mut5 were used to infect DBT cells at an MOI of 0.1 PFU/ml. Following 30 min of attachment, cells were rinsed three times and incubated in medium. Samples were obtained at the postinfection (p.i.) times indicated, and titers were determined by plaque assay on DBT cells at 37°C. Error bars indicate standard deviations of plaque assay replicates. (B) Comparison of growth at low and high MOIs. A59 and mut5 viruses were used to infect DBT cells at MOIs of 5 and 0.1 PFU/ml. Virus growth assay was performed as described for panel A, with samples taken at the postinfection times indicated and titers determined by plaque assay. The values shown are averages of duplicate plaque assays.
During single-cycle growth experiments, wild-type A59 and assembled wild-type MHV (icwt), as well as predicted cleaving mutants (mut8 and mut9), had peak viral titers of 2 × 107 to 5 × 107 PFU/ml (Fig. 4A). In contrast, predicted noncleaving mutants (mut3, mut4, and mut5) had peak viral titers of 1 × 106 to 3 × 106 PFU/ml, consistently 0.5 to 1.5 logs less than the wild type and the predicted cleaving mutants. Interestingly, predicted cleaving mutant mut8 showed lower titers at 12 h but achieved wild-type titers by 24 h, while the predicted noncleaving mutants never attained wild-type titers (Fig. 4A). On repeat experiments, mut8 and mut9 occasionally showed this intermediate growth phenotype.
To determine if there was a direct difference in mutant virus growth at a higher MOI, A59 and putative noncleaving mutant mut5 were directly compared for growth in culture following infections at MOIs of 0.1 and 5 PFU/cell (Fig. 4B). At an MOI of 0.1 PFU/cell, wild-type A59 had a peak titer of 2 × 107 PFU/ml while mut5 had a peak titer of 9 × 105 PFU/ml, consistent with the experiment in Fig. 4A. When the MOI was increased 50-fold to 5 PFU/cell, A59 had a peak titer of 3 × 107 PFU/ml while mut5 had a peak titer of 6 × 106 PFU/ml. A similar result was seen with an MOI of 0.001, where even with delayed growth kinetics, mut5 never achieved wild-type titers (data not shown). Thus, there was a clear viral growth defect in the predicted noncleaving mutants compared with the wild type and the predicted CS1 cleaving mutants.
Cleavage at CS1 is abolished by mutations at residues P2 and P1.
To determine if CS1 mutants were capable of cleaving p28 from p65 in the replicase polyprotein, immunoprecipitation assays were performed. DBT cells were either mock infected or infected with A59, icwt, or the CS1 mutants and labeled with [35S]Met-Cys from 6 to 7 hpi. Cytoplasmic extracts were prepared and immunoprecipitated with antiserum directed against p28 and p65 (UP102) (Fig. 5). The predicted CS1-cleaving mutants, mut8 and mut9, demonstrated patterns of protein expression and processing identical to those of A59 and icwt, with distinct p28 and p65 proteins detected (Fig. 5A). In cells infected with the predicted noncleaving mutants, mut3, mut4, and mut5, the pattern of detected proteins was quite different, with no p28 or p65 detected following 1 h of labeling. A much less prominent protein band with a mobility of approximately 28 kDa was detected in the mut3-, mut4-, and mut5-infected lysates; however, it also was detected in mock-infected cells by immune serum in infected cells and by preimmune serum (data not shown), indicating that it was a comigrating, nonspecifically precipitated product. A new, discrete band of 93 kDa was immunoprecipitated only in mut3-, mut4-, and mut5-infected cells by immune serum. This 93-kDa protein was consistent with the size of a putative uncleaved p28-p65 precursor. We also observed several heterogeneous bands of label, most prominently one of approximately 70 kDa that was unique to cells infected with the noncleaving mutants (Fig. 5A). This 70-kDa protein could not be accounted for by putative uncleaved precursors. While additional studies are required to identify this protein and its source, possibilities include alternate folding or degradation of p93, oligomerization, or alternative cleavage in the p28 or p65 protein (see Discussion).
FIG. 5.
Processing of p28 and p65 in CS1 mutants. (A) Pulse-labeling of replicase proteins p28 and p65. DBT cells were infected for 6 h and labeled with [35S]Met-Cys for 60 min, and lysates of infected cells were immunoprecipitated with UP102, followed by electrophoresis in sodium dodecyl sulfate-5 to 18% polyacrylamide gels. Molecular mass (kilodaltons) markers are to the left of the gel; A59, icwt, and mutant viruses are indicated above the lanes; and masses of specific proteins (in kilodaltons) are shown to the right of the gel, with p28 and p65 also indicated by filled circles. The novel 93- and 70-kDa products are indicated by arrows. (B) Pulse-chase labeling. The experiment was performed the same way as the pulse-labeling experiment, except that following the 60-min labeling period (indicated by the number 60 above lanes), the radiolabel was removed and cells were incubated in medium with excess unlabeled Met-Cys for an additional 90-min chase (c90). Viruses, markers, and proteins are the same as in panel A.
To determine if any delayed processing at CS1 was occurring during infection with mut3, mut4, and mut5, pulse-chase experiments were performed. Infected DBT cells were labeled for 60 min at 6 hpi and followed by a chase with excess unlabeled Met-Cys for an additional 90 min (Fig. 5B). p28 and p65 were readily detectable at 60 min in cells infected with icwt but not in cells infected with noncleaving mutants. Even following the additional 90-min chase, there was no disappearance of p93 linked to the concomitant appearance of p28 and p65 in noncleaving mutants, supporting the conclusion that processing did not occur at CS1 in the noncleaving mutants. Together, the label and chase experiments demonstrated that P2-R246 and P1-G247 are critical residues for CS1 cleavage during replicase polyprotein expression and processing in infected cells, while P1′-V248 could be replaced with both conservative (Ala) and nonconservative (His) residues and still allow cleavage at CS1. The results were consistent with the previous in vitro analyses (10, 14). The finding that replication occurred in the presence of the P1′ substitutions was interesting, but what was remarkable was that even P2 and P1 mutations that appeared to abolish cleavage at CS1 allowed recovery of viable virus. Thus, these results suggest that cleavage of p28 from p65 is not required for virus viability; however, the absence of CS1 processing was associated with impairment of virus growth.
Deletion of CS1 P2-P1′ RGV allows recovery of viable virus.
To confirm that MHV was capable of replicating in the absence of CS1 cleavage, we generated mutant viruses that contained an in-frame 9-nt deletion of residues 246RGV248 at CS1 (mutΔCS1) (Fig. 2 and 3). Following electroporation of mutΔCS1 transcripts into BHK-MHVR cells, CPE was observed that was identical to that caused by mut5, with delayed appearance of CPE and small, plaque-like foci of infection with limited distal spread. During plaque purification, plaques were delayed in growth and reduced in size. Stocks were obtained for sequencing, and the virus was passaged as with the point mutations. Sequencing confirmed the in-frame 9-nt deletion in the recovered virus, and no additional changes, insertions, or deletions were noted across the first ∼5 kb of the genome following 10 passages (Fig. 3).
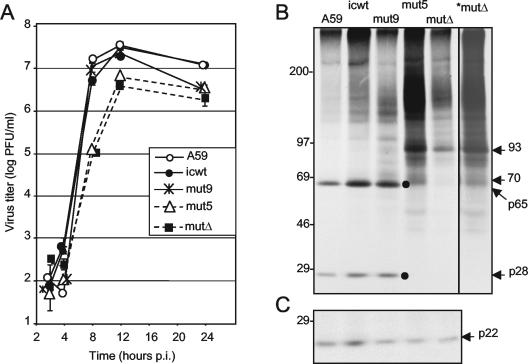
The mutΔCS1 virus was compared with A59, icwt, cleaving mut9, and noncleaving mut5 for virus growth and protein expression (Fig. 6). When mutΔCS1 was directly compared for virus growth with wt, icwt, cleaving mut9, and noncleaving mut9, mutΔCS1 was identical to noncleaving mut5 in growth kinetics (Fig. 6A). Peak titers of mutΔCS1 and mut5 were 5.5 × 106 to 6 × 106 PFU/ml, in comparison with peak titers of 2 × 107 to 4 × 107 for A59, icwt, and cleaving mut9, completely consistent with experiments in Fig. 4 in demonstrating a 0.5-log reduction in peak titers and a lack of the ability to achieve wild-type titers at any point during infection.
FIG. 6.
Growth and protein expression of CS1 deletion mutant mutΔCS1. (A) Viral growth. Following infection at an MOI of 0.1 PFU/cell, virus growth was determined by plaque assay as described in the legend to Fig. 4 for the viruses indicated in the inset. mutΔ indicates mutΔCS1 throughout the figure. p.i., postinfection. (B) Protein expression. DBT cells were infected for 6 h and labeled with [35S]Met-Cys for 60 min, and lysates of infected cells were immunoprecipitated with UP102, followed by electrophoresis on sodium dodecyl sulfate-5 to 18% polyacrylamide gels. Molecular mass markers are to the left of the gel, and specific products are to the right (masses are in kilodaltons). Viruses are indicated above the lanes. mutΔ indicates mutΔCS1. *mutΔ is the same gel lane as mutΔ but exposed to film four times as long as the other lanes. (C) Lysates from panel B immunoprecipitated with α-p22 antiserum. A marker protein is shown to the left, and the location of p22 is shown to the right of the gel.
When infected cells were analyzed for protein expression and CS1 cleavage, mutΔCS1 had the same pattern of proteins as mut5, with no detectable p28 or p65 and with detection of the novel 93-kDa protein and the less prominent, heterogeneous 70-kDa protein (Fig. 6B). Both virus viability and detection of the 93-kDa protein strongly suggested that PLP1, PLP2, and 3CLpro activities and cleavage were intact and unaffected by CS1 mutations. To determine the activity of 3CLpro at downstream cleavage sites, cell lysates for all viruses were probed for p22, a well-characterized protein known to be cleaved by 3CLpro (Fig. 6C). p22 was readily and equivalently detected in A59, icwt, cleaving mut9, and noncleaving mut5 and mutΔCS1, demonstrating intact 3CLpro function and cleavage at known sites. Thus, by growth and protein expression and processing, mutΔCS1 behaved identically to the noncleaving point mutant, mut5. The recovery and growth of mutΔCS1 further confirmed the lack of CS1 cleavage of mut3, mut4, and mut5 by demonstrating the ability of MHV to replicate in the absence of the three-amino-acid RGV core CS1 cleavage site. Further, the viability of mutΔCS1 demonstrated that amino acids, specifically, the three-residue cleavage site, could be deleted from the polyprotein and still permit virus growth.
Diminished viral RNA synthesis with noncleaving CS1 mutant viruses.
Inhibition of MHV replicase polyprotein processing by proteinase inhibitors leads to rapid shutoff of viral RNA synthesis (15). To determine if inhibition of CS1 cleavage affects viral RNA synthesis, DBT cells were either mock infected or infected with the wild type or the CS1 virus mutants at an MOI of 5 PFU/cell. Cells were metabolically labeled with 100 μCi of [3H]uridine/ml in the presence of actinomycin D from 5 to 7 hpi, and TCA-precipitable RNA was measured by liquid scintillation (Fig. 7A). All CS1-cleaving viruses (A59, icwt, mut8, and mut9) had equivalent amounts of incorporation of label into viral RNA in comparison with the background measurement in the mock-infected, actinomycin D-treated cells. In contrast, noncleaving mutants (mut3, mut4, mut5, and mutΔCS1) had significantly decreased total RNA synthesis, from 50 to 75% reduced compared to the wild type and the cleaving mutants. While there was some variability in the precise amount of incorporation between different noncleaving mutants in replicates of the experiment (data not shown), the noncleaving mutants were significantly reduced in total RNA synthesis in every experiment.
FIG. 7.
RNA synthesis in CS1 mutants. (A) Metabolic labeling of viral RNA. DBT cells were mock infected (M) or infected with A59, icwt, or the indicated CS1 mutants. At 4.5 hpi, actinomycin D was added to a final concentration of 20 μg/ml. Cells were radiolabeled with [3H]uridine from 5 to 7 hpi and lysed, and TCA-precipitated viral RNA was quantitated by liquid scintillation counting. Error bars represent standard deviations of duplicate measurements. mutΔ indicates mutΔCS1 throughout the figure. (B) Northern blot analysis. DBT cells were mock infected or infected with A59, icwt, or the indicated CS1 mutants in parallel with the infections shown in panel A. Cells were lysed in Trizol at 10 hpi, and RNA from approximately 4 × 105 cells was separated on a 0.8% agarose-formaldehyde gel. RNA was transferred to a nylon membrane, UV cross-linked, and probed with a 32P-labeled negative-polarity primer complementary to the 3′ UTR to detect positive-strand RNA species. Top, 6-h exposure of genomic RNA (RNA 1); bottom, 1-h exposure of all positive-strand RNA species. Genomic RNA and sgRNA species are indicated by number to the right.
Having demonstrated an inhibition of viral RNA synthesis with the noncleaving CS1 mutants, we next sought to determine if these viruses exhibit specific defects in genome replication or subgenomic RNA (sgRNA) synthesis. DBT cells were mock infected or infected at an MOI of 5 PFU/cell. At 10 hpi, total cellular RNA was harvested and probed by Northern blot assay with a radiolabeled probe complementary to the 3′ UTR to detect all MHV positive-strand full-length genomic RNA and sgRNA (Fig. 7B). All seven species of viral RNA were detected for all viruses, both cleaving and noncleaving. The CS1-noncleaving viruses exhibited reduced levels of both full-length genome RNA and sgRNA species compared with A59, icwt, and the CS1-cleaving mutants; however, the measured ratios of genomic RNA to sgRNA were the same for the wild type and the cleaving and noncleaving mutants (data not shown). These results indicate that the global reduction in RNA synthesis measured in Fig. 7A was due to overall reduction in RNA synthesis rather than inhibition of either transcription of sgRNAs or replication of genomic RNA alone.
DISCUSSION
Proteolytic cleavage of p28 and p65 at CS1 is not required for, but enhances, viral replication.
In the present study, we determined that cleavage of p28 from p65 is not necessary for MHV replication; however, abolition of cleavage resulted in a global reduction in all species of viral RNA, with an associated reduction in viral growth and spread. The fact that CS1 cleavage is required for efficient replication is interesting for several reasons. CS1 is conserved in group 1 and 2 coronaviruses but not in group 3 coronavirus (IBV), suggesting that either introduction or retention of CS1 is critical to the replication or pathogenesis of these viruses (27). The ability of the virus to survive abolition of the cleavage site suggests that the cleavage site may have evolved after fundamental proteins and functions. If this is so, then the genetic experiments in this study may have recapitulated an earlier stage of coronavirus evolution in which group 1, 2, and 3 coronaviruses may have had in common more expression and possibly functional characteristics in the amino-terminal replicase proteins. Moreover, the possibility that cleavage sites evolved after protein function suggests that the 93-kDa uncleaved precursor resulting from inhibition at CS1 could serve functions similar to those of the p87 protein of the group 3 coronavirus IBV, which does not undergo further processing during IBV replication. However, no 93-kDa p28-p65 precursor has ever been detected in MHV-infected cells, with cleavage at CS1 clearly demonstrated to be an early, rapid, cotranslational event both in in vitro translation studies and during virus infection, suggesting an early regulatory process (9). In addition, there is no amino acid identity, similarity, or predicted structural similarity (data not shown) between IBV p87 and the engineered MHV p93 protein, suggesting that these are in fact different proteins. Generation of chimeric viruses in the 5′ end of the replicase gene would likely be necessary to determine any functional similarities in these proteins. In summary, all of the data support the conclusion that MHV has carefully orchestrated the separation of p28 from p65 at CS1 as the first step after initiation of replicase polyprotein translation.
Noncleaving CS1 mutants have defects in viral growth and RNA synthesis: implications for p28 and p65 functions.
The effects of mutations at CS1 on viral replication suggest possible functional roles for p28 and p65 in the MHV life cycle. Previous cell imaging and biochemical results have indicated that p28 and p65 may function during viral RNA synthesis. Specifically, both p28 and p65 localize to replication complexes and are associated with the same membranous complex as the putative RdRp (Pol) protein (4, 20; Brockway et al., submitted). It is clear from the results of this report that separation of p28 from p65 likely contributes to optimal RNA synthesis and virus growth. If p28 and p65 are integral components of viral replication complexes, how is MHV able to survive such a profound alteration in the expression of these proteins caused by abolition of CS1 cleavage? The most straightforward explanation is that the functions of p28 and/or p65 are partially retained in the uncleaved p28-p65 precursor (p93). In this model, the demonstrated inhibition of virus growth and RNA synthesis in the noncleaving mutants could be due to alterations in membrane or protein interactions, intracellular localization, or possible altered conformation of the p28 or p65 domain within p93. In essence, the defects in virus growth could be due either to a single population of p93 performing both p28 and p65 functions at suboptimal levels or to an apportioning of the p93 protein into two populations: one performing the function(s) of p28 and the other fulfilling the functional requirements of p65.
Another possibility is that an alternative cleavage of the 93-kDa precursor could generate proteins capable of at least the minimal activity required for virus replication. The heterogeneous ∼70-kDa product was detected by immunoprecipitation with UP102 (anti-p28 and -p65) from all of the CS1-noncleaving mutants, including mutΔCS1. Since CS2 was efficiently cleaved in these mutants, a p70 protein would most likely be produced by a cleavage upstream of CS1 in the polyprotein. It is interesting in this regard that MHV contains a Gly-Ala dipeptide upstream of CS1 in the p28 domain, which would produce an ∼70-kDa protein if cleaved (data not shown). Further, such a cleavage would result in proteins of 20 and 70 kDa, which are similar to the predicted sizes of nsp1 and nsp2 of SARS-CoV. Thus, an alternative 70-kDa protein might at least partially compensate for p65 function. However, if this is normally an unused site, then it is also possible that such a protein is not preferred by the virus and that an alternative cleavage product might actually function in a dominant-negative fashion, thus contributing to the RNA synthesis and growth defects in mut3, mut4, mut5, and mutΔCS1. Studies to determine the possibility of such an alternative cleavage event, as well as possible requirements within the coding sequences of p28 and p65 necessary to allow such an event, will be the focus of a future study.
Processing determinants within CS1 and stability of mutations.
Our results confirmed and extended the previously published in vitro results (10, 14) by demonstrating that substitutions at P2-R246 and P1-G247 abolished cleavage at CS1, while in contrast, P1′-V248 could be replaced with both small noncharged residues, such as Ala, and with bulkier charged residues, such as His, and still retain complete CS1 cleavage. Thus, the requirements for P2 and P1 appear to be strict, whereas P1′ appears quite tolerant of substitutions. Furthermore, the recovery of mutant viruses with P2-P1′ deleted (mutΔCS1) demonstrated that substantial changes are tolerated within P2-P1′ of CS1. When mutant viruses were passed in culture under conditions to select for more rapid replication and CPE, no changes were noted in the replication phenotype of the virus by growth, CPE, or plaque morphology, and sequence comparison of the “parental” and passaged viruses identified no primary- or secondary-site changes within the first ∼5 kb of gene 1. It is possible that the growth defect in the CS1-noncleaving mutants of a 0.5- to 1.5-log reduction in the peak titer was not enough to allow the emergence of revertants over the passages in this experiment. Increased selective pressure, such as temperature changes or the presence of proteinase inhibitors, may be required to further drive down the replication of the noncleaving mutants and favor selection for viruses with compensating mutations. Another possibility is that there may be unique constraints on changes to the nucleotides or amino acids at and flanking the cleavage sites, or more broadly in the 5′ 5 kb of the RNA genome. At the protein level, the determinants for CS1 cleavage may be so local (P5 through P1) that more distal mutations cannot compensate for the strict requirements for recognition and cleavage of CS1. If the viruses retain their RNA synthesis and growth phenotype over extended passage or under increased pressure for reversion, it will be important to sequence the entire genome of one or more of these viruses to determine whether changes in downstream replicase, accessory, or structural proteins can compensate for the introduced changes.
In conclusion, we have engineered and established mutant viruses that either allow or abolish cleavage between p28 and p65 at CS1. These viruses maintain their introduced mutations and growth phenotypes upon repeated passage in culture, despite defects at the levels of RNA synthesis and viral titer, suggesting that there may be significant genetic stability of viable cleavage site mutants. These viruses provide a panel of mutants with distinct phenotypes in growth, RNA synthesis, and protein processing, but the viruses are readily grown and analyzed in culture. Thus, they will provide powerful models for determination of the roles of p28 and p65 in replication complex formation and RNA synthesis. Further, the viruses will form the basis for studies of the functions and requirements of PLP1 and PLP2 function during replication. Since the engineered defects are localized to p28 and p65, it will be possible to determine which of the proteins is responsible for the defect or if it results from the unique fusion of the proteins. Finally, they will form the basis for studies of replicase proteins in viral pathogenesis and in possible mechanisms of attenuation of replication and virulence that might be broadly applicable to studies of animal and human coronaviruses.
Acknowledgments
We thank Hung-Yi Wu (University of Tennessee, Knoxville) for assistance with Northern blot analysis and Jennifer Sparks for critical reading of the manuscript.
Support for this work was provided by National Institutes of Health grants AI26603 (M.R.D.) and AI23946 (R.S.B.). S.M.B. is also supported by the Training Grant in Mechanisms of Vascular Disease, Department of Pathology, Vanderbilt University School of Medicine (5T32HL007751). R.L.G. is also supported by the Training Grant in Cellular, Biochemical, and Molecular Sciences, Vanderbilt University School of Medicine (5T32GM00855).
REFERENCES
- 1.Baker, S. C., C.-K. Shieh, L. H. Soe, M.-F. Chang, D. M. Vannier, and M. M. C. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 63:3693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. [DOI] [PubMed] [Google Scholar]
- 3.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W., and R. S. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, W., V. J. Madden, C. J. Bagnell, and R. S. Baric. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228:318-332. [DOI] [PubMed] [Google Scholar]
- 7.Denison, M., and S. Perlman. 1987. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology 157:565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denison, M. R., S. A. Hughes, and S. R. Weiss. 1995. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology 207:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, S., and S. C. Baker. 1994. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology 204:541-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold, J., A. E. Gorbalenya, V. Thiel, B. Schelle, and S. G. Siddell. 1998. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 72:910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2): plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219-225. [PubMed] [Google Scholar]
- 14.Hughes, S. A., P. J. Bonilla, and S. R. Weiss. 1995. Identification of the murine coronavirus p28 cleavage site. J. Virol. 69:809-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. C., R. A. Spence, P. F. Currier, X. T. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor e64dd. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. (Pt. 81 10):2491-2496. [DOI] [PubMed]
- 18.Perlman, S., D. Reese, E. Bolger, L. J. Chang, and C. M. Stoltzfus. 1987. MHV nucleocapsid synthesis in the presence of cycloheximide and accumulation of negative strand MHV RNA. Virus Res. 6:261-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicki, D. L., and S. G. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 22.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 23.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 24.Wu, H. Y., J. S. Guy, D. Yoo, R. Vlasak, E. Urbach, and D. A. Brian. 2003. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81(Pt. 4):853-879. [DOI] [PubMed] [Google Scholar]
- 27.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]