SUMMARY
The Fanconi Anemia (FA) core complex provides the essential E3 ligase function for the FA pathway activation through the spatially defined FANCD2 ubiquitination. Of the seven FA gene products forming the core complex, FANCL possesses a RING domain with demonstrated E3 ligase activity. The other six components have no clearly defined roles. Through epistatic analyses, we identified three functional modules in the FA core complex: a catalytic module consisting of FANCL, FANCB, and FAAP100 is absolutely required for the E3 ligase function; the FANCA-FANCG-FAAP20 module and the FANCC-FANCE-FANCF module provide non-redundant and ancillary functions supporting the chromatin and DNA damage association of the catalytic module. Disruption of the catalytic module renders total loss of the core complex function whereas loss of any ancillary module component does not. Our work revealed the roles of several FA gene products with previously undefined functions and a modularized assembly of the FA core complex.
INTRODUCTION
Fanconi anemia (FA) is a complex genetic disorder encompassing 16 tumor suppressor genes that act together to protect cells against genotoxic stress, particularly complexed DNA lesions such as DNA interstrand crosslinks (Bogliolo et al., 2013; D'Andrea, 2010) and potentially DNA-protein crosslinks created by endogenous metabolites (Langevin et al., 2011; Rosado et al., 2011). Classical manifestations of FA include pancytopenia, chromosomal abnormalities, congenital abnormalities, and a high predisposition to a broad spectrum of cancers. Despite the identification of genetic defects in patients with FA, the molecular mechanism underpinning FA pathway functions remains unclear.
A group of classical FA genes is connected by a DNA damage-induced monoubiquitination reaction in the nucleus (Garcia-Higuera et al., 2001; Smogorzewska et al., 2007; Taniguchi et al., 2002). Monoubiquitination of the FANCD2/I complex has the presumed functions of recruiting DNA lesion-processing endonucleolytic activities (Knipscheer et al., 2009; Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010; Smogorzewska et al., 2010) and transcriptional activation of tumor suppressor genes (Park et al., 2013). The E3 ligase activity of this reaction resides in the FA core complex consisting of seven FA proteins (A, B, C, E, F, G, and L) and two FA-associated proteins (FAAP20 and FAAP100), with the RING domain protein FANCL bearing the E3 ligase activity (Alpi et al., 2008; Meetei et al., 2003). Aside from FANCL and FAAP20, most other components of the core complex have neither recognizable motifs nor clearly defined functions as to how they contribute to the DNA damage-mediated FANCD2/I monoubiquitination.
Studies of protein-protein interactions within the FA core complex have suggested the existence of three sub-complexes (Fig. 1A). FANCA, FANCG, and FAAP20, form a subcomplex (A-G-20) (Ali et al., 2012; Garcia-Higuera et al., 1999; Kruyt et al., 1999; Reuter et al., 2000; Waisfisz et al., 1999). The UBZ domain of FAAP20 is suggested to bind to ubiquitinated histone (Leung et al., 2012; Yan et al., 2012). FANCG contains seven TPR repeats and is considered a possible scaffold for the subcomplex (Blom et al., 2004; Léveillé et al., 2004). The FANCB-FANCL-FAAP100 sub-complex (B-L-100) contains the E3 ligase FANCL (Ling et al., 2007; Medhurst et al., 2006). Given that FANCL alone acts sufficiently in reconstituted ubiquitination reactions (Alpi et al., 2008; Longerich et al., 2009; Sato et al., 2012), whether FANCB and FAAP100 contribute to the E3 activity is unclear. A third sub-complex is formed by FANCC, FANCE, and FANCF (C-E-F). FANCF has been shown to interact with FANCM (Deans and West, 2009) and was also suggested to act as an adaptor protein (Léveillé et al., 2004). Depite these observations, the function of each sub-complex and how they integrate together in the context of the FA core complex remain largely unclear.
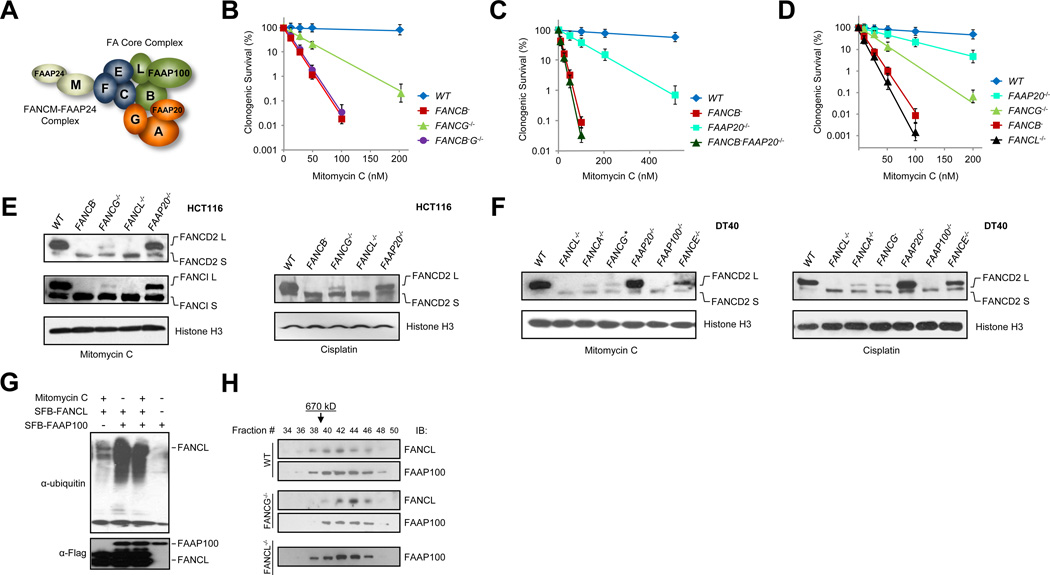
Fig. 1. Various sensitivities of FA knockout mutants identify the FA core complex catalytic module.
(A) Depiction of three protein interaction modules within the FA core complex and the FANCM-FAAP24 complex. See also Fig. S1A-C.
(B) Clonogenic survival of wild-type HCT116 (WT), FANCB−, FANCG−/−, and FANCB− G−/− mutants treated with mitomycin C. See also Fig. S1D-E and S2.
(C) Clonogenic survival of wild-type HCT116 (WT), FANCB−, FAAP20−/−, and FANCB− FAAP20−/− mutants treated with mitomycin C. See also Fig. S2.
(D) Clonogenic survival of wild-type HCT116 (WT), FAAP20−/−, FANCG−/−, FANCB−, and FANCL−/− mutants treated with mitomycin C.
(E) Immunoblots detecting MMC- or cisplatin-induced monoubiquitination of chromatin-bound FANCD2 and FANCI in wild-type HCT116 cells (WT) and the indicated knockout mutants. See also Fig. S3A-B.
(F) Immunoblot detecting MMC- or cisplatin-induced monoubiquitination of chromatin-bound FANCD2 in wild-type chicken DT40 (WT) and indicated knockout mutants. *FANCG gene localizes in the single Z chromosome and only one allele exists in DT40 cells. See also Fig. S3C-D.
(G) Immunoblots detecting FANCL auto ubiquitination in protein extracts prepared from 293T cells stably expressing SPB-FAAP100 and/or SFB-FANCL with or without MMC treatment.. Protein extracts from indicated cell lines were subjected to S beads pull-down and immunoblotted by anti-ubiquitin or Flag antibodies.
(H) Superose 6 gel filtration profiling of the FA core complex in HCT116 WT, FANCG−/−, and FANCL−/− mutants. Nuclear extracts were fractionated by Superose 6 gel filtration and the indicated fractions were immunoblotted (IB:) with FANCL or FAAP100 antibodies. The arrow indicates the elution position of the FA core complex (670kD). Fraction 21 marks the void.
Error bars in survival curves were derived from SDs from 4–6 independent experiments with triplicates.
Because that conservation of the classical FA pathway is primarily within vertebrates, typical genetic platforms such as yeast or Drosophila could not be employed to reveal the interaction among FA genes. In this study, we undertook an epistatic analysis approach with mammalian and chicken DT40 cells to elucidate the functions of the FA core components. By generating a series of isogenic loss-of-function single and double mutants of key core complex genes, we find that loss of different FA core components gave rise to variable impacts on the activation of the FA pathway and correspondingly variable cellular sensitivities to crosslinking reagents. Here, we present evidence that the differential sensitivities reflect distinct functions of the FA core complex components, suggesting a modularized functional assembly of the core complex that enables it to carry out the spatially defined monoubiquitination reaction upon DNA damage.
RESULT
Differential Damage Sensitivities of FA Core Component Mutants
FA core complex components form three protein interaction modules: A-G-20, B-L-100, and C-E-F, as suggested by several groups and us (Léveillé et al., 2004; Ling et al., 2007; Medhurst et al., 2006; Yan et al., 2012) (Fig. 1A and Fig. S1A-C). To address the functionality of each protein interaction module in cellular survival against DNA damage, we created FANCG−/−, FANCB−, and a FANCG−/−FANCB− double knockout mutants via homologous replacement targeting (Fig. S2A-B, D) in the HCT116 parental cell background. The strict isogenicity among these knockout mutants allows direct comparison of cellular survival outcomes as a direct reflection of protein function. Using clonogenic survival assay, we found that the FANCG−/− mutant was significantly more resistant than FANCB− to crosslinking DNA damage (Fig. 1B and Fig. S1D-E), but deletion of both genes did not produce any additional sensitivity over that of FANCB−. These differential sensitivities suggest that FANCB has an epistatic yet more important role than FANCG in cellular resistance to crosslinking DNA damage. Because FANCG and FAAP20 reside in the same module, we next tested FAAP20−/− and FAAP20−/− FANCB− knockout mutants (Fig. 1C and Fig. S2E) and observed a similar survival profile, which implicates an epistatic but more essential function of FANCB in cellular resistance to crosslinking reagents.
Loss of FANCB led to marked decrease of FANCL protein levels (Fig. S1A). Thus it is plausible that the strong damage sensitivity of the FANCB− mutant is attributable to its presence in the same protein-association module with FANCL and to its ability to stabilize FANCL. To test this premise, we compared four isogenic HCT116 knockout mutants (Fig. 1D) for their crosslinking damage sensitivity and found that loss of FANCL or FANCB rendered comparable but much more severe phenotypes than loss of FANCG or FAAP20. The different degrees of sensitivities from distinct components of the FA core complex seems to indicate distinct functions executed by the two different protein modules.
FANCD2 monoubiquitination is a critical determinant of cell survival against crosslinking agents. We reasoned that the differential sensitivities of the different FA core component mutants might reflect their ability to influence the level of FANCD2 monoubiquitination. To verify this notion, we examined a panel of HCT116 (human) and DT40 (chicken) somatic cellular knockout mutants for DNA damage-induced FANCD2 monoubiquitination, using chromatin fractions prepared from cells exposed to mitomycin C or cisplatin. We found that mutants devoid of FANCL, B, or FAAP100 exhibited complete loss of FANCD2 and FANCI monoubiquitination, whereas loss of FANCA, G, E, or FAAP20 showed readily detectable levels of FANCD2 and FANCI monoubiquitination (Fig. 1E-F). To ascertain that the absence or reduction of FANCD2 monoubiquitination was not a result of differential FANCD2 protein loss in the knockout mutants, chromatin-bound and soluble protein fractions were tested for their FANCD2 levels. We found (Fig. S3) that each knockout mutant maintained an amount of total FANCD2 comparable to that in the parental HCT116 or DT40 cells.
To explore the potential function of FAAP100 in the FA pathway activation, we co-expressed SFB-tagged FANCL and/or FAAP100 in 293T cells and analyzed FANCL auto ubiquitination in the presence of absence of mitomycin C (Fig. 1G). We found that overexpression of FAAP100 increased strongly FANCL autoubiquitination, suggesting that FAAP100 may have functions in helping FANCL engaging its targets in a cellular context. These results collectively suggest that FANCL, B, and FAAP100 constitute a catalytic core that is absolutely essential for the E3 ligase activity of the FA core complex. These three proteins most likely form a highly integrated module as demonstrated by a strong interdependency of protein stabilities where loss of FANCB is accompanied by drastic decreases of both FANCL and FAAP100 (Fig. S1C) (Ling et al., 2007; Medhurst et al., 2006).
On the other hand, the A-G-20 and C-E-F modules seem to have auxiliary roles in FA core complex function. However, complete loss of the E3 ligase activity of the core complex may also arise from disruption of the entire core complex. To examine this possibility, we performed gel filtration profiling of FA core component knockout mutants and found that deletion of FANCG and FANCL did not result in a gross disruption of the core complex (Fig. 1H), which is reflected by the similar elution positions of the mutant core complex to that of the wild type FA core complex (670 kD) (Fig. 1H).
Chromatin and DNA Damage Association of the Core Complex Depend on both the A-G-20 and the C-E-F Modules
E3 ligases are either monomeric, such as HECT domain-containing E3 ligases, or multi-subunit such as cullin-based E3s with adaptor and substrate-recognition components in addition to the catalytic subunit (Metzger et al., 2012; Zimmerman et al., 2010). Even though the FA core complex contains nine subunits, FANCL was found to be singly competent in monoubiquitinating FANCD2/I in vitro together with Ube2t (Alpi et al., 2008; Hodson et al., 2013; Longerich et al., 2009; Sato et al., 2012), suggesting that the A-G-20 and C-E-F modules may have functions autonomous to the E3 catalytic core module. Because the FA core complex is recruited to chromatin upon DNA damage, monoubiquitination of FANCD2/I most likely takes place in a spatially defined fashion which entails the core complex to be localized to the site of DNA lesions.
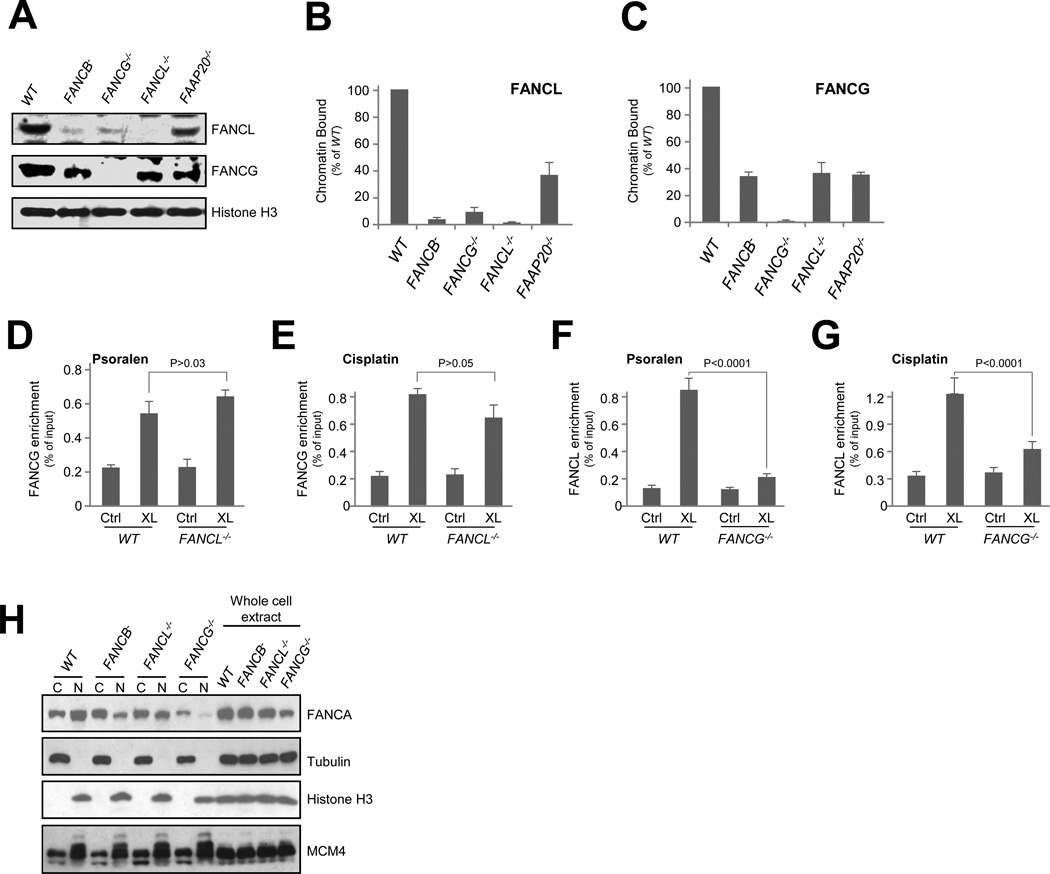
To search for such modular activities, we measured damage-induced chromatin loading of FANCL in FANCB−, FANCG−/−, and FANCL−/− mutants (Fig. 2A-B). As expected, much reduced FANCL chromatin presence was found in FANCB− mutant because of its reduced FANCL protein level. However, despite normal FANCL protein abundance, FANCL chromatin binding in FANCG−/− was drastically reduced to approximately 10% that of the wild type cells. This result suggests that chromatin retention of the E3 ligase activity relies on the presence of FANCG. Deletion of FAAP20 led to partial loss of FANCL chromatin binding, indicating a minor role of FAAP20 in FANCL chromatin association. Conversely, FANCG chromatin binding, albeit weakened compared to wild type cells, maintained a significant level in FANCB− and FANCL−/− mutant cells (Fig. 2A, C). Thus, the A-G-20 module seems to be able to associate with chromatin in the absence of the FANCL-based catalytic module, raising the possibility that the A-G-20 module supports the localization of the catalytic core to the site of DNA lesions.
Fig. 2. The A-G-20 module is required for FANCL association with DNA damage.
(A) Chromatin binding of FANCG and FANCL in wild-type HCT116 cells (WT) and the indicated mutants exposed to mitomycin. Histone H3 served as a loading control for chromatin-bound protein fractions.
(B) Quantification of chromatin-bound FANCG in FANCB−, G−/−, L−/−, and FAAP20−/− knockout mutants.
(C) Quantification of chromatin-bound FANCL in FANCB−, G−/−, L−/−, and FAAP20−/− knockout mutants.
(D) eChIP analysis of FANCG enrichment to a defined psoralen lesion in wild-type HCT116 cells (WT)and the FANCL−/− mutant.
(E) eChIP analysis of FANCG enrichment to a cisplatin-adducted plasmid substrate in wild-type HCT116 cells (WT) and the FANCL−/− mutant.
(F) eChIP analysis of FANCL enrichment to a defined psoralen lesion in wild-type HCT116 cells (WT) and the FANCG−/− mutant.
(G) eChIP analysis of FANCL enrichment to a cisplatin-adducted plasmid substrate in wild-type HCT116 cells (WT) and the FANCG−/− mutant.
(H) Immunoblotting of FANCA in cytoplasmic (C), nuclear (N), and whole cell protein extracts from FANCB−, FANCL−/−, and FANCG−/− mutant cells. Tubulin, Histone H3, and MCM4 are controls for loading and extraction.
Error bars for chromatin fraction and eChIP analyses represent SDs derived from three independent experiments.
To validate this notion, we used the eChIP (Shen et al., 2009) assay with psoralen- or cisplatin-damaged DNA substrates to assess the DNA damage-dependent enrichment of FANCG and FANCL in FANCL−/− and FANCG−/− mutants, respectively. Consistent with the chromatin fractionation results, we found that damage-dependent FANCG enrichment does not require FANCL (Fig. 2D-E), whereas enrichment of FANCL strongly depends on the presence of FANCG (Fig. 2F-G). Together, these observations support that the A-G-20 module likely acts as a chromatin-binding component of the FA core complex.
Cell biology evidence suggests that FANCA nuclear localization relies on its interaction with FANCL or FANCB for nuclear localization (Medhurst et al., 2006). Thus, it is plausible that FANCB−/− and FANCL−/− mutant phenotypes and the composition of the FA core complex may also be complicated by a potential absence of FANCA from the nucleus. To determine how FANCA localization is affected in the FANCB− and FANCL−/− mutants, we analyzed whole cell, cytoplasmic, and nuclear extracts prepared from FANCG−/−, FANCB−/−, and FANCL−/− cells. As shown in Fig. 2H, significant nuclear presence of FANCA is found in both FANCB−/−, and FANCL−/− cells despite moderately reverted cytoplasmic/nuclear ratios when compared to WT cells. Loss of FANCG resulted in reduced FANCA in both nuclear and cytoplasmic fractions. These observations suggest that loss of FANCB or FANCL do not eliminate the A-G-20 module, consistent with the positive detection of FANCA-FANCG interaction in FANCB−/− and FANCL−/− cells (Fig. S1B).
Upon DNA damage, the FA core complex is initially loaded onto the chromatin by the FANCM/FAAP24 complex via interacting with FANCF (Fig. S4A) (Deans and West, 2009; Wang et al., 2013b), suggesting that the FANCF-containing module (C-E-F) may serve as a separate chromatin-binding element within the FA core complex. Moreover, the residual level of FANCL chromatin binding in the FANCG−/− mutant (Fig. 2A-B) also indicates that, aside from FANCG, a separate factor acts in the damage-induced chromatin enrichment of FANCL. To test whether the C-E-F module contributes independently to the full extent of FANCD2 ubiquitination, we depleted FANCF in wild type HCT116 cells (Fig. S4B-F) and found that loss of FANCF alone resulted in a moderate reduction of FANCD2 monoubiquitination and chromatin-bound FANCL (Fig. 3A, B), indicating a chromatin-association function for FANCF. When FANCF was depleted in the FANCG−/− mutant background, FANCD2 monoubiquitination as well as FANCL chromatin enrichment were completely abolished. However, FANCF chromatin-binding was not affected in FANCG−/− cells treated with mitomycin C (Fig. S4G). Together, these results suggest that the A-G-20 and C-E-F modules can act independently to facilitate the chromatin binding of the catalytic module.
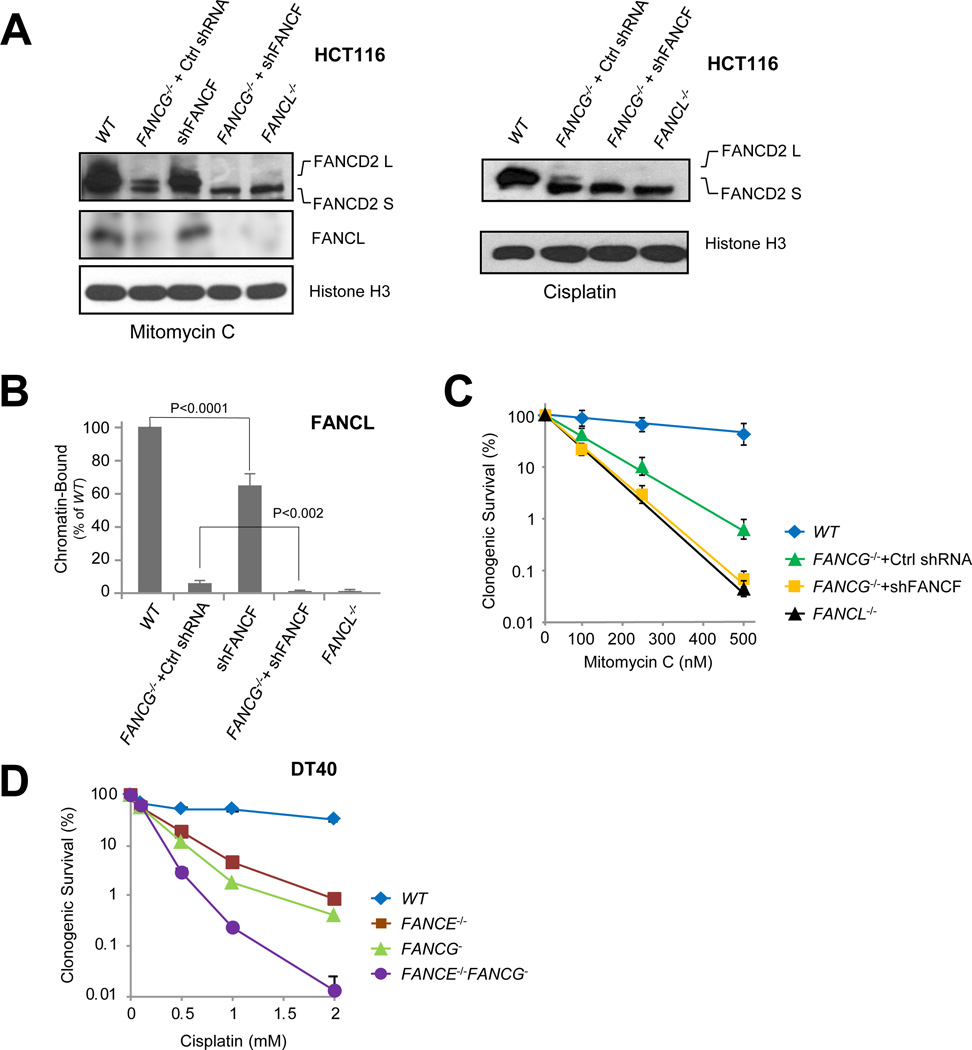
Fig. 3. The A-G-20 and C-E-F modules exhibit non-redundant functions in the chromatin recruitment of the FA core complex and resistance to crosslink damage.
(A) Immunoblot detecting monoubiquitination of FANCD2 and chromatin-bound FANCL in cells exposed to mitomycin C (left panel) or cisplatin (right panel) in wild-type HCT116 (WT) and the indicated knockout/knockdown mutant cells. See also Fig. S4.
(B) Quantification of chromatin-bound FANCL (left panel of A) in wild-type HCT116 (WT), FANCG−/− + Ctrl shRNA, WT + shFANCF alone (shFANCF), FANCG−/− + shFANCF, and FANCL−/− cells exposed to mitomycin C. See also Fig. S4.
(C) Clonogenic survival of parental HCT116 (WT), FANCG−/− + Ctrl shRNA, FANCG−/− + shFANCF, and FANCL−/− cells treated with mitomycin C. See also Fig. S4.
(D) Clonogenic survival of parental DT40 (WT), FANCE−/−, FANCG−, and FANCE−/− FANCG− cells treated with cisplatin. See also Fig. S2C.
Error bars for chromatin-bound FANCL quantification (B) and clonogenic survivals (C and D) were derived from SDs from three or more independent tests.
Consistently, cells devoid of both FANCG and FANCF (FANCG−/− + shFANCF) exhibited much profound sensitivity to mitomycin C as opposed to loss of FANCG or FANCF alone, comparable to that of the FANCL−/− mutant (Fig. 3C). The much heightened sensitivity of FANCG−/− + shFANCF cells most likely reflects a complete loss of FANCD2 monoubiquitination resulting from the complete absence of the catalytic module at the site of DNA damage. Given that FANCE and FANCF reside in the same module, we further validated the non-redundant functions of the A-G-20 and C-E-F modules by generating a FANCE−/−FANCG− double knockout mutant in DT40 cells and compared its clonogenic survival against that of FANCE−/− and FANCG− single mutants (Fig. 3D, Fig. S2C). Loss of both FANCG and FANCE led to drastically increased cellular sensitivity, implicating independent functions of A-G-20 and C-E-F modules, possibly through promoting the recruitment of the core complex to the site of DNA damage.
FANCM Functions Epistatically with the C-E-F Module in FA Core Complex Chromatin Association
The above results suggest that FANCF acts as a chromatin-enrichment factor for the FA core complex via its interaction with FANCM. As such, depletion of both FANCM and FANCF is expected to yield no additional phenotype than loss of FANCM alone as loss of one or both proteins lead to disrupted core complex loading. To test this premise, we analyzed FANCD2 monoubiquitination in FANCM−/− cells stably depleted of FANCF. As shown in Figure 4A-B, loss of FANCF in FANCM−/− cells produced no additional loss of FANCD2 or FANCI monoubiquitination upon DNA damage. Consistently, loss of both FANCM and FANCF (FANCM−/− + shFANCF) also failed to acquire additional cellular sensitivity over that of the FANCM−/− mutant alone (Fig. 4C). Therefore, FANCM and FANCF functions appear epistatic in supporting the chromatin association of the core complex.
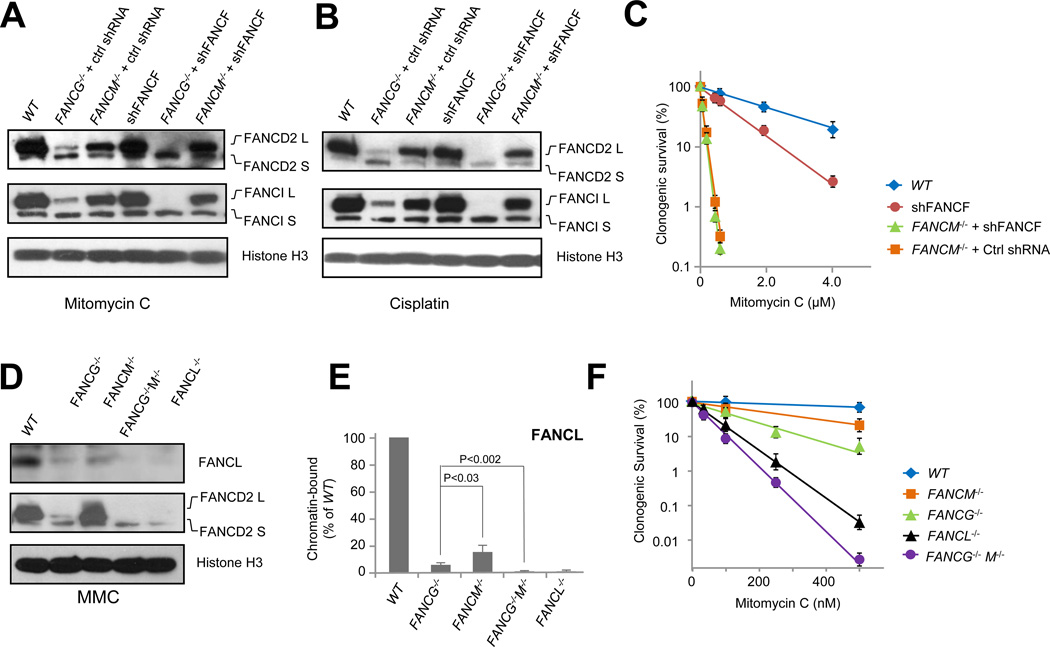
Fig. 4. FANCM functions epistatically with the C-E-F module in FA core complex chromatin association.
(A) Immunoblot detecting mitomycin C-induced monoubiquitination of FANCD2 and FANCI in wild-type HCT116 cells (WT) and the indicated knockout/knockdown mutant cells. See also Fig. S4.
(B) Immunoblot detecting cisplatin-induced monoubiquitination of FANCD2 in wild-type HCT116 cells (WT) and the indicated knockout/knockdown mutant cells. See also Fig. S4.
(C) Clonogenic survival of HCT116 FANCF knockdown alone (shFANCF), FANCM−/− +shFANCF, and FANCM−/− +Ctrl shRNA mutants treated with mitomycin C. See also Fig. S4.
(D) Immunoblot detecting MMC-induced monoubiquitination of FANCD2 and chromatin binding of FANCL in wild-type HCT116 cells (WT) and the indicated mutant cells exposed to mitomycin C. Histone H3 serves as a loading control for chromatin-bound protein fractions. See also Fig. S2F.
(E) Quantification of (C) - chromatin-bound FANCL in FANCG−/−, FANCM−/−, and FANCG−/− M−/− double mutants. See also Fig. S2F.
(F) Clonogenic survival of wild-type HCT116 cells (WT), FANCM−/−, FANCG−/−, FANCL−/−, and FANCG−/− M−/− mutants treated with mitomycin C. See also Fig. S2F.
Error bars for chromatin fraction represent SDs derived from three independent experiments. Error bars for clonogenic survival are derived from SDs from four ndependent tests done in triplications.
In contrast, the A-G-20 and C-E-F modules seems to possess independent functions in supporting chromatin binding of the catalytic module. To completely abolish DNA/chromatin binding of the core complex and FA pathway-mediated cell survival, both modules have to be disrupted. As shown in Figure 3, disruption of the C-E-F and A-G-20 modules led to compromised FA pathway activation and ICL resistance. To further verify the non-redundant recruitment functions of the A-G-10 and the C-E-F modules, we constructed a FANCG−/−FANCM−/− double mutant (Fig. S2F). Although FANCG−/− and FANCM−/− both retained significant levels of chromatin-bound FANCL, the FANCG−/−FANCM−/− double mutant completely lost chromatin-bound FANCL (Fig. 4D, E). Congruent with this observation, only the ablation of both FANCG and FANCM yielded complete loss of FANCD2 monoubiquitination (Fig. 4D), further suggesting that the A-G-20 module and the FANCM-dependent C-E-F module provide a combined recruitment for the FA core complex to execute the spatially-defined FANCD2/I monoubiquitination reaction. Consistently, the DNA damage sensitivity of the FANCG−/−FANCM−/− double mutant was much more pronounced than that of each single mutant (Fig. 4F), demonstrating the functional impact of eliminating both recruiting elements of the FA core complex. The additional sensitivity of the FANCG−/−FANCM−/− mutant over that of the FANCL−/− mutant is most likely attributable to FANCM functions independent of the FA mechanism (Huang et al., 2010; Wang et al., 2013b).
DISCUSSION
Through creation of isogenic single and double mutants of the FA genes, we came to the unexpected finding that not all the components of the core complex contribute equally to cellular resistance against DNA damage. This observation deviates from a general paradigm that losing any one of the core FA proteins leads to complete elimination of FANCD2 activation and disintegration of the core complex. Instead, our results suggest that different functional modules exist in the core and that the overall integrity of the core complex is sustained when certain FA proteins are removed.
Our work establishes a modularized functional assembly of the FA core complex consisting of a catalytic module and two modules with non-redundant functions in the chromatin recruitment of the core complex. The coordinated actions of these three modules enable the E3 ligase activity to be localized to the sites of DNA damage and carry out the spatially-defined FANCD2/I monoubiquitination with maximum efficiency to counter DNA damage. The catalytic core module is the most critical component functionally, as reflected by the most severe phenotypes of the FANCL and FANCB mutants. The reason why the FANCL E3 ligase needs to form a catalytic core with FANCB and FAAP100 is unclear. A plausible scenario could be that FANCB and FAAP100 play a role in substrate engagement or stimulating the E3 ligase activity of FANCL as reflected by the fact that overproduction of FAAP100 leads to much elevated levels of FANCL activity (Fig. 1G). Gel filtration profiling of FA core component knockout mutants showed that deletion of FANCG and FANCL did not result in a gross disruption of the core complex (Fig. 1H). However, it is possible that certain core components are critical for retaining the overall integrity of the complex (Gordon et al., 2005). Additional studies may be required to verify whether such structural components for the core complex exist.
We have demonstrated that FANCF and FANCM have epistatic roles in recruiting the core complex to the site of DNA damage. The upstream role of FANCM in chromatin-binding of the FA core complex suggests that the C-E-F sub complex may serve as an initial loading module for the core complex. When the A-G-20 or the C-E-F module is individually defective, the monoubiquitination is weakened but not fully diminished. Disruption of both, however, eliminates the chromatin binding and DNA damage localization of the core complex. As a result, the FANCL E3 ligase activity is no longer in the vaccinity of DNA-bound FANCD2/I, which is a much more robust substrate than the unbound form.
Our model of nonredundant recruitment functions of the A-G-20 and the C-E-F modules may also explain the enhanced phenotypes of the FANCC-FANCG double knockout mice (Pulliam-Leath et al., 2010). How the A-G-20 module promote the chromatin E3 ligase activity remains unclear. FANCA has been shown to exhibit DNA binding affinity in vitro (Yuan et al., 2012). FAAP20 can bind to ubiquitinated forms of H2A (Yan et al., 2012), although the weak phenotypes of FAAP20 mutant suggest a secondary role for FAAP20 in chromatin association of the core complex. These biochemical properties could afford the A-G-20 module the ability to bind to DNA/chromatin.
FANCM is known to be important for the chromatin association of the core complex. While deletion of FANCM results in substantial loss of steady state levels of FANC core complex binding to chromatin, the decrease in FANCD2 monoubiquitination is moderate (Wang et al., 2013a; Wang et al., 2013b). It is likely that the A-G-20 module, in the absence of the FANCM-mediated recruitment, is sufficient to support a transient presence of the E3 ligase activity at the site of DNA damage. This notion is substantiated by the complete elimination of FANCD2 monoubiquitination in the FANCG−/−M−/− double mutant and in the FANCG−/− FANCF-knockdown cells.
The modular model of the FA core complex offers novel insights into several FA genes that had no previously defined functions. However, further validaton of this model requires biochemical and structural investigations. The varying importance of different core components may also contribute to the skewed prevalence of gene mutations in FA patients. Mutations in FANCA, C, and G together account for nearly 90% of all patient mutations, whereas FANCB and FANCL patients, account for fewer than 1% (Neveling et al., 2009). This biased distribution may reflect the functional importance of each gene in the FA pathway. Our results refine and advance the molecular understanding of this important DNA damage response and tumor suppressor pathway by revealing the functional divergence among the FA core complex components.
EXPERIMENTAL PROCEDURES
Cell culture and antibodies
HCT116 cells, somatic knockout mutants, and HEK293T cells were all cultured in DMEM plus 10% fetal calf serum (FBS) at 37°C. DT40 wild type and knockout cells were generated via published procedures and cultured in RPMI-1640 medium plus 10% FBS, 1% chicken serum, 5 µM 2-mercaptoethanol, and 10mM HEPES, at 39.5°C.
Antibodies against FANCL, FAAP100 (affinity purified), FAAP20, FANCM, and anti-chicken FANCD2 were a kind gift from Weidong Wang’s laboratory (NIA, NIH). Commercial antibodies used are as follows: anti-human FANCD2 (Abcam ab2187, Santa Cruz SC-20022, Novus NB100-182); anti-human FANCI (Bethyl A300-254A); anti-human FANCA (Bethyl A301-980A); anti-human FANCG (Novus NB100-2566); anti-histone H3 (Millipore 05-928); anti-β-tubulin (Sigma-Aldrich T4026); anti-FLAG M2 (Sigma-Aldrich F3165).
Construction of somatic cellular knockout mutants
Targeting vectors for human HCT116 cells were constructed using the USER system as described previously (Wang et al., 2013b). Each mutant was validated by complementation with a wild type cDNA of the targeted gene. FANCE DT40 mutant was constructed by replacing a 5.3 kb coding area (emcompassing 10 exons) with Bsr resistance gene cassette. Positivie clones were identified by Southern blotting with HindIII-digested genomic DNA. Targeting of FANCG in DT40 cells was done as previously described (Yamamoto et al., 2003). The FANCE−/−G− double mutant was constructed sequentially by targeting FANCG in a FANCE−/− mutant.
Clonogenic survival assay
1–3×105 HCT 116 or derivative mutants were seeded in 100 mm culture plate and allowed to recover overnight. Cells were exposed to various doses of DNA damage agents for 1 hr, washed, trypsinized, and re-seeded with appropriate densities in triplicate 100 mm plates. Colonies were fixed with glutaraldehyde 6.0% (v/v) and visualized with 0.5% crystal violet (w/v). Clonogenic survival of DT40 and derivative mutants against cisplatin was performed in medium contain 1.4% methylcellulose with continuous exposed to indicated concentrations of cisplatin (Nippon Kayaku, Tokyo, Japan).
Chromatin fractionation and Chromatin IP
To obtain chromatin-bound protein fractions, cells were lysed and centrifuged to collect insoluble pellet. The insoluble chromatin pellet was treated with 0.2 N HCl and centrifuged to collect the supernatant. The eChIP assay was carried out as previously described (Shen et al., 2009). Cisplatin-adducted plasmid substrate was prepared by incubating with 15 µM cisplatin for 3 hrs at 37°C in the dark, which produces on average 3 ICLs per kb.
Supplementary Material
Highlights.
-
-
Identification of FA gene products which form the catalytic core module
-
-
Revealing the role of FANCG-associated chromatin binding module
-
-
Implicating the role of the FANCF-mediated chromatin-association module
-
-
A novel model of modularized functional assembly of the FA core complex
Acknowledgments
This work was supported by NIH grants (CA127945 and CA97175-Project 3 to L.L; CA157448 to J.C; and Cancer Center Support Grant CA016672 to The University of Texas MD Anderson Cancer Center) and Grants-in aid from the Ministry of Education, Science, Sports, and Culture of Japan (JSPS KAKENHI 21390094 and MEXT KAKENHI 23114010, to M.T.). Yaling Huang is supported by the Hubert L. and Olive Stringer endowed professorship to L.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, Grassman E, Auerbach AD, Pang Q, Meetei AR. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119:3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic Insight into Site-Restricted Monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Molecular Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Blom E, van de Vrugt HJ, de Vries Y, de Winter JP, Arwert F, Joenje H. Multiple TPR motifs characterize the Fanconi anemia FANCG protein. DNA repair. 2004;3:77–84. doi: 10.1016/j.dnarep.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo Juan P, Minguillón J, Ramírez María J, Pujol R, et al. Mutations in ERCC4, Encoding the DNA-Repair Endonuclease XPF, Cause Fanconi Anemia. The American Journal of Human Genetics. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. The New England journal of medicine. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol. 1999;19:4866–4873. doi: 10.1128/mcb.19.7.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gordon SM, Alon N, Buchwald M. FANCC, FANCE, and FANCD2 form a ternary complex essential to the integrity of the fanconi anemia DNA damage response pathway. Journal of Biological Chemistry. 2005;280:36118–36125. doi: 10.1074/jbc.M507758200. [DOI] [PubMed] [Google Scholar]
- Hodson C, Purkiss A, Miles JA, Walden H. Structure of the Human FANCL RING-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection. Structure. 2013 doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D'Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Kruyt FA, Abou-Zahr F, Mok H, Youssoufian H. Resistance to mitomycin C requires direct interaction between the Fanconi anemia proteins FANCA and FANCG in the nucleus through an arginine-rich domain. The Journal of biological chemistry. 1999;274:34212–34218. doi: 10.1074/jbc.274.48.34212. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand crosslink repair. Proc Natl Acad Sci U S A. 2012;109:4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé F, Blom E, Medhurst AL, Bier P, Laghmani EH, Johnson M, Rooimans MA, Sobeck A, Waisfisz Q, Arwert F, et al. The Fanconi Anemia Gene Product FANCF Is a Flexible Adaptor Protein. Journal of Biological Chemistry. 2004;279:39421–39430. doi: 10.1074/jbc.M407034200. [DOI] [PubMed] [Google Scholar]
- Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 Acts with FANCI-FANCD2 to Promote DNA Interstrand Cross-Link Repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. The Journal of biological chemistry. 2009;284:23182–23186. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Déclais A-C, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA Repair Nuclease Recruited to DNA Damage by Monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhurst AL, Laghmani el H, Steltenpool J, Ferrer M, Fontaine C, de Groot J, Rooimans MA, Scheper RJ, Meetei AR, Wang W, et al. Evidence for subcomplexes in the Fanconi anemia pathway. Blood. 2006;108:2072–2080. doi: 10.1182/blood-2005-11-008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. Journal of Cell Science. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Endt D, Hoehn H, Schindler D. Genotype–phenotype correlations in Fanconi anemia. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2009;668:73–91. doi: 10.1016/j.mrfmmm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Park E, Kim H, Kim Jung M, Primack B, Vidal-Cardenas S, Xu Y, Price Brendan D, Mills Alea A, D'Andrea Alan D. FANCD2 Activates Transcription of TAp63 and Suppresses Tumorigenesis. Molecular cell. 2013;50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam-Leath AC, Ciccone SL, Nalepa G, Li X, Si Y, Miravalle L, Smith D, Yuan J, Li J, Anur P, et al. Genetic disruption of both Fancc and Fancg in mice recapitulates the hematopoietic manifestations of Fanconi anemia. Blood. 2010;116:2915–2920. doi: 10.1182/blood-2009-08-240747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter T, Herterich S, Bernhard O, Hoehn H, Gross HJ. Strong FANCA/FANCG but weak FANCA/FANCC interaction in the yeast 2-hybrid system. Blood. 2000;95:719–720. [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nature structural & molecular biology. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Research. 2012;40:4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, Wang W, Li L. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Molecular cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase–specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Waisfisz Q, de Winter JP, Kruyt FA, de Groot J, van der Weel L, Dijkmans LM, Zhi Y, Arwert F, Scheper RJ, Youssoufian H, et al. A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc Natl Acad Sci U S A. 1999;96:10320–10325. doi: 10.1073/pnas.96.18.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han X, Wu F, Leung JW, Lowery MG, Do H, Chen J, Shi C, Tian C, Li L, et al. Structure analysis of FAAP24 reveals single-stranded DNA-binding activity and domain functions in DNA damage response. Cell research. 2013a doi: 10.1038/cr.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung JW, Jiang Y, Lowery MG, Do H, Vasquez KM, Chen J, Wang W, Li L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell. 2013b;49:997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ishiai M, Matsushita N, Arakawa H, Lamerdin JE, Buerstedde J-M, Tanimoto M, Harada M, Thompson LH, Takata M. Fanconi Anemia FANCG Protein in Mitigating Radiation- and Enzyme-Induced DNA Double-Strand Breaks by Homologous Recombination in Vertebrate Cells. Molecular and Cellular Biology. 2003;23:5421–5430. doi: 10.1128/MCB.23.15.5421-5430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Guo R, Paramasivam M, Shen W, Ling C, Fox D, 3rd, Wang Y, Oostra AB, Kuehl J, Lee DY, et al. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell. 2012;47:61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Qian L, Zhao X, Liu JY, Song L, D'Urso G, Jain C, Zhang Y. Fanconi anemia complementation group A (FANCA) protein has intrinsic affinity for nucleic acids with preference for single-stranded forms. The Journal of biological chemistry. 2012;287:4800–4807. doi: 10.1074/jbc.M111.315366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Current Opinion in Structural Biology. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.