Summary
B cells produce a diverse antibody repertoire by undergoing gene rearrangements. Pathogen exposure induces the clonal expansion of B cells expressing antibodies that can bind the infectious agent. To assess human B cell responses to trivalent seasonal influenza and monovalent pandemic H1N1 vaccination, we sequenced gene rearrangements encoding the immunoglobulin heavy chain, a major determinant of epitope recognition. The magnitude of B cell clonal expansions correlates with an individual’s secreted antibody response to the vaccine and the expanded clones are enriched for those expressing influenza-specific mAbs. Additionally, B cell responses to pandemic influenza H1N1 vaccination and infection in different people show a prominent family of convergent antibody heavy chain gene rearrangements specific to influenza antigens. These results indicate that microbes can induce specific signatures of immunoglobulin gene rearrangements and that pathogen exposure can potentially be assessed from B cell repertoires.
Introduction
Human B cells generate a vast diversity of antibodies by rearranging immunoglobulin V (variable), D (diversity) and J (joining) genes in their genomes (Tonegawa, 1983). For decades, most monitoring of human antibody responses to infections or vaccines has been performed by serological measurements that can evaluate antibody specificities, while giving only limited insight into the underlying changes in clonal populations of B cells, or the gene rearrangements responsible for the antibodies. More recently, single-cell sorting and antibody gene cloning, as well as optimized culture systems and hybridoma generation, have given greater insight into the specificity and breadth of reactivity of the antibodies produced by influenza-specific B cells, and molecular understanding of the genes encoding such antibodies (Li et al., 2012; Wrammert et al., 2011; Wrammert et al., 2008; Yu et al., 2008). High-throughput DNA sequencing methods now permit detailed monitoring of B cell repertoires in humans, and are starting to be applied extensively to the study of vaccine responses (Boyd et al., 2009; DeKosky et al., 2013; Jiang et al., 2013; Krause et al., 2011; Liao et al., 2011; Wu et al., 2011).
It is largely unknown whether different people use similar antibody genes in their responses to common pathogen-associated antigens. With a few exceptions, such as the antibody responses to repetitive polysaccharide antigens (Ademokun et al., 2011; Park et al., 1996; Scott et al., 1989; Silverman and Lucas, 1991), there has been little evidence of similarity between different humans’ responses to most pathogens. Indeed, antibodies would themselves be expected to exert a selection pressure upon the pathogens they target, causing pathogens to avoid expressing antigens that are recognized by human antibody genes.
Here, we conduct a detailed study of B cell clonal expansions in response to influenza vaccination, and use deep sequencing to identify clonal expansion signatures within a week of vaccination that correlate with the magnitude of the serological response in vaccinated individuals. Comparison of expanded clones to influenza-specific plasmablasts identified by single cell sorting from the same subjects demonstrates substantial overlap between these populations. More surprisingly, we identify convergent antibody responses to the H1N1 2009 influenza strain that are shared among different people, both in response to vaccination and infection. These results represent an example of a signature in immunoglobulin gene rearrangements specific to the pathogen that elicited them, and suggest that features of an individual’s history of pathogen exposure can be identified by sequence analysis.
Results
Deep sequencing of rearranged IGH from the trivalent inactivated seasonal influenza vaccine response
To take an overview of B cell responses induced by vaccination, we carried out deep sequencing of IGH from the peripheral blood B cells of 14 healthy young individuals vaccinated with the 2007 or 2008 trivalent inactivated seasonal influenza vaccine (TIV) (Moody et al., 2011). Seven individuals were ‘seroconverters’ who raised at least a 4-fold increase in titer above baseline to 2 or more vaccine antigens as measured by ELISA against purified hemagglutinins (HA). The other 7 were ‘non-seroconverters’ that failed to increase their vaccine-specific antibody to meet these criteria (Table S1) (Moody et al., 2011).
Twelve replicate IGH libraries were prepared from independent genomic DNA template aliquots from cryopreserved peripheral blood mononuclear cells for each individual at each of 3 time points: pre-vaccination, day 7 and day 21 post-vaccination (Figure 1A). On average, 35,436 IGH sequences were analyzed for each individual. Sequencing depth was relatively evenly distributed across the time points with an average 11,661 IGH sequences pre-vaccination, 12,200 at day 7 and 11,564 at day 21.
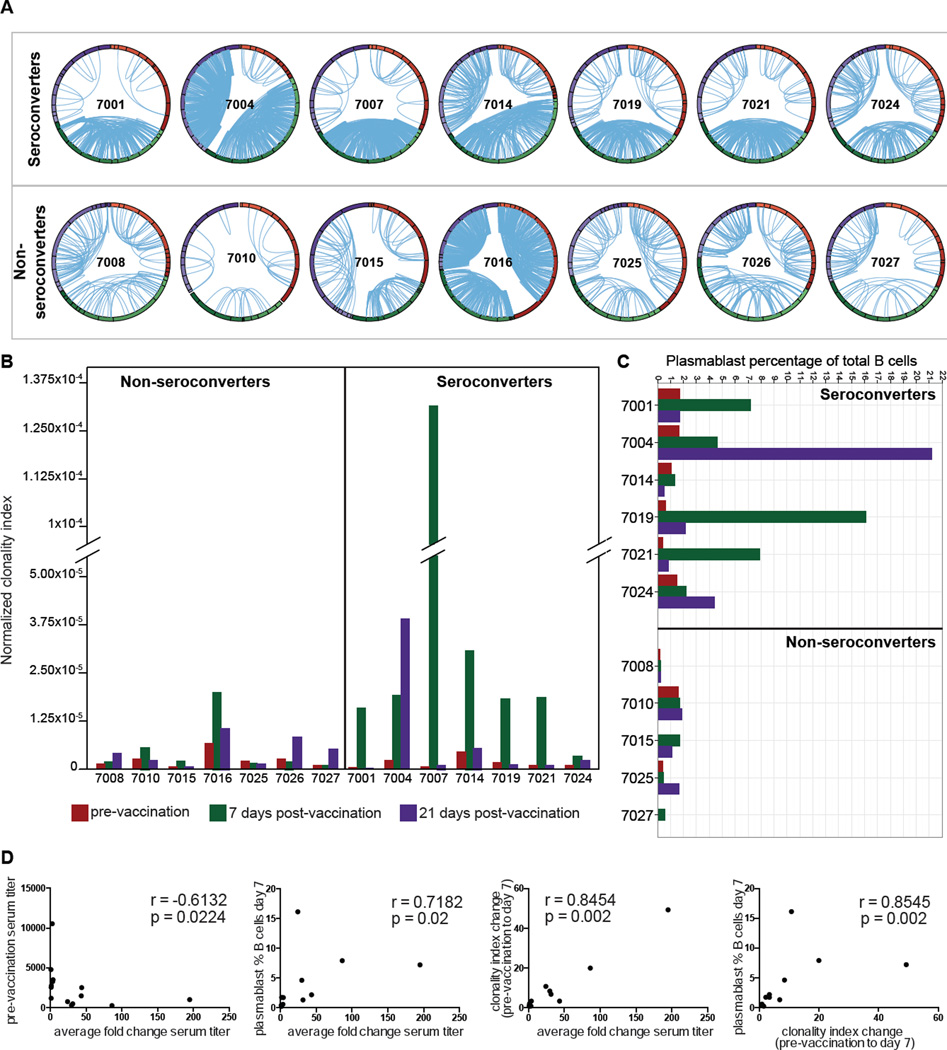
Figure 1. Quantitation of clonal B cells in the blood following vaccination predicts seroconversion.
(A) Replicate IGH libraries were generated from peripheral blood B cells for 14 individuals (Table S1) at three time points: pre-vaccination (red arc), day 7 (green arc) and day 21 post-vaccination (purple arc). Replicates are shown as bands within each arc and lines connect clonally related VDJs from independent replicates. Detailed IGH repertoires for each individual are shown in Figure S1A. Figure S1B presents criteria for definition of clonal lineages. (B) Normalized clonality index scores; pre-vaccination (red), 7 days (green) and 21 days post-vaccination (purple). (C) Plasmablast percentages of total B cells (Table S3); pre-vaccination (red) and at days 7 (green) and 21 (purple) post-vaccination. (D) Correlation between metrics and serological antibody response.
B cell clonal signatures from deep sequencing correspond to serological measures of vaccine response
Clonally related B cell lineages were identified by the presence of identical, or near-identical, IGH in independent replicate sequence libraries from genomic DNA for each time point. This approach ensures that high expression of antibody gene mRNA in individual cells, or amplification biases, are not misinterpreted as evidence for clonal B cell populations. Most seroconverters showed a response with 1 to 3 larger clones and variable numbers of smaller clones, although subject 7024 was an exception with predominance of smaller lineages. The median number of expanded clones for seroconverters at day 7 was 69 (range 39–92) compared to 25 (range 8–85) for the non-seroconverters.
To compare the clonal signatures between samples we used a clonality index previously described (Wang et al.). The clonality index is a scale-independent normalized measure that reflects the probability that two independent rearrangements derive from clonally-related B cells (Figure 1B). Each of the seroconverters showed a prominent increase in clonality at day 7 compared to pre-vaccination (Figure S1A). Measured changes in clonality from pre-vaccination to day 7 ranged from 3.38- to 247.57-fold among seroconverters. Non-seroconverters showed a mixture of modestly increased (4 individuals) and modestly decreased (3 individuals) indices (Figure 1B).
The change in B cell clonality by day 7 was positively correlated to the fold change in antibody titer against vaccine HA antigens as measured by ELISA 21 days post-vaccination (Spearman r = 0.8454, p-value = 0.002, Figure 1D). Non-seroconverter 7016 showed a partial response, increasing their titer 1.58-fold for combined TIV antigens, and showing a 2.98-fold increase in clonality index, but with a strong response to one vaccine component (5.39-fold change to A/Brisbane/10/2007/H3).
Plasmablast counts in blood following vaccination have been reported to correlate with serological responses (Liao et al., 2011) (Sasaki et al.). Day 7 plasmablast frequencies (Table S3) in the samples studied here showed significant correlation with the clonality index (r=0.8545, p-value=0.002, Figure 1D), and a slightly weaker correlation with changes in serum antibody titer (r=0.7182, p=0.02, Figure 1D). The difference in correlation of plasmablast frequencies and the clonality index to the change in vaccine specific antibody titer did not however rise to statistical significance (p=0.1214, Steiger’s dependent variable correlation) (Steiger, 1980). Consistent with prior literature, pre-vaccination titers were negatively correlated with vaccine-stimulated titer changes (Figure 1D) (Sasaki et al.).
Clones from IGH deep sequencing comprise a subset of antigen-specific plasmablasts
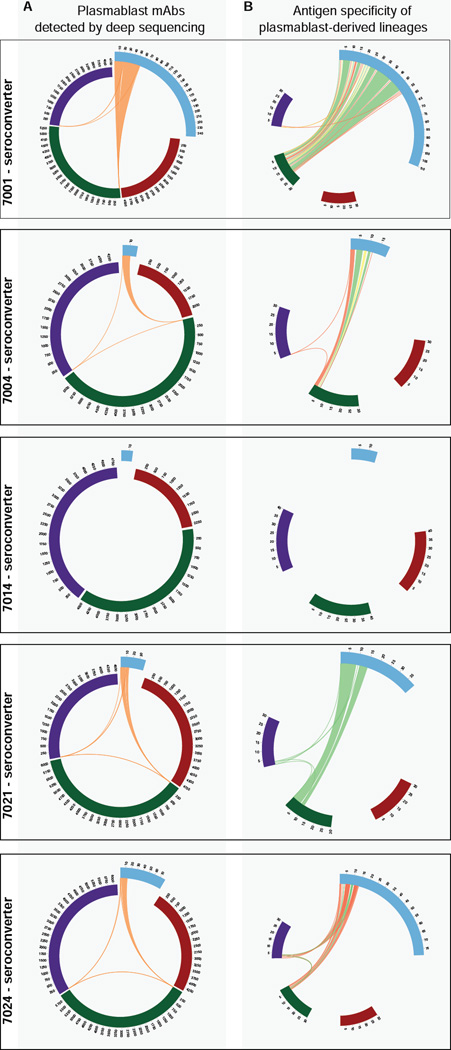
To directly evaluate the influenza specificity of the detected B cell expansions, and assess if these represent members of the plasmablast pool, the data was compared to IGH from flow-sorted plasmablasts isolated from the same day 7 samples for 5 of the seroconverters (7001, 7004, 7014, 7021, 7024). A total of 398 sorted plasmablasts were expressed as recombinant monoclonal antibodies (mAbs) and their antigen specificities were evaluated as part of a previous study (Krause et al., 2011; Moody et al., 2011; Wrammert et al., 2011; Wrammert et al., 2008) with 59.8% being influenza-reactive.
Analysis revealed that 24.4% of sorted plasmablasts belonged to clonally-expanded lineages containing IGH identified with sequencing (Figure 2A; Table S2). Sixty-six percent of the plasmablast-derived IGH in these lineages were from influenza-HA binding mAbs (Figure 2B; Table S2). Conversely, 10.2% of the day 7 expanded clones were in lineages that included plasmablast IGHs. Subject 7001, who contributed 247 mAbs (Moody et al.), had 19.6% of day 7 clonally-expanded B cell lineages shared with plasmablast IGHs, and 82.4% of these included influenza-specific mAbs (Figure 2B, Table S2). This indicates that at day 7 post-vaccination there is partial overlap of clonally-expanded IGH and plasmablasts sorted based on their immunophenotype.
Figure 2. Antigen specificity of vaccine-induced B cell clonal populations.
(A) B cell lineages with members from mAbs derived from day 7 sorted plasmablasts (blue arc) and deep sequenced IGH from peripheral B cells prior to (red) and at days 7 (green) and 21 (purple) post TIV vaccination. Lines join lineage members. (B) Antigen binding of plasmablast-derived mAbs (Table S2); influenza antigen (green), unknown/untested (yellow), non-influenza antigen (red). Arcs are ‘zoomed’ to shared lineages. 7014 had no shared lineages.
Somatic hypermutation of vaccine-stimulated B cell repertoires
Seroconverters and non-seroconverters differed in the somatic mutation of expanded B cell clones at day 7 post-vaccination (mean mutation 6.4% and 4.3%, respectively, p=0.0041, t-test). Prior to vaccination, the groups shared similar levels of mutation in expanded clones (3.2% and 3.4%, p=1.00) and non-clonal IGHV sequences (1.5% and 1.3%, p=0.8048). At day 21, the expanded lineages of the seroconverters retained a higher somatic point mutation (5.7%), while non-seroconverters returned to the pre-vaccination mutation frequency of 3.5% (p=0.1649).
Time course of response to H1N1 single antigen vaccination
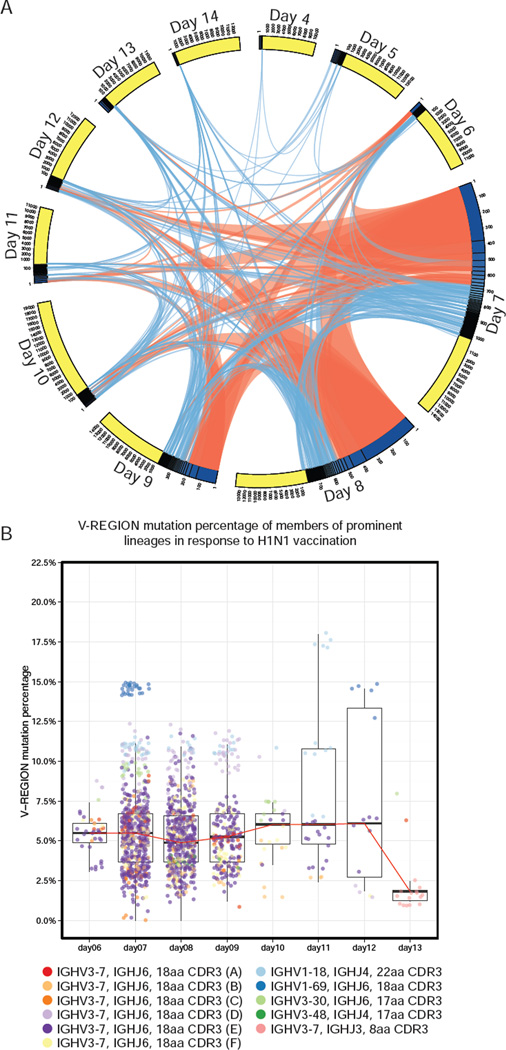
To obtain a more detailed view of the dynamics of the B cell response to influenza vaccine in a somewhat simpler vaccination context, IGH sequencing from a daily time course of the response to the single-agent pandemic H1N1 2009 influenza vaccination was undertaken, in an additional healthy subject (BFI-278) (Figure 3A, Figure S2). Prior to vaccination, few clonally-expanded B cell lineages were detected (5 expanded clones, on average 0.06% of total). After vaccination, a clonal response was seen that peaked at day 7 and decreased toward pre-vaccination levels by day 10 (Figure 3A). Overall, 256 clonal lineages were detected post-vaccination, with 11 lineages strongly induced by vaccination, each contributing more than 0.1% of total rearrangements. Vaccine-induced clonal lineages reached their peak at day 7, when they accounted for 7.1% of total IGH. Many of the vaccine-induced expansions continued to be detected by day 9, although some were observed only at a single time point (Figure 3A). Clonal lineages utilizing IGHV3-7 paired with IGHJ6 were over-represented in the vaccine-induced proliferation, with 6 of the 11 prominent lineages using these genes with an 18 amino acid CDR3 (Figure 3B).
Figure 3. Time course of B cell clonal expansions induced by inactivated H1N1 vaccine.
(A) Vaccine induced B cell clonal lineage relationships during H1N1 vaccine response (complete IGH repertoires at Figure S2). Each arc represents a post-vaccination time point sub-divided into expanded (blue) and unexpanded (yellow) compartments. Lines join lineage members at later time points. Orange lines indicate that a lineage was strongly induced at the originating time point (> 0.1% of total IGH). (B) IGHV percentage mutation for strongly induced lineages. Median (red line), quartiles shown as box and whiskers and points are jittered to prevent over-plotting.
The post-vaccination clonally expanded IGH were somatically mutated, with members of the 11 prominent lineages showing an average of 5.68% IGHV mutation (Figure 3B). The full IGH repertoire detected in this individual had a mean IGHV mutation of 1.18%.
Identification of convergent IGH rearrangements in the H1N1 vaccine response
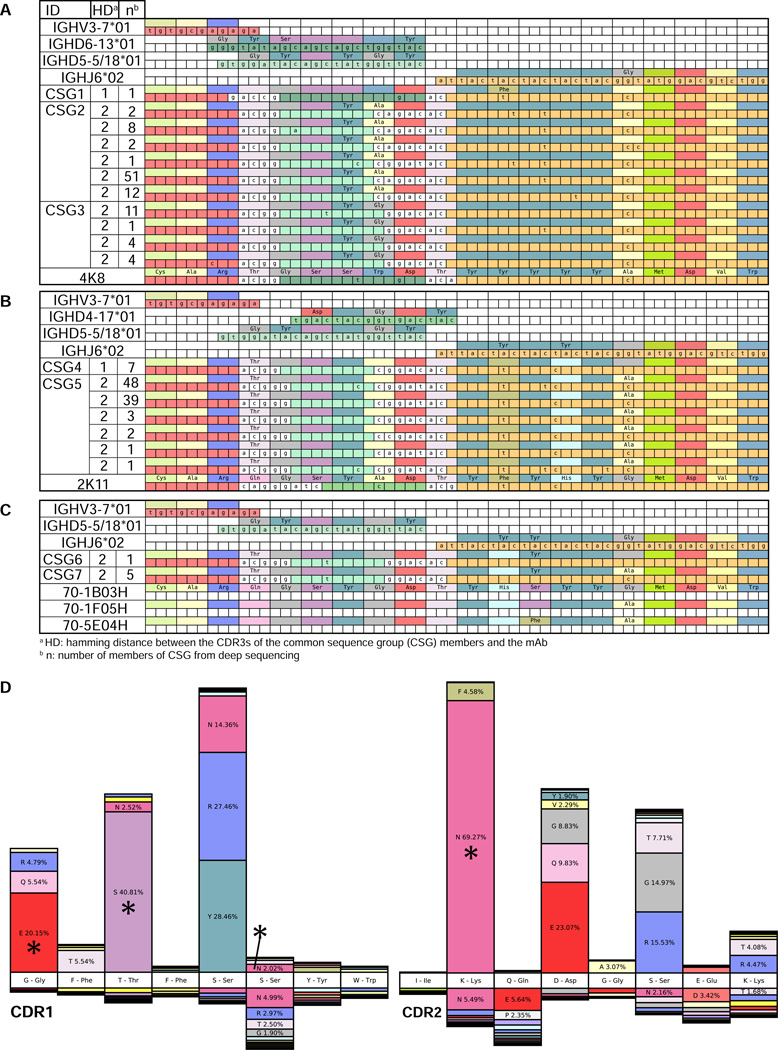
The dominant IGH lineages in BFI-278’s response to single antigen H1N1 vaccination (Figure 3) had stereotyped features: IGHV3-7, IGHJ6, and an 18 amino acid CDR3. These lineages were compared to previously reported H1N1-responding B cell repertoires. Surprisingly, we observed response convergence with constrained CDR3 sequences observed in two other studies (Wrammert et al.) (Krause et al., 2011). The CDR3 sequences of 4K8 and 2K11 (Krause et al., 2011) differed at a single amino acid residue from members BFI-278’s dominant expansion (Figure 4A, 4B, Table S4). 48K and 2K11 were isolated from the same individual post-pandemic H1N1 vaccination and both had activity against 2009 H1N1 as well as human and swine influenza strains from 1918, 1930, 1976 and 1977 by hemagglutination activation inhibition (HAI) (Krause et al., 2011). The HAI activity of a mAb generated from the IGH isolated from BFI-278 paired with the light chain of antibody 4K8 showed HAI with a titer of 20 in triplicate assays against the Influenza A H1N1 California/7/09 strain, but no detectable activity against the other strains in the panel.
Figure 4. Convergent immunoglobulin heavy chain rearrangements identified in H1N1 vaccine responses in different individuals.
Multiple sequence alignment of closely related stereotypic H1N1 clones isolated from difference sources. Nucleotide positions are labeled when they differ from the germline genes (top of each block). Nucleotides inferred to be derived from non-templated nucleotide addition during V(D)J rearrangement are not colored. Amino acids positions are labeled when they differ from those in the mAb (bottom of each block). Each block is labeled with the germline, the conserved sequence group (CSG) and the mAb. Members of CSGs are detailed in Table S4. (A) IGH related to 4K8 mAb isolated (Krause et al., 2011) (B) IGH related to 2K11 (Krause et al., 2011) (C) IGH related to CDR3s previously identified by Wrammert et al. (Wrammert et al., 2011). (D) The percentage of total sequences with CDR1 and CDR2 somatic mutation amino acid substitutions in convergent H1N1 clones (upper) compared to 4,784 IGH using IGHV3-7 from the pre-vaccination repertoire of 14 subjects pre-pandemic (lower). The germline sequence is depicted in the white boxes (middle). Asterisks (*) mark amino acid substitutions shared with mAbs 48K or 2K11 (Krause et al., 2011).
Stereotypic IGH were also present in H1N1-stimulated clones from a single donor 15 days postacute pandemic H1N1 infection (Wrammert et al., 2011). One mAb, 70-1B03, differed by 2 CDR3 residues from IGH isolated from BFI-278, and was reported to be cross-reactive between pandemic H1N1 and the 2009 annual TIV vaccine antigens (Figure 4C) (Wrammert et al., 2011). Two further IGH, 70-5E04H and 70-1F05H, were derived from H1N1 infection induced plasmablasts but had no detectable binding to the panel of antigens tested (Wrammert et al., 2011).
We examined somatic mutations in IGHV to assess additional evidence of molecular convergence in these antibodies. Overall, the IGHV of convergent H1N1-specific lineage members from BFI-278 showed an average of 5.24% mutation, with the most highly mutated sequence showing 12.27% mutation. Somatically mutated positions in the CDR1 and CDR2 were shared with 4K8 and 2K11 mAbs (Figure 4D). Shared substitutions could result from intrinsic mutation hotspots favoring mutation at these locations. This is very unlikely for the observed convergent mutations, as the CDR1 and CDR2 mutations in the BFI-278 H1N1 antibody sequences were uncommon in two unrelated, non-vaccinated datasets; IGHV3-7 IGH from pre-vaccination of 14 TIV 2007 and 2008 subjects (Figure 4D), and IGHV3-7 IGH from 27 healthy subjects who had not been recently vaccinated (Wang et al., 2014) (both p < 0.0001, Pearson’s dependent groups). Importantly, the two non-vaccination datasets were highly similar in their mutations (r=0.9311) and both were distinct from the H1N1 lineages (p=0.1794). Evaluation of mutated positions in mAbs 48K and 2K11 for whether they were more likely to have been drawn from the mutation distribution of the H1N1 lineage, or from the two non-vaccination datasets, showed that 4K8 carries mutations similar to the H1N1 lineage (log likelihood ratio, H1N1: −7.98, IGHV3-7 background: −22.59) while 2K11 did not show higher similarity to either distribution (H1N1: −41.97, IGHV3-7 background: −42.52).
The response of BFI-278 to the next year’s 2010 TIV, which included pandemic H1N1, was also examined. The 2010 TIV response included 28 IGH with stereotypic features at day 7 post-vaccination (0.36%). These differed by at least 4 CDR3 residues to the prior year’s stereotypic clones, and differ at more than 5 positions to the H1N1-stimulated B cells from (Wrammert et al., 2011) and 6 positions to the H1N1-specific mAbs from (Krause et al., 2011). The 2010 TIV IGH also lacked convergent somatic mutations and therefore appear to represent distinct B cell clones.
IGH rearrangements with stereotypic features prior to 2009
To evaluate whether the convergent B cell clones were present at high frequencies in BFI-278’s repertoire prior to pandemic H1N1 antigen exposure, we studied previously reported samples collected 14 months apart in 2006 and 2008 (Boyd et al., 2009). Only 4 IGH from 2006 (0.03%), and 10 IGH from 2008 (0.06%) used IGHV3-7 and IGHJ6 with an 18-amino acid CDR3. The CDR3s of the pre-2009 IGH differed by at least 6 amino acids from the H1N1 response clones, and appear to be unrelated.
The contribution of stereotypic IGH chains in independent populations was also analyzed in 151,217 unique IGH pooled from IGH sequencing of 27 additional adults (2008 and 2009) (Wang et al., 2014). IGH using IGHV3-7 and IGHJ6 represented 1.01% of total rearrangements, with 115 IGH of these having an 18 residue CDR3. The 115 rearrangements did not share similar CDR3 sequences with IGH from BFI-278’s H1N1 vaccination, with all having less than 85% sequence identity. Among 496,104 IGH sequences from the 14 TIV 2007 and 2008 subjects a single IGH was found (seroconverter 7014), 2008 TIV) that differed by 1 amino acid residue from a rearrangement from individual BFI-278’s H1N1 2009 response. Examination of over 500,000 IGH sequences collected from 41 individuals prior to 2009 therefore revealed only a single example of the convergent IGH seen in 2009 in BFI-278 and in the (Wrammert et al.) and (Krause et al., 2011) studies, indicating that antibody lineages capable of binding the pandemic H1N1 strain were indeed present in human B cell repertoires, but were relatively rare.
Discussion
Deep sequencing of immunoglobulin libraries enables tracking of known antigen-specific B cell populations (Liao et al., 2011; Liao et al., 2009; Wu et al., 2011). In addition, replicate libraries of B cells from an individual can identify clonally-expanded populations arising in response to immune stimuli such as vaccination or infection. Recently, influenza vaccine responses in humans have been associated with increased numbers of plasmablast immunophenotype B cells in the blood, peaking at approximately 7 days post-exposure, with the majority of these B cells expressing antibodies specific for influenza antigens, and often featuring significant clonal expansion of particular plasmablast lineages (Brokstad et al., 1995; Cox et al., 1994; Halliley et al., 2010; Sasaki et al., 2007; Wrammert et al., 2008). Prior deep sequencing analysis of influenza vaccination responses has identified sequences present at elevated levels in libraries amplified from cDNA of peripheral blood B cells (Jiang et al., 2013). Our results reveal a strong correlation between the levels of B cell clonal expansion detected in the blood, and the serological response to vaccination. Vaccine-induced clonal lineages measured by replicate library sequencing peak at day 7 having increased 118-fold over background lineages, and persist at detectable levels until about two weeks post vaccination.
To evaluate the antigen specificity of the clones detected by sequencing, we compared our data to the sequences of mAbs generated from plasmablasts at day 7 post-vaccination individuals who seroconverted to seasonal TIV (Moody et al., 2011). Almost 25% of the IGH utilized by the plasmablast derived mAbs shared clonal lineages with those we identified and 66% of these were specific for influenza. This cross identification confirms that a relatively simple evaluation of clonal expansions in the vaccine response by IGH repertoire sequencing highlights antigen-specific lineages. In addition, the incomplete overlap suggests that there are low frequency plasmablast lineages are not identified as clonally-expanded, and that there may be other clonally-expanded B cell populations that do not have a plasmablast immunophenotype, such as influenza-specific memory B cells, which have been previously reported to significantly increase their levels in the blood at day 7 after influenza vaccination (Wrammert et al.).
Unexpectedly, the response to monovalent H1N1 vaccination showed striking stereotyped features (use of IGHV3-7, IGHJ6, and an 18-amino acid CDR3) in the most prominent post-vaccination clonally-expanded B cell lineages. We looked for evidence of this stereotyped IGH response in other individuals responding to influenza vaccination or infection, expecting that the likelihood of finding ‘convergent evolution’ in antibody responses to influenza was low, given the very large potential size of the immunoglobulin repertoire, the still relatively shallow sampling of each individual’s repertoire that can be conducted with deep sequencing (Boyd et al., 2009; Glanville et al., 2011), and the selection pressures on influenza strains to avoid common immune responses. To our surprise, IGH from two independent H1N1 response studies (Krause et al., 2011; Wrammert et al., 2011) shared the stereotypic features. The CDR3s of these rearrangements differed by only one of eighteen amino acids, highlighting a striking degree of convergence in the antibody responses in these unrelated individuals. A recombinant mAb of the BFI-278 stereotypic IGH lineage, paired with the light chain from the previously reported 48K mAb (Wrammert et al., 2011), showed HAI specific for the pandemic H1N1 strain. Notably, in a search of over 500,000 IGH sequences from the B cell repertoires of 41 subjects studied from 2006 to 2009, we identified a single IGH with the same convergent features, highlighting the low prior frequency of such clones that are preferentially stimulated by the 2009 pandemic H1N1 influenza strain (Brokstad et al., 1995; Sasaki et al., 2007).
The use of IGH involving IGHV3-7, IGHJ6 and an 18 amino acid CDR3 was a recurrent pattern of the response of BFI-278 (Figure 3) to successive years of H1N1 influenza vaccination, but importantly, the stereotyped B cell clones in 2009 were distinct from those that appeared in 2010 following vaccination with TIV containing the same pandemic H1N1 antigens. This suggests the recruitment of new B cells with stereotypic IGH chains, rather than a recall response dominated by the highest-frequency members of the prior year’s clones. This favoring of new clones could be the consequence of serum antibody derived from the 2009 vaccination decreasing restimulation of the dominant 2009 B cell clones, but not preventing stimulation of B cells that recognize somewhat different epitopes of the viral antigens.
In summary, high-throughput DNA sequencing of peripheral blood B cells provides a highly informative measure of clonal expansions that correlates with serological vaccine responses. B cell clones that expand following vaccination show substantial but incomplete clonal overlap with vaccine-specific single-cell plasmablasts. Responses to vaccination with the 2009 pandemic H1N1 influenza strain revealed a dominant pattern of antibody responsiveness that was convergent in different individuals. Antibodies with convergent features were rare prior to 2009. Overall, if the detection of convergent antibody signatures can be generalized to other antigens and infectious diseases, it may be feasible to use IGH repertoire sequencing to assess an individual’s history of antigenic exposures and infections more broadly (Glanville et al., submitted). The results also lend some support to the idea that evolutionary selection of the germline IGH genes may predispose the adaptive immune response to follow common paths of response to common pathogens. Further evaluation in the context of different vaccines and pathogen infections will be able to determine whether these findings are the exception, or the rule, in human antibody responses.
Experimental Procedures
Collection of specimens
All subject recruitment was performed using informed consent. The Duke University Institutional Review Board approved protocols for the study of response to 2007 and 2008 seasonal TIV. The Stanford IRB approved study of the 27 additional pre-vaccination subjects. Subjects recruited at Duke University were given either 2007-2008 or 2008-2009 FluZone® TIV (Sanofi Pasteur, Swiftwater PA) as described in Table S1 and detailed in Supplemental Information. Blood was drawn before vaccination and on days 7 and 21 after challenge. Subjects recruited at Stanford University were 27 healthy individuals aged 20 to 89 years (Wang et al., 2014). Peripheral blood samples from these patients were collected prior to vaccination in two successive years (2008 and 2009).
BFI-278 recruited at Stanford University was administered Influenza A (H1N1) 2009 monovalent inactivated vaccination in January 2010. Blood was drawn before vaccination and daily on days 4 to 14 after challenge. Ten months after receiving the monovalent H1N1 vaccine, BFI-278 was administered a seasonal TIV that included the Influenza A/California/07/2009-like pandemic H1N1 antigenic component. Blood was drawn prior to 2010 TIV vaccination, and at days 7 and 21 post-vaccination.
Sample preparation and PCR amplification for deep sequencing
Peripheral blood mononuclear cells were isolated by centrifugation of blood layered over Histopaque-1077 (Sigma-Aldrich), and cryopreserved (Moody et al.). Column purification (Qiagen, Valencia, CA) was used to isolate genomic DNA template. IGH were amplified from genomic DNA template using the previously described primer design for 454 instrument sequencing as detailed in Supplemental Information (Boyd et al., 2009).
Deep sequencing of immunoglobulin heavy chain
Amplicon library pools were quantified by real-time PCR (Roche, Connecticut). Sequencing data presented here are derived from the 454 instrument using Titanium chemistry, with long-range amplicon pyrosequencing beginning from the "B" primer in the manufacturer's protocol (Roche, Connecticut). The complete data sets from the samples generated are available at phs000760.v1.p1.
Sequence data analysis
Sequences from each input specimen were demultiplexed using the sample and replicate library barcodes as detailed in Supplemental Information. Alignment of rearranged IGH sequences to germline V, D and J, and determination of V-D junctions and D-J junctions was performed using iHMMune-align (Gaëta et al., 2007). IGH were assigned to clonal lineages by clustering on CDR3 nucleotide similarity for IGHs that shared the same IGHV, IGHJ and CDR3 length. CDR3 sequence similarity was measured by Hamming distance and clusters were assigned using a Hamming distance of 95% identity to any existing sequence and at least 80% identity within the cluster (Figure S1B).
Scale-independent normalized measure of overall clonality
To compare the degree of B cell clonal expansion detected in data sets with varying sequencing depth, we used a scale-independent metric we have previously described (Wang et al.). This clonality metric can be considered as the probability that any two randomly selected sequences drawn from independent replicates are members of the same clonal lineage.
Analysis of hypermutation spectra
CDR1 and CDR2 sequences, as defined by IMGT criteria, were extracted from iHMMune-align results and translated to amino acid sequences. CDR1 and CDR2 position count matrices of mutations were converted to substitution frequency vectors. Pearson’s correlation and Pearson’s test for dependent correlations were used to assess whether there were differences between mutation spectra for different datasets. Log-likelihood ratios were used to explore single sequence mutation distributions allowing comparison of the likelihood that the observed substitutions were drawn from the mutation distributions of different datasets.
Single-cell plasmablast sorting, mAb expression and quantification of plasmablast frequencies
PBMC were cryopreserved with standard methods, and single-cell sorting was performed as previously described (Moody et al., 2011). Flow cytometry was carried prior to single cell IGH V(D)J PCR and expression of recombinant IgG1 mAbs as previously described (Liao et al., 2009; Moody et al., 2011) and detailed in Supplemental Information. In addition, samples collected prior to vaccination, and days 7 and 21 post-vaccination were assessed for plasmablast and B cell phenotypes.
Serum antibody binding tests
Plasma samples were evaluated by ELISA with purified HAs and split vaccine preparations as detailed in Supplemental Information. Five-parameter curve fits were used for data analysis. Endpoint titers were calculated as 3-fold above assay background, and the assay cutoff was a 1:25 dilution. Expressed mAbs were tested for binding to influenza antigen by ELISA, as previously described (Moody et al., 2011). Reactivity to influenza antigens was also studied using a standardized custom Luminex® assay (Luminex, Austin, TX).
Hemagglutination inhibition assay of monoclonal antibody
HAI assays of mAbs were performed as described elsewhere (Davtyan et al., 2011; Wang et al., 2006) and are detailed in Supplemental Information. Working stocks of influenza were standardized to a HA titer of 8 HA units/50μL for assays for A/California/4/2009 (H1N1), A/Brisbane/59/2007 (H1N1), A/Brisbane/10/2007 (H3N2), B/Brisbane/60/2008, and B/Florida/4/2006. The HAI titer was defined as the reciprocal of the highest dilution of antibody that inhibits red blood cell hemagglutination by influenza virus.
Supplementary Material
Highlights.
Human antibody gene repertoire sequencing identifies vaccine-specific B cell clones
Quantified B cell clonal expansions correlate with serological vaccine responses
Humans show convergent antibody responses to pandemic H1N1 2009 influenza vaccination
Acknowledgements
The metadata and sequences reported in the paper can be accessed at dbGAP, with accession number phs000760.v1.p1. The authors thank Sally Mackey for project, regulatory and data management; Research Nurses Sue Swope and Cynthia Walsh; Phlebotomist Michele Ugur and Research Assistant Kyrsten Spann for scheduling and conducting the study visits. This work was supported by NIH grants U19AI090019, P01AI089618, U19AI067854, 1U19AI089987, and grants from the Ellison Medical Foundation to Mark Davis and Scott Boyd. We would like to thank Thomas Kepler for helpful discussions about data analysis of sequences derived from the single cell sorting experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ademokun A, Wu YC, Martin V, Mitra R, Sack U, Baxendale H, Kipling D, Dunn-Walters DK. Vaccination- induced changes in human B- cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging cell. 2011;10:922–930. doi: 10.1111/j.1474-9726.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Science translational medicine. 2009;1:12ra23–12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokstad KA, Cox RJ, Olofsson J, Jonsson R, Haaheim LR. Parenteral influenza vaccination induces a rapid systemic and local immune response. Journal of Infectious Diseases. 1995;171:198–203. doi: 10.1093/infdis/171.1.198. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Brokstad KA, Zuckerman MA, Wood JM, Haaheim LR, Oxford JS. An early humoral immune response in peripheral blood following parenteral inactivated influenza vaccination. Vaccine. 1994;12:993–999. doi: 10.1016/0264-410x(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Davtyan H, Ghochikyan A, Cadagan R, Zamarin D, Petrushina I, Movsesyan N, Martinez-Sobrido L, Albrecht RA, García-Sastre A, Agadjanyan MG. The immunological potency and therapeutic potential of a prototype dual vaccine against influenza and Alzheimer’s disease. J Transl Med. 2011;9:127–127. doi: 10.1186/1479-5876-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nature biotechnology. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaëta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- Glanville J, Kuo TC, von Büdingen H-C, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proceedings of the National Academy of Sciences. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliley JL, Kyu S, Kobie JJ, Walsh EE, Falsey AR, Randall TD, Treanor J, Feng C, Sanz I, Lee F. Peak frequencies of circulating human influenza-specific antibody secreting cells correlate with serum antibody response after immunization. Vaccine. 2010;28:3582–3587. doi: 10.1016/j.vaccine.2010.02.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He X-S, Dekker CL, Zheng N-Y, Huang M, Sullivan M, et al. Lineage Structure of the Human Antibody Repertoire in Response to Influenza Vaccination. Science Translational Medicine. 2013;5:171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, Crowe JE. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. The Journal of Immunology. 2011;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-M, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng N-Y, Lee J-H, Huang M, Qu X, Edupuganti S. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proceedings of the National Academy of Sciences. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu J-S, Hwang K-K. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. The Journal of Experimental Medicine. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang K-K. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of Virological Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MK, Sun V, Olander JV, Hoffmann JW, Nahm MH. The repertoire of human antibodies to the carbohydrate capsule of Streptococcus pneumoniae 6B. Journal of Infectious Diseases. 1996;174:75–82. doi: 10.1093/infdis/174.1.75. [DOI] [PubMed] [Google Scholar]
- Sasaki S, He X-S, Holmes TH, Dekker CL, Kemble GW, Arvin AM, Greenberg HB. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, Arvin AM, Greenberg HB. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. Journal of Virology. 2007;81:215–228. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MG, Tarrand JJ, Crimmins DL, McCourt DW, Siegel N, Smith C, Nahm M. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. II. IgG antibodies contain VH genes from a single VH family and VL genes from at least four VL families. The Journal of Immunology. 1989;143:293–298. [PubMed] [Google Scholar]
- Silverman G, Lucas A. Variable region diversity in human circulating antibodies specific for the capsular polysaccharide of Haemophilus influenzae type b. Preferential usage of two types of VH3 heavy chains. Journal of Clinical Investigation. 1991;88:911. doi: 10.1172/JCI115394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger JH. Tests for comparing elements of a correlation matrix. Psychological Bulletin. 1980;87:245. [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Y, Xu LT, Jackson KJL, Roskin KM, Pham TD, Laserson J, Marshall EL, Seo K, Lee J-Y, et al. Effects of Aging, Cytomegalovirus Infection, and EBV Infection on Human B Cell Repertoires. The Journal of Immunology. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Taaffe J, Parker C, Solórzano A, Cao H, García-Sastre A, Lu S. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codonoptimized HA DNA vaccines. Journal of Virology. 2006;80:11628–11637. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of Experimental Medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.