Abstract
The cytotoxic T-cell response in chronic hepatitis B virus (HBV) infection has been described as weak and mono- or oligospecific in comparison to the more robust virus-specific T-cell response present in resolved infection. However, chronic hepatitis B is a heterogeneous disease with markedly variable levels of virus replication and liver disease activity. Here we analyzed (both directly ex vivo and after in vitro stimulation) the HBV-specific CD8 T-cell responses against structural and nonstructural HBV proteins longitudinally in patients with different patterns of chronic infections. We found that the profiles of virus-specific CD8+-T-cell responses during chronic infections are highly heterogeneous and influenced more by the level of HBV replication than by the activity of liver disease. An HBV DNA load of <107 copies/ml appears to be the threshold below which circulating multispecific HBV-specific CD8+ T cells are consistently detected. Furthermore, CD8+ T cells with different specificities are differentially regulated during chronic infections. HBV core-specific CD8+ T cells are associated with viral control, while CD8+ T cells specific for envelope and polymerase epitopes can occasionally be found in the setting of high levels (>107 copies) of HBV replication. These findings have implications for the design of immunotherapy for chronic HBV infections.
Hepatitis B virus (HBV) is a hepatotropic, noncytopathic DNA virus which can cause chronic hepatitis, cirrhosis, and liver cancer (19).
The ability to clear HBV after infection has been associated with the presence of a strong virus-specific T-cell response. Multispecific antiviral CD4+- and CD8+-T-cell responses with a type 1 profile of cytokine production are detectable in subjects with self-limited acute hepatitis B (5, 10, 12, 13, 29, 32, 40). Due to the lack of suitable animal models, it has been difficult to establish a causative effect, but recently CD8+-T-cell deletion experiments performed with HBV-infected chimpanzees showed the essential role of HBV-specific CD8+ T cells in HBV control (41).
In contrast to the robust virus-specific CD8+- and CD4+-T-cell responses present in patients with resolved HBV infections, patients with chronic infections are usually characterized by weak virus-specific T-cell responses (2, 10, 12, 13, 28, 40). However, the difference between T-cell responses present in acute or resolved versus chronic hepatitis B has obscured the diversity present within chronic hepatitis B. Chronic hepatitis B is clearly a highly heterogeneous disease, and the levels of virus replication, liver disease activity (19), and humoral responses (23) can differ markedly in patients with chronic hepatitis B. Patients can be characterized by levels of HBV replication ranging from 103 to 109 HBV DNA copies/ml in the presence or in the absence of liver inflammation.
Furthermore, in some patients, the profile of liver disease and HBV replication is stable, whereas other patients experience episodic flares of disease, with fluctuating levels of HBV DNA. All of these different profiles of disease (in particular, high levels of alanine transaminase [ALT], an enzyme released by lysed hepatocytes) have traditionally been associated with different magnitudes of HBV-specific CD8+-T-cell responses (30), but convincing data demonstrating this association are still lacking. Episodes of acute flares during chronic HBV infection are associated with a recovery of HBV-specific T-helper responses (36, 42), but increased levels of HBV-specific CD8+-T-cell responses have been demonstrated only following the resolution of chronic infection (33), after the inhibition of HBV replication during lamivudine therapy (6, 22), and in patients with a low level of HBV replication and no signs of liver inflammation (20).
Here we carried out a longitudinal study to analyze the dynamic profiles of HBV-specific CD8+-T-cell responses according to the level of virus replication and liver inflammation by using immunological methods (HLA tetramers and intracellular cytokine staining [ICCS]) that allow a precise quantification of virus-specific CD8+ T cells. We also evaluated whether CD8+ T cells specific for different antigenic determinants may differently contribute to viral control. During HBV infection, HBV-specific CD8+ T cells can target different epitopes located within the HBV core (28, 39), envelope (25), polymerase (32), and X (11) proteins. Differences between CD8+ T cells specific for different HBV antigens were recently demonstrated for patients with chronic HBV infection. Envelope-specific CD8+ T cells are not visualized by HLA tetramers (34), and CD8+ T cells specific for structural and nonstructural HBV antigens are not equally effective in the recognition of HBV-infected hepatocytes in the transgenic mouse model of HBV infection (14). Whether CD8+ T cells specific for different HBV antigens are differentially regulated during chronic hepatitis B, however, has never been evaluated.
MATERIALS AND METHODS
Patients.
A total of 25 HLA-A2-positive patients with HBV infection were selected for this study. Screening for HLA-A2 positivity was performed by staining peripheral blood mononuclear cells (PBMC) from patients with a fluorescent anti-HLA-A2.01 antibody (Serotec); selected patients were confirmed to have the HLA-A2.01 allele by PCR DNA typing. A total of 12 patients had clinical, biochemical, and virological evidence of resolved acute HBV infection (recovered from acute hepatitis B between 6 months and 3 years before, normal ALT levels, positive for antibody to the HBV core [anti-HBc], and negative for HBV surface antigen [HBsAg]); the remaining 13 patients had clinical, biochemical, and virological evidence of chronic HBV infection (HBsAg positive, anti-HBc positive, and negative for antibodies to hepatitis C virus, delta virus, human immunodeficiency virus type 1 [HIV-1], and HIV-2) (Table 1). Patients C5 and C12 had completed a course of antiviral therapy 9 and 13 months before entering the study, with no benefit, but no other patients had been treated. These 13 patients also had no other causes for chronic liver damage, such as alcohol use, drug use, congestive heart failure, or autoimmune diseases. A longitudinal analysis of antiviral T-cell immunity in the periphery, with a cross-sectional analysis of responses within the liver, was approved by the local ethics committee, and all patients provided written informed consent.
TABLE 1.
Characteristics of patients with chronic HBV infections (HBsAg positive and anti-HBc positive)a
| Patient | Age (yr) | Sex | Route of infection | Presence (+) or absence (−) of:
|
HBV genotype | No. of HBV DNA copies/ml | ALT level (U/liter) | Liver histologic feature | |
|---|---|---|---|---|---|---|---|---|---|
| HBeAg | Anti-HBe | ||||||||
| C1 | 22 | F | Neonatal | − | + | D | 2.4 × 104 | 36 | ND |
| C2 | 35 | F | ? | − | + | D | 2 × 104 | 28 | ND |
| C3 | 37 | F | ? | − | + | ND | 1.3 × 104 | 31 | ND |
| C4 | 56 | F | Sexual (?) | − | + | B | 2.7 × 106 | 74 | Min NI, sev fibrosis |
| C5 | 61 | F | ? | − | + | D | 1.5 × 106 | 61 | Min NI, sev fibrosis |
| C6 | 44 | M | Neonatal | − | + | D | 1.0 × 106 | 57 | Min NI, min fibrosis |
| C7 | 43 | M | ? | + | − | D | 1.0 × 108 | 227 | Mod NI, mod fibrosis |
| C8 | 36 | M | Neonatal | + | − | A | 4.0 × 108 | 2,635 | Mod NI, min fibrosis |
| C9 | 25 | M | ? | + | − | A | 5.3 × 108 | 291 | Cirrhosis |
| C10 | 49 | F | ? | + | − | C | 5.9 × 108 | 20 | Min NI, min fibrosis |
| C11 | 25 | M | Neonatal | + | − | A | 5.8 × 108 | 66 | Min NI, min fibrosis |
| C12 | 32 | M | Neonatal | + | − | E | 6.8 × 108 | 60 | Mod NI, no fibrosis |
| C13 | 25 | M | ? | + | − | D | 10 × 108 | 54 | Mod NI, no fibrosis |
F, female; M, male; ?, unknown; ND, not determined; min, minimal; mod, moderate; sev, severe; NI, necroinflammation.
Virological assessments.
HBsAg, antibody to the HBV surface, total and immunoglobulin M anti-HBc, HBV envelope antigen (HBeAg), antibody to the HBV envelope (anti-HBe), and antibodies to delta virus, hepatitis C virus, HIV-1, and HIV-2 were determined by using commercial enzyme immunoassay kits (Abbott Laboratories, North Chicago, Ill.; Ortho Diagnostics, Raritan, N.J.; and Sanofi Diagnostic Pasteur, Marnes la Coquette, France). Serum HBV DNA was quantified by using an Amplicor Monitor assay (Roche Pharmaceuticals Ltd., Branchburg, N.J.) with a DNA detection limit of 400 copies/ml (0.0014 pg/ml).
PCR and HBV DNA sequencing.
DNA was extracted from serum samples taken at the time of liver biopsy by using a QIAamp DNA blood minikit (Qiagen, Crawley, United Kingdom). HBV DNA was amplified with primers specific for the HBV core and envelope genes as described previously (24). The amplicons were purified, and the core or envelope regions were sequenced directly by using an ABI 377 automated sequencer (Applied Biosystems). The HBV genotype of the predominant virus population in chronically infected patients was determined by analyses of sequenced portions of the core and surface antigens (26).
Synthetic peptides.
Peptides corresponding to various HBV-specific HLA-A2-restricted CD8 epitopes were purchased from Chiron Mimotopes (Clayton, Victoria, Australia) or from Primm (Milano, Italy). The purity of the peptides was determined to be greater than 90% by high-pressure liquid chromatography analysis. The peptides were based on HBV genotype D (serotype ayw); the degree of homology among the various genotypes is shown in Table 2.
TABLE 2.
Peptides used for analysis of HBV-specific CD8+-T-cell responses
| Peptide | Sequence | Change in epitope sequence in the following HBV genotypea:
|
HLA-A2.01 binding affinityb (nM) | |||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |||
| Core 18-27 | FLPSDFFPSV | = | I27c | I27 | = | = | I27 | 3.3 |
| Core 107-115 | CLTFGRETV | = | = | = | = | = | = | ND |
| Pol 455-463 | GLSRYVARL | = | = | = | = | = | = | 71 |
| Pol 502-510 | KLHLYSHPI | = | = | = | = | = | = | 17 |
| Pol 575-583 | FLLSLGIHL | = | = | = | = | = | V579 | 10 |
| Pol 655-663 | ALMPLYACI | = | = | = | = | T661 | N661 | 10 |
| Pol 816-824 | SLYADSPSV | = | = | = | = | V820 | = | 14 |
| Env 183-191 | FLLTRILTI | = | K185 | = | = | K185 | K185 | 7 |
| Env 335-343 | WLSLLVPFV | = | = | A340 | = | = | Q341 | 7 |
| Env 338-347 | LLVPFVQWFV | = | = | A340 | = | A347 | Q341 | 3.2 |
| Env 348-357 | GLSPTVWLSV | A357 | = | = | = | = | L356 | 19 |
NCBI accession numbers of the HBV genotypes used to analyze amino acid variability in the HBV epitopes were as follows: genotype A, X51970; genotype B, M54923; genotype C, D00630; genotype D, J02203; genotype E, X75664; and genotype F, X75658. =, the sequence of the HBV epitope was identical to that of the peptide.
Binding is expressed as the dose of the tested peptide yielding 50% inhibition of radiolabeled peptide binding to HLA-A2 molecules. The data reported are from the study of Bertoni et al. (4). ND, not determined.
The NCBI accession number for HBV genotype B with isoleucine at position 27 (I27) is the same as that listed in footnote a. The NCBI accession number for HBV genotype B with valine instead of isoleucine at position 27 is D00329.
Isolation of PBMC and production of T-cell lines.
PBMC were isolated from fresh heparinized blood by Ficoll-Hypaque density gradient centrifugation and suspended at concentrations of 1.5× 106 to 2 × 106/ml in RPMI 1640 (Gibco)-10% fetal calf serum (FCS). Cells were stimulated with 1 μM concentrations of various peptides in a 96-well plate. Recombinant interleukin 2 (R&D Systems, Abingdon, United Kingdom) was added on day 4 of culturing, and cells were analyzed after 10 to 12 days of culturing. PBMC stimulation was performed with a panel of 11 peptides (Table 2) corresponding to HBV sequences previously identified as HLA-A2-restricted cytotoxic T-lymphocyte epitopes (4).
Purification of T cells from liver biopsy specimens.
Mononuclear cells were purified from biopsy specimens. Liver tissue not needed for diagnostic purposes was extensively washed in RPMI 1640 and then digested with collagenase (1 mg/ml; Sigma Chemical Co., St. Louis, Mo.) and DNase (25 μg/ml; Sigma Chemical Co.) for 1 h at 37°C. The cell suspension was washed twice, and mononuclear cells were recovered by centrifugation over a Ficoll-Hypaque density gradient.
Intracellular IFN-γ production.
For analysis of gamma interferon (IFN-γ) production, ex vivo-purified PBMC or short-term T-cell lines were stimulated at 2 × 106 to 3 × 106/ml in RPMI 1640-10% FCS with HBV peptides (1 μM) or with HepG2 cell lines (HLA-A2+) supporting full HBV replication (37) for 6 h at 37°C in the presence of brefeldin A (10 μg/ml; Sigma-Aldrich, Poole, Dorset, United Kingdom). Cells were washed, stained with Cy-chrome-conjugated anti-CD8 antibodies (Sigma Chemical Co.), permeabilized, and fixed with Cytofix/Cytoperm (Pharmingen, San Diego, Calif.) according to the manufacturer's instructions. Fluorescein isothiocyanate-conjugated anticytokine antibodies or isotype-matched control antibodies were added (30 min, 22°C), and the cells were washed twice and analyzed by flow cytometry.
Staining with HLA tetramer complexes.
HLA class I tetramers were produced as described previously (21) and lately purchased commercially (Proimmune, Oxford, United Kingdom). Directly purified circulating cells or T-cell lines or T cells purified from liver biopsy specimens were incubated for 30 min at 37°C with 1 μg of phycoerythrin-labeled tetramer complex in RPMI 1640-10% FCS in round-bottom polystyrene tubes (Becton Dickinson, Paramus, N.J.). Cells were washed in phosphate-buffered saline and incubated at 4°C for 30 min with saturating concentrations of Cy-chrome-conjugated anti-CD8 antibodies. After further washings, cells were analyzed by using FACSort (Becton Dickinson) and CELLQuest software immediately or after the addition of 1% paraformaldehyde.
RESULTS
Longitudinal analysis of HBV-specific CD8+ T cells in patients with chronic hepatitis B.
Thirteen patients with chronic hepatitis B were studied longitudinally for at least 6 months. The clinical and virological features of the patients at the time of selection are shown in Table 1. Patients were initially selected for their levels of ALT and HBV DNA, HBeAg status (presence or absence of HBeAg or anti-HBe), and presence or absence of hepatitis flares. We divided them into four categories: (i) group A—patients C1, C2, and C3, anti-HBe positive and stable levels of ALT (<40 U/liter) and HBV DNA (<106 copies/ml); (ii) group B—patients C4, C5, and C6, anti-HBe positive, ALT level of >40 U/liter, and HBV DNA level of <107 copies/ml; (iii) group C—patients C7, C8, and C9, HBeAg positive, anti-HBe negative, ALT level of >100 U/liter, HBV DNA level fluctuating from 103 to >107 copies/ml, and episodes of hepatic flares; and (iv) group D—patients C10, C11, C12, and C13, HBeAg positive, anti-HBe negative, ALT level of <70 U/liter, HBV DNA level of >107 copies/ml, and stable profile of disease. Two patients who recovered from acute hepatitis B were also monitored longitudinally for 3 years after recovery.
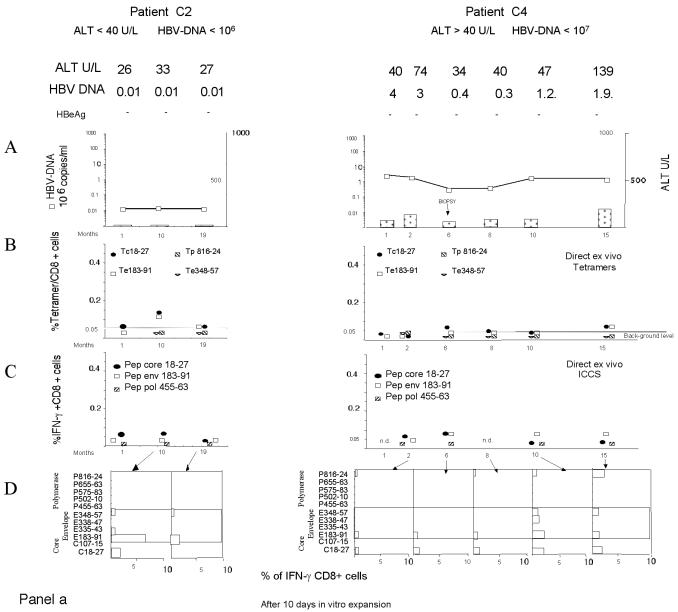
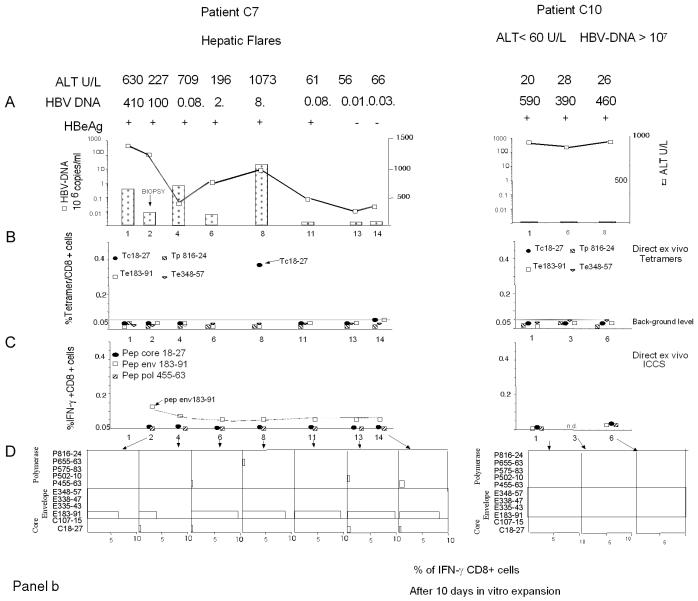
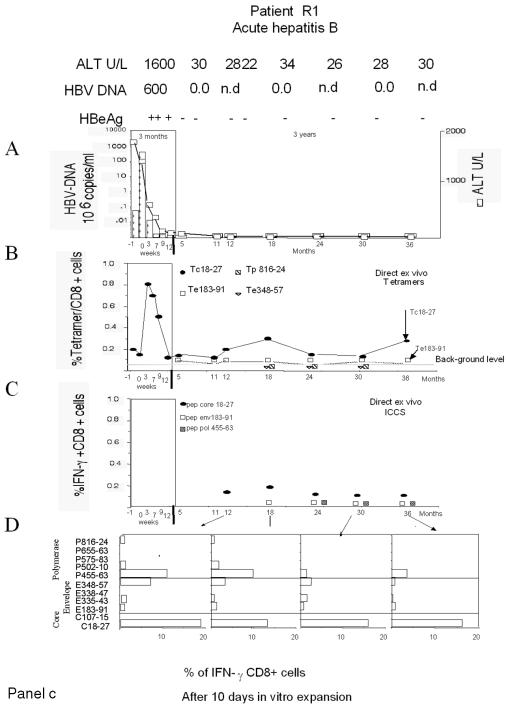
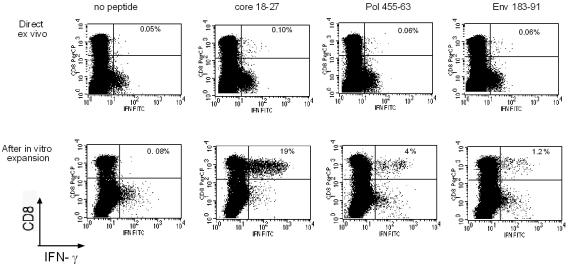
Figure 1 shows the complete set of HBV-specific CD8+-T-cell analyses performed for a representative patient from each chronic HBV infection group (Fig. 1a and b) and for one patient who recovered from acute hepatitis (Fig. 1c). The frequency of HBV-specific CD8+ T cells was calculated directly ex vivo with HLA-A2 tetramers (specific for core positions 18 to 27 [core 18-27], envelope positions 183 to 191 [env 183-191] and 348 to 357 [env 348-357], and polymerase positions 816 to 824 [pol 816-824]) (20) or by ICCS with the following peptides: core 18-27, env 183-191, and pol 455-463 (polymerase positions 455 to 463). The presence of HBV-specific CD8+ cells was also analyzed after in vitro expansion of PBMC stimulated with a larger set of HBV peptides corresponding to known HLA-A2-restricted epitopes (Table 2). The short-term lines produced after in vitro stimulation of PBMC with individual peptides were tested for the presence of peptide-specific IFN-γ-producing cells by ICCS. The frequency of HBV-specific CD8+ T cells was quantified by calculating the frequency of IFN-γ CD8+ T cells restimulated with the stimulatory peptides minus the frequency of IFN-γ CD8+ T cells stimulated with an irrelevant peptide or with no peptide.
FIG. 1.
Longitudinal analysis of HBV-specific CD8+-T-cell responses. Patients with different clinical profiles of HBV infection were studied longitudinally for ALT and HBV DNA levels and the frequencies of HBV-specific CD8+ T cells directly ex vivo and after in vitro expansion. A representative patient for each clinical group is shown: chronically infected patients C2, C4, C7, and C10 in panels a and b and patient R1, with resolved acute hepatitis B, in panel c. (A) Time course analysis of HBV DNA and ALT levels. Serum ALT levels (units per liter [U/L]) (bars) and HBV DNA levels (copies per milliliter) (squares) were analyzed longitudinally. Time zero in patient R1 is the day of clinical onset. n.d., not determined. (B) Percentages of CD8+ T cells that were tetramer positive at each time point. PBMC were tested directly ex vivo with the indicated tetramers (Tc18-27, core 18-27 tetramer; Te183-91, env 183-191 tetramer; Tp816-24, pol 816-824 tetramer; Te348-57, env 348-357 tetramer). The background level of direct ex vivo tetramer staining (0.05%, indicated by a horizontal line) was calculated in HLA-A2-positive, noninfected subjects and in HLA-A2-negative, HBV-infected patients (21). (C) Percentages of peptide-specific IFN-γ-producing CD8+ T cells determined directly ex vivo at different time points. PBMC were stimulated directly ex vivo with the indicated peptides (Pep). The percentages of IFN-γ-producing CD8+ T cells were calculated after subtraction of doubly positive cells obtained from nonstimulated PBMC. (D) Percentages of peptide-specific IFN-γ-producing CD8+ T cells after in vitro expansion (bars). PBMC were stimulated with the indicated peptides. After 10 days of in vitro expansion, the percentages of peptide-specific IFN-γ-producing CD8+ T cells were calculated.
Direct ex vivo frequency of circulating HBV-specific CD8+ T cells.
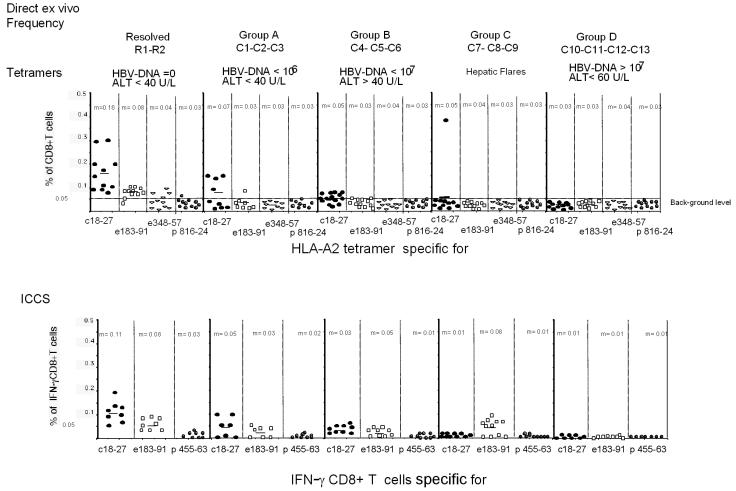
Results of the analyses of the direct ex vivo frequency of HBV-specific CD8+ T cells with HLA-A2 tetramers and by ICCS for 13 HLA-A2-positive chronically infected patients and 2 patients who recovered from acute HBV infection are summarized in Fig. 2. Differences in the profiles of virus-specific CD8+ T cells were found among the patients in the chronic hepatitis B groups.
FIG. 2.
Direct ex vivo frequencies of HBV-specific CD8+ T cells. The direct ex vivo frequencies of CD8+ T cells stained with HLA tetramers (Tc18-27, Te183-191, Te348-357, and Tp816-824) or producing IFN-γ after stimulation with core 18-27, env 183-191, and pol 455-463 peptides are shown for the indicated groups of patients. The frequency of IFN-γ-producing CD8+ T cells stimulated by the single peptides was subtracted from the frequency obtained in non-peptide-stimulated PBMC. All of the data obtained at different time points for each patient are shown. m, mean. HBV DNA levels are given in copies per milliliter; ALT levels are given in units per liter (U/L).
HBV-specific CD8+ T cells were not detected directly ex vivo in patients with stable high levels of HBV DNA replication (group D; HBV DNA level of >107 copies/ml, HBeAg positive) and either normal ALT levels (<35 U/liter) or elevated ALT levels (∼70 U/liter). Four patients were analyzed at 11 different times (a total of 59 different tests), but HBV-specific CD8+ T cells were not visualized either with HLA-tetramers or by ICCS. In contrast, HBV-specific CD8+ T cells could be visualized directly ex vivo in other groups of chronic hepatitis B patients characterized by lower levels of HBV replication.
CD8+ cells specific for core 18-27 (core 18-27-specific CD8+ cells) could be visualized only in patients with levels of HBV replication of <107 copies/ml either with tetramers or by ICCS. Frequencies were low (never higher than 0.15% with the core 18-27 tetramer) but occasionally were comparable to those found in patients with resolved hepatitis (Fig. 2).
We also detected a high frequency of core 18-27-specific CD8+ cells in one chronically infected patient during an episode of a hepatic flare (Fig. 1b, patient C7). This patient was characterized by fluctuating levels of HBV DNA (from 8 × 104 to 4 × 108 copies/ml), and the high frequency of core 18-27-specific CD8+ T cells was visualized with tetramers only at the time of the flare characterized by levels of HBV DNA of <107 copies/ml, which preceded HBeAg seroconversion. Note that these core 18-27-specific CD8+ T cells were unable to produce IFN-γ or to expand (Fig. 1a, patient C7). Importantly, the other two hepatic flares present in patient C7 (Fig. 1b), as well as two other distinct flares present in patients C8 and C9 (data not shown), were not coupled with an increase in the frequency of HBV-specific CD8+ T cells.
Envelope-specific CD8+ cells could be detected directly ex vivo in four out of six chronically infected patients with levels of HBV DNA of <107 copies/ml but only in two out of seven patients with levels of HBV DNA of >107 copies/ml (patient C7, env 183-191-specific CD8+ cells; and patient C8, env 348-357-specific CD8+ cells) (data not shown). However, envelope-specific CD8+ cells could be detected at a very low frequency (often <0.05%) only by ICCS and not with the specific HLA-A2 tetramer (Fig. 2). A characterization of tetramer-negative CD8+ cells specific for env 183-191 and env 348-357 epitopes was recently reported in a cross-sectional study (34). Here we were able to monitor the frequencies of env 183-191-specific CD8+ T cells for more than 1 year in at least three patients (patients C4, C5, and C7). The frequencies were remarkably stable over time (Fig. 1, patients C4 and C7). The results obtained for patient C7 (Fig. 1a), who displayed episodes of acute disease flares, were particularly interesting. Despite fluctuating levels of HBV DNA, the absence of mutations in the relevant env 183-191 epitope (Table 3), and ALT levels falling from 1,073 to 60 U/liter, the direct ex vivo frequency (and the capacity for in vitro expansion) of env 183-191-specific CD8+ T cells remained unchanged. This stability contrasts with the behavior of core 18-27-specific CD8+ cells present in the same patients and with that observed during acute hepatitis, where numbers of circulating CD8 T cells increased in proportion to liver enzyme levels and decreased sharply after recovery (21).
TABLE 3.
Viral mutations in selected HLA-A2-restricted epitopes
| Patient | CD8+-T-cell response of and mutation in the following peptide (sequence)a:
|
|||
|---|---|---|---|---|
| Core 18-27 (FLPSDFFPSV) | Env 183-191 (FLLTRILTI) | Env 348-357 (GLSPTVWLSV) | Pol 455-463 (GLSRYVARL) | |
| C2 | + | + | + | − (ND) |
| C4 | + (I to V 27) | + | + | − |
| C5 | + | + | + | − |
| C6 | + | + | − (V to A 352) | + |
| C7 | + | + | − | − |
| C8 | − | − | − (A to V 357) | + |
| C9 | − | − | − (A to V 357) | + |
| C10 | − (V to I 27) | − | − | − (P to S 457) |
| C11 | − | − | − (ND) | − |
| C12 | − | + | − (ND) | − |
| C13 | − | − | − (ND) | − (ND) |
Presence (+) or absence (−) of an epitope-specific CD8+-T-cell response. Mutations are shown in parentheses; e.g., I to V 27 represents a mutation from isoleucine to valine at position 27. ND, not determined.
Frequency of HBV-specific CD8+ T cells after in vitro expansion.
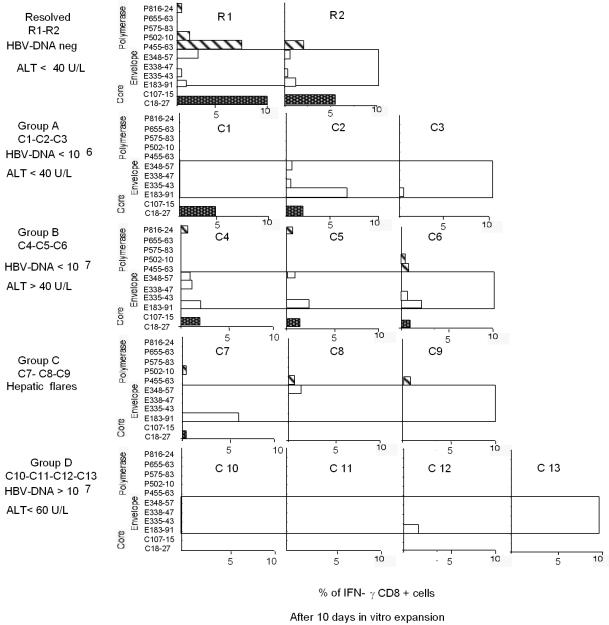
The different sensitivities of direct ex vivo analysis and analysis after in vitro expansion for HBV-specific CD8+-T-cell responses are shown in Fig. 3. For a representative subject (R1), HBV-specific CD8+ T cells were demonstrated after a round of in vitro expansion but would have been missed by direct ex vivo analysis. The profiles of HBV-specific CD8+ T cells after in vitro expansion in all patients are shown in Fig. 4. The results obtained by analysis after in vitro expansion confirmed the patterns observed after direct ex vivo analysis. In the absence of mutations in the epitope (Table 3), circulating core 18-27-specific CD8+ T cells were detectable only when HBV DNA levels were <107 copies/ml (Fig. 1 and 4, patients C1, C4, and C7). In contrast, efficient expansion of envelope- and polymerase-specific CD8+ T cells was seen even with HBV DNA levels of >107 copies/ml (Fig. 1 and 4, patient C7).
FIG. 3.
Analysis of IFN-γ-producing CD8+ T cells directly ex vivo or after in vitro expansion. IFN-γ-producing CD8+ T cells were tested directly ex vivo in PBMC (upper dot plots) or after 10 days of in vitro expansion with the corresponding peptides (lower dot plots). The results shown are from experiments performed with PBMC from patient R1. PBMC were stimulated with the indicated individual peptides either for direct ex vivo analysis or for experiments performed after 10 days of in vitro expansion. In the experiments performed after in vitro expansion, the growing cells were restimulated either with no peptide or with the initial stimulatory HBV peptide. Subdominant peptides (pol 455-463 and env 183-191) tested positive only after 10 days of in vitro expansion.
FIG. 4.
Profiles of HBV-specific CD8+ responses obtained after in vitro expansion in various patients. Peptide-specific IFN-γ-producing CD8+ T cells were quantified after 10 days of in vitro expansion with the indicated peptides. One representative experiment for each patient is shown. Results for patients R1 and R2 were obtained, respectively, at 12 (R1) and 18 (R2) months after the resolution of acute hepatitis. Results for patients C2, C4, C7, and C10 represent the results shown in Fig. 1a and b at month 10 (C2 and C4), month 8 (C7), and month 3 (C10). HBV DNA levels are given in copies per milliliter; ALT levels are given in units per liter (U/L).
No correlation between the level of ALT and the detection of HBV-specific CD8+ T cells was found.
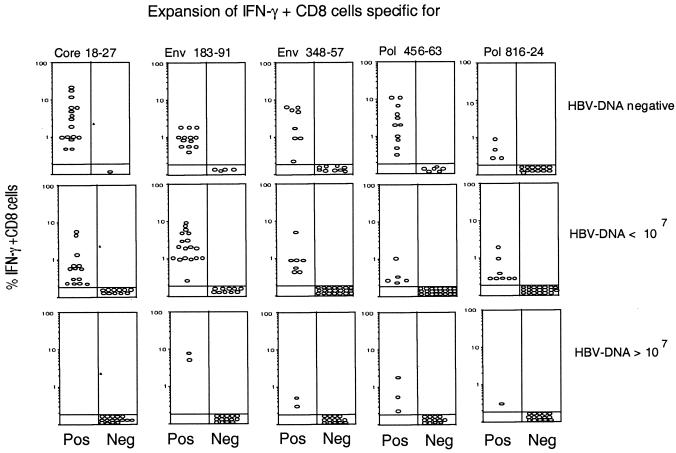
The different relationships of the core-, envelope-, and polymerase-specific CD8+ T cells to the level of HBV DNA replication are evident in Fig. 5. The frequency of epitope-specific CD8+ cells after in vitro expansion at all times was plotted against the level of HBV DNA present at the time of the analysis for the various patients. The in vitro expansion of circulating core 18-27-specific CD8+ T cells was inversely proportional to the quantity of HBV DNA. A large expansion of IFN-γ-producing core 18-27-specific CD8+ T cells was present in patients with resolved acute hepatitis (HBV DNA undetectable), a lower level of expansion was present in patients with HBV DNA levels of <107 copies/ml, and no expansion was present in patients with HBV DNA levels of >107 copies/ml. In contrast, the expansion of pol 455-463-, pol 816-824-, env 183-191-, and env 348-357-specific CD8+ T lymphocytes was less influenced by HBV DNA replication and was also seen in some patients with HBV DNA levels of >107 copies/ml and without amino acid mutations in the viral epitope (Table 3).
FIG. 5.
Expansion of epitope-specific IFN-γ-producing CD8+ T cells in relation to HBV DNA levels. The percentages of IFN-γ-producing CD8+ T cells specific for the indicated HBV epitopes present in the T-cell lines generated after in vitro peptide stimulation of PBMC were grouped in relation to the HBV DNA levels present at the time of the assays. Data include “positive” (Pos) experiments (presence of peptide-specific IFN-γ-producing CD8+ T cells) and “negative” (Neg) experiments (absence of peptide-specific IFN-γ-producing CD8+ T cells) performed for patients studied longitudinally. HBV DNA levels are given in copies per milliliter.
Patients with chronic HBV infections and with HBV DNA levels of <107 copies/ml demonstrate multispecific CD8+-T-cell responses.
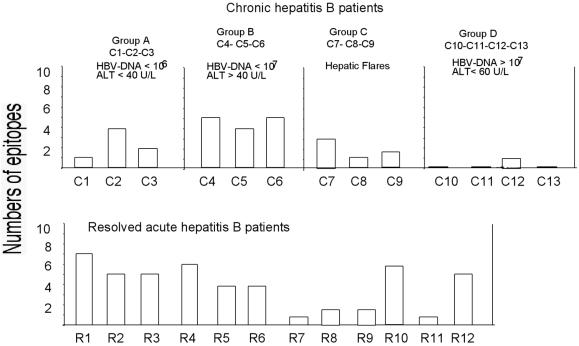
The analysis of HBV-specific CD8+ T cells after a round of in vitro expansion with the 11 different HBV peptide epitopes showed that several chronically infected patients possessed CD8+ T cells specific for multiple HBV determinants (Fig. 4). We compared the extents of multispecificity present in patients with resolved infections and persistently infected patients. PBMC from 12 HLA-A2-positive patients who had recovered from acute HBV infections were stimulated with the 11 different peptides used in the longitudinal study. Figure 6 shows that the numbers of HBV epitopes that could be recognized by cells from patients with resolved acute and chronic HBV infections were similar. In particular, cells from all three chronically infected patients with HBV DNA levels of <107 copies/ml and signs of inflammatory liver disease could recognize at least 4 of the 11 HBV epitopes tested. This degree of multispecificity was higher than or identical to that seen in 6 of 12 patients with resolved acute HBV infections.
FIG. 6.
Numbers of HBV epitopes recognized by patients with chronic or resolved HBV infections. Bars indicate the numbers of peptides recognized by each patient for the 11 HBV epitopes tested. PBMC from the indicated patients with chronic or resolved HBV infections were stimulated with 11 HBV peptides representing known HLA-A2-restricted epitopes (Table 2). After 8 to 10 days of in vitro expansion, the short-term cell lines were tested for the presence of peptide-specific IFN-γ-producing CD8+ T cells. HBV DNA levels are given in copies per milliliter; ALT levels are given in units per liter (U/L).
Influence of intrahepatic sequestration and of epitope mutations on core 18-27-specific CD8+ T cells.
The failure to detect circulating core 18-27-specific CD8+ T cells in patients with HBV DNA levels of >107 copies/ml might be explained by intrahepatic localization of these cells. To address this possibility, we analyzed the direct ex vivo frequency of core 18-27-specific CD8+ cells in the liver of patients with chronic hepatitis B. The frequency of intrahepatic T cells found by HLA tetramer staining in relation to the HBV DNA levels at the time of liver biopsy clearly showed that core 18-27-specific CD8 +T cells do not accumulate in the liver in proportion to HBV DNA levels (Fig. 7). On the contrary, and in keeping with previous results (20), the frequency of core 18-27-specific CD8+ T cells was inversely proportional to the level of HBV replication (P value, <0.0001, as determined by the Spearman correlation test), and these cells were not found in the liver of patients with serum HBV DNA levels of >107 copies/ml. The only exception was patient C7, who had core 18-27-specific CD8+ T cells at a frequency of 0.3% of total intrahepatic CD8 T cells. Thus, the absence of circulating core 18-27-specific CD8+ T cells in patients with a high level of HBV replication cannot be explained by preferential intrahepatic compartmentalization.
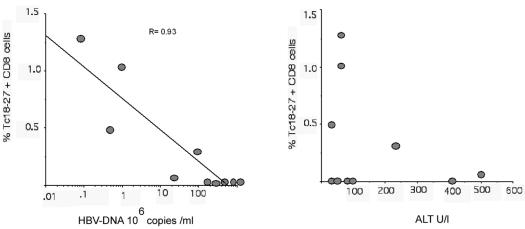
FIG. 7.
Frequencies of intrahepatic core 18-27-specific CD8+ T cells in relation to HBV DNA and ALT levels. Percentages of tetramer core 18-27-specific (Tc18-27) CD8+ T cells present in lymphocytes purified from liver biopsy specimens were plotted in relation to the HBV DNA and ALT levels (units per liter [U/l]) present at the time of liver biopsy. Results obtained for 10 different patients are shown. There was a strong negative correlation between HBV DNA and core 18-27-specific CD8+ T cells (r value determined by the Spearman correlation test, −0.93; P = 0.0001) but no correlation between ALT and core 18-27-specific CD8+ T cells (r = 0.23; P = 0.53).
The absence of core 18-27-specific CD8+ T cells is also not caused by viral mutations resulting in specific epitope inactivation (27). The core protein of HBV strains involved in chronic infections was sequenced. Except for patient C10, infected with an HBV strain carrying an isoleucine-to-valine mutation at position 27, viral mutations could not be found within the core 18-27 epitope or within flanking residues in patients with HBV DNA levels of >107 copies/ml (Table 3). Note that this isoleucine-to-valine mutation is not associated with core 18-27 unresponsiveness, since this amino acid mutation was also detected in patient C4, who showed a CD8+-T-cell response against this epitope (Table 3).
Functional avidity of HBV-specific CD8+ T cells present in patients with resolved or chronic HBV infections.
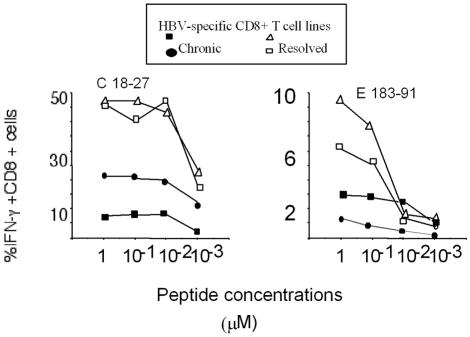
The persistence of virus-specific CD8 T cells during chronic infection can depend on the avidity of T cells for their target or on the different quantities of virus antigens displayed by infected cells. High-affinity T cells are more likely to be deleted than low-affinity ones, and viral epitopes that are displayed at higher levels appear to drive faster T-cell deletion (1, 24). We investigated whether chronic HBV infection could have selected HBV-specific CD8+ T cells with a low avidity. The activation of core 18-27- and env 183-191-specific CD8+-T-cell lines from both patients with resolved HBV infections and patients with chronic HBV infections was analyzed by measuring IFN-γ production induced by target cells pulsed with decreasing peptide concentrations. T-cell lines were generated from patients with different levels of HBV DNA. Although T-cell lines from patients with resolved infections had a higher frequency of core- or envelope-specific CD8+ T cells, we did not find any evidence of selection of low-avidity HBV-specific T cells in chronically infected patients. Core 18-27-specific CD8+ T cells produced in both patients with chronic infections and patients with resolved infections efficiently recognized target cells pulsed with 10−3 μM peptide. There was also no detectable difference in T-cell avidity between env 183-191-specific CD8+ T cells from patients with resolved infections and those from patients with chronic infections, both of which recognized target cells pulsed with 10−2 μM peptide (Fig. 8). Core- and envelope-specific CD8+ cell lines from patients with chronic infections and patients with resolved infections were both able to recognize HepG2 cells transfected with the entire HBV genome (37) (data not shown).
FIG. 8.
Functional T-cell avidity of HBV epitope-specific CD8+ T cells. Various T-cell lines specific for the indicated HBV epitopes and produced in patients with resolved infections or chronic infections were stimulated with various concentrations of peptides. Core 18-27- and env 183-191-specific CD8+ T cells were derived from patients R1 and R2 (with resolved infections). Core 18-27-specific CD8+ T cells were derived from chronically infected patients C4 and C5; env 183-191-specific CD8+ T cells were derived from patients C5 and C7. Frequencies of IFN-γ-producing CD8+ T cells activated by the indicated peptide concentrations are shown.
DISCUSSION
The HBV-specific CD8+-T-cell response in patients with chronic HBV infections has been studied to date by using cytotoxic experiments performed after several rounds of in vitro expansion (2, 28, 31, 32, 38). Few studies have attempted to evaluate this response directly ex vivo (12, 20, 40). Here we used a combination of methods (HLA tetramers and ICCS) directly ex vivo and after in vitro expansion to increase our ability to study virus-specific CD8+ T cells. Due to the functional alterations of CD8+ T cells caused by chronic viral infection (34), immunological studies performed with single detection methods might underestimate the quantity of virus-specific CD8+ T cells.
The results obtained here confirm that the HBV-specific CD8 T-cell response is weak in the blood of patients with chronic HBV infections (9), with the direct ex vivo frequency of HBV-specific cells usually being <0.1 to 0.2% of total CD8+ T cells. Despite this low frequency, the majority of patients with chronic hepatitis B possess HBV-specific CD8+ T cells, in line with the results of previous investigations of the helper T-cell response (18, 23). Circulating HBV-specific CD8+ T cells were demonstrated in 10 out of 13 patients with chronic hepatitis B (76%), even though analysis of the CD8+-T-cell response was measured with a selected repertoire of HLA-A2-restricted epitopes which is not fully representative of the overall CD8 response (16).
An important finding of this study is that we did not observe a correlation between the profile of circulating HBV-specific CD8+ T cells and the degree of liver damage. Higher frequencies of HBV-specific CD8+ T cells were not present in patients with higher levels of transaminases (Fig. 2). This lack of correlation between HBV-specific CD8+ T cells and liver damage was also supported by results obtained during episodes of acute clinical flares (i.e., ALT levels of >400 U/liter). In four out of five flares, studied in three different patients, we were unable to detect any increase in the frequency of circulating HBV-specific CD8+ T cells. Only in a single case, in which the flare preceded HBeAg seroconversion and occurred with HBV DNA levels of <107 copies/ml, were we able to detect a sharp increase in the frequency of core 18-27-specific CD8+ T cells. It is possible that our inability to correlate CD8+-T-cell frequency with the extent of liver damage results from the fact that events in the periphery poorly reflect those occurring within the liver. We cannot exclude the possibility that recovery of the HBV-specific T-cell response takes place exclusively within the liver during flares. Furthermore, CD8+-T-cell epitopes other than those analyzed in this study might play a role in the pathogenesis of hepatic flares. Nevertheless, we did not find any consistent association between the HBV-specific CD8+-T-cell response and the extent of liver damage, in contrast to the clear association between the presence of these cells and viral load. We consistently detected a higher frequency of HBV-specific CD8+ T cells in patients with a low level of HBV replication than in those with a high level of HBV replication. This finding is in agreement with the results of previous studies carried out with fewer cytotoxic T-lymphocyte epitopes (20, 40) and in those performed after spontaneous (33) or treatment-induced (6, 33) reduction of HBV replication.
A serum HBV DNA concentration of about 107 copies/ml appears to be the dividing threshold for consistent detection of circulating HBV-specific CD8+ T cells. Below this concentration, HBV-specific CD8+ T cells could be detected both directly ex vivo and after in vitro expansion; in addition, the CD8 response was often multispecific, with CD8+ T cells specific for epitopes being present in both structural (core and envelope) and nonstructural (polymerase) proteins. Above this concentration, the detection of HBV-specific CD8+ cells in the circulation of chronic hepatitis B patients was more difficult; CD8+ T cells displayed phenotypic alterations (i.e., tetramer negativity) (34), and the frequency of direct detection was very low, with CD8+ T cells occasionally being detectable only after a round of in vitro expansion. More importantly, differences were detectable among CD8+ T cells specific for different epitopes. Core 18-27-specific CD8 cells, which are often dominant in patients with acute hepatitis B (Fig. 1) (21, 43), are most affected by increased levels of HBV replication. These cells cannot be detected in the circulation (either directly ex vivo or after in vitro expansion) when HBV DNA levels are >107 copies/ml, and we demonstrated that this lack of detection within the circulatory compartment is not caused by preferential intrahepatic localization of core 18-27-specific CD8+ T cells. The frequency of core 18-27-specific CD8+ T cells within the liver is also inversely proportional to the level of HBV replication. Except for a single case (patient C10), HBV infecting patients at concentrations of >107 copies/ml did not show within the core 18-27 sequence mutations which might abolish core 18-27-specific CD8+-T-cell expansion. Furthermore, we did not find any evidence for a progressive selection of low-avidity core 18-27-specific CD8+ T cells in chronically infected patients with HBV DNA levels of <107 copies/ml. Thus, the absence of circulating core 18-27-specific CD8+ T cells associated with HBV DNA levels of >107 copies/ml might be due to deletion caused by a high level of antigen (14, 35) or by an inherent inability to mount a CD8+-T-cell response against this epitope.
Envelope- and polymerase-specific CD8+ T cells are instead the only cells which can be demonstrated in patients with chronic hepatitis B and concentrations of HBV DNA of >107 copies/ml. Their ability to persist in the presence of a high level of HBV replication is associated with an apparent inability to exert antiviral function. We did not find viral mutations in the relevant envelope and polymerase epitopes in patients who had envelope- or polymerase-specific CD8+ T cells. More importantly, envelope-specific CD8+ cells are characterized by an altered phenotype (tetramer negativity) (34), and their indifference to the dynamic fluctuations of HBV DNA levels (Fig. 1a, patient C7) is suggestive of a tolerant state. The persistence of polymerase-specific CD8+ T cells could be the result of the low quantity of polymerase epitopes expressed in vivo by infected hepatocytes, as suggested by recent results obtained with the transgenic mouse model of HBV infection (14). Interestingly, polymerase-specific CD8+ T cells do not recognize target cells transfected with the HBV genome and producing whole HBV virions (2.2.15 cells) (N. Naoumov et al., unpublished data), while core- and envelope-specific CD8+ T cells are efficiently activated by them.
We cannot rule out the possibility that the different behaviors of the distinct CD8 T-cell epitopes are due not to their HBV protein derivation but to the intrinsic features of the epitopes (such as affinity of HLA-A2 binding, efficiency of presentation, availability of a T-cell repertoire, and cross-reactivity) (7, 8, 17). Nevertheless, we think that a number of pieces of evidence support the former possibility. First, the different behaviors of HBV-specific CD8+ T cells, according to their antigenic derivation, were shown recently for transgenic mice (14), and our results are in line with these findings. Second, the persistence of envelope- and polymerase-specific CD8+ T cells in patients with HBV DNA levels of >107 copies/ml is not restricted to a single epitope but encompasses different epitopes present in the same HBV proteins (34). Third, CD8+-T-cell responses specific for core epitopes and restricted to non-HLA-A2 molecules display behaviors identical to those of HLA-A2-restricted core 18-27-specific CD8+ T cells. They are usually dominant in non-HLA-A2-positive patients with self-limited infection (4) and are only present in chronically infected patients with low HBV DNA levels (40).
We think that this extensive analysis of HBV-specific CD8+ T cells in patients with chronic hepatitis B might be relevant to the tailoring of new immunotherapeutic approaches for hepatitis B treatment. It remains difficult to select the best HBV proteins for inclusion in a therapeutic vaccine. The fact that core-specific CD8+ cells are associated, as shown both here and in other work (10, 15), with the control of HBV replication strongly suggests that core antigen should be included in a vaccine formulation. Nevertheless, association is not proof of a causative effect, and we should not forget that this association might be the consequence and not the cause of low HBV DNA levels. However, demonstration that the extent of the peripheral repertoire of HBV-specific CD8+ cells is inversely proportional to the level of HBV replication reinforces the therapeutic approach of inhibiting HBV by using antiviral drugs before using vaccines to boost HBV-specific T-cell immunity (3). Our data suggest that HBV replication should be reduced to a level lower than 107 copies/ml in order to maximize the chances of vaccines to expand a broad repertoire of HBV-specific CD8+ cells.
Acknowledgments
We thank Mala K. Maini and Nikolai Naoumov for helpful comments and Annette Ives for manuscript preparation.
This work was supported by a National Lottery Board grant awarded through the Digestive Disorders Foundation and by EU grant QLK2-CT-2002-00700.
REFERENCES
- 1.Alexander-Miller, M. A., G. R. Leggatt, A. Sarin, and J. A. Berzofsky. 1996. Role of antigen, CD8, and CTL avidity in high dose antigen induction of apoptosis of effector CTL. J. Exp. Med. 184:485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoletti, A., A. Costanzo, F. V. Chisari, M. Levrero, M. Artini, A. Sette, A. Penna, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1994. Cytotoxic T lymphocyte response to a wild-type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med. 180:933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoletti, A., and N. V. Naoumov. 2003. Translation of immunological knowledge into better treatments of chronic hepatitis B. J. Hepatol. 39:115-124. [DOI] [PubMed] [Google Scholar]
- 4.Bertoni, R., J. Sidney, P. Fowler, R. Chesnut, F. Chisari, and A. Sette. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Investig. 100:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bocher, W., S. Herzog-Hauff, J. Schlaak, K. Meyer zum Buschenfelde, and H. Lohr. 1998. Kinetics of hepatitis B surface antigen-specific immune responses in acute and chronic hepatitis B or after HBs vaccination: stimulation of the in vitro antibody response by interferon gamma. Hepatology 29:238-244. [DOI] [PubMed] [Google Scholar]
- 6.Boni, C., A. Penna, G. Ogg, A. Bertoletti, M. Pilli, A. Cavalli, S. Urbani, R. Boheme, R. Panebianco, F. Fiaccadori, and C. Ferrari. 2001. Lamivudine treatment can overcome cytotoxic T cell hyporesponsiveness in chronic hepatitis B: new perspective for immune therapy. Hepatology 33:963-971. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, M. A., A. K. Pinto, K. A. Daniels, J. P. Schneck, R. M. Welsh, and L. K. Selin. 2002. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat. Immunol. 3:627-634. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W., L. C. Anton, J. R. Bennink, and J. W. Yewdell. 2000. Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity 12:83-93. [DOI] [PubMed] [Google Scholar]
- 9.Chisari, F. 1997. Cytotoxic T cells and viral hepatitis. J. Clin. Investig. 99:1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, C., A. Penna, A. Bertoletti, A. Valli, A. Degli Antoni, T. Giuberti, A. Cavalli, M. A. Petit, and F. Fiaccadori. 1990. Cellular immune response to hepatitis B virus encoded antigens in acute and chronic hepatitis B virus infection. J. Immunol. 145:3442-3449. [PubMed] [Google Scholar]
- 11.Hwang, Y. K., N. K. Kim, J. M. Park, K. Lee, W. K. Han, H. I. Kim, and H. S. Cheong. 2002. HLA-A2 1 restricted peptides from the HBx antigen induce specific CTL responses in vitro and in vivo. Vaccine 20:3770-3777. [DOI] [PubMed] [Google Scholar]
- 12.Jung, M., B. Hartmann, J. Gerlach, H. Diepolder, R. Gruber, W. Schraut, N. Gruner, R. Zachoval, R. Hoffmann, T. Santantonio, and G. Pape. 1999. Virus-specific lymphokine production differs quantitatively but not qualitatively in acute and chronic hepatitis B infection. Virology 261:165-172. [DOI] [PubMed] [Google Scholar]
- 13.Jung, M., U. Spengler, W. Schraut, R. Hoffman, R. Zachoval, J. Eisemburg, D. Eichenlaub, G. Riethmuller, G. Paumgartner, H. W. L. Ziegler-Heitbrock, and G. R. Pape. 1991. Hepatitis B virus antigen-specific T-cell activation in patients with acute and chronic hepatitis B. J. Hepatol. 13:310-317. [DOI] [PubMed] [Google Scholar]
- 14.Kakimi, K., M. Isogawa, J. Chung, A. Sette, and F. V. Chisari. 2002. Immunogenicity and tolerogenicity of hepatitis B virus structural and nonstructural proteins: implications for immunotherapy of persistent viral infections. J. Virol. 76:8609-8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau, G. K., D. Suri, R. Liang, E. I. Rigopoulou, M. G. Thomas, I. Mullerova, A. Nanji, S. T. Yuen, R. Williams, and N. V. Naoumov. 2002. Resolution of chronic hepatitis B and anti-HBs seroconversion in humans by adoptive transfer of immunity to hepatitis B core antigen. Gastroenterology 122:614-624. [DOI] [PubMed] [Google Scholar]
- 16.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 76:6104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitsky, V., Q. J. Zhang, J. Levitskaya, and M. G. Masucci. 1996. The life span of major histocompatibility complex-peptide complexes influences the efficiency of presentation and immunogenicity of two class I-restricted cytotoxic T lymphocyte epitopes in the Epstein-Barr virus nuclear antigen 4. J. Exp. Med. 183:915-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohr, H. F., W. Weber, J. Schlaak, B. Goergen, K. H. Meyer zum Buschenfelde, and G. Gerken. 1995. Proliferative response of CD4+ T cells and hepatitis B virus clearance in chronic hepatitis with or without hepatitis B e-minus hepatitis B virus mutants. Hepatology 22:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Lok, A. S., and B. J. McMahon. 2001. Chronic hepatitis B. Hepatology 34:1225-1241. [DOI] [PubMed] [Google Scholar]
- 20.Maini, M. K., C. Boni, C. K. Lee, J. R. Larrubia, S. Reignat, G. S. Ogg, A. S. King, J. Herberg, R. Gilson, A. Alisa, R. Williams, D. Vergani, N. V. Naoumov, C. Ferrari, and A. Bertoletti. 2000. The role of virus-specific CD8+ cells in viral control and liver damage during persistent hepatitis B virus (HBV) infection. J. Exp. Med. 191:1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maini, M. K., C. Boni, G. S. Ogg, A. S. King, S. Reignat, C. K. Lee, J. R. Larrubia, G. J. M. Webster, A. J. McMichael, C. Ferrari, R. Williams, D. Vergani, and A. Bertoletti. 1999. Direct ex vivo analysis of hepatitis B virus-specific CD8+ T cells associated with the control of infection. Gastroenterology 117:1386-1396. [DOI] [PubMed] [Google Scholar]
- 22.Malacarne, F., G. J. Webster, S. Reignat, J. Gotto, S. Behboudi, A. K. Burroughs, G. M. Dusheiko, R. Williams, and A. Bertoletti. 2003. Tracking the source of the hepatitis B virus-specific CD8 T cells during lamivudine treatment. J. Infect. Dis. 187:679-682. [DOI] [PubMed] [Google Scholar]
- 23.Maruyama, T., A. McLachlan, S. Iino, K. Koike, K. Kurokawa, and D. Milich. 1993. The serology of chronic hepatitis B infection revisited. J. Clin. Investig. 91:2586-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan, D. J., H. T. Kreuwel, and L. A. Sherman. 1999. Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J. Immunol. 163:723-727. [PubMed] [Google Scholar]
- 25.Nayersina, R., P. Fowler, S. Guilhot, G. Missale, A. Cerny, H. J. Schlicht, A. Vitiello, R. Chesnut, J. L. Person, A. J. Redeker, and F. V. Chisari. 1993. HLA-A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J. Immunol. 150:4659-4671. [PubMed] [Google Scholar]
- 26.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 27.Nowak, M. A., R. M. May, R. E. Phillips, S. Rowland-Jones, D. G. Lalloo, S. McAdam, P. Klenerman, B. Koppe, K. Sigmund, C. R. Bangham, et al. 1995. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature 375:606-611. [DOI] [PubMed] [Google Scholar]
- 28.Penna, A., F. V. Chisari, A. Bertoletti, G. Missale, P. Fowler, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1991. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J. Exp. Med. 174:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penna, A., G. Del Prete, A. Cavalli, A. Bertoletti, M. M. D'Elios, R. Sorrentino, M. D'Amato, C. Boni, M. Pilli, F. Fiaccadori, and C. Ferrari. 1997. Predominant T-helper 1 cytokine profile of hepatitis B virus nucleocapsid-specific T cells in acute self-limited hepatitis B. Hepatology 25:1022-1027. [DOI] [PubMed] [Google Scholar]
- 30.Perrillo, R. P. 2001. Acute flares in chronic hepatitis B: the natural and unnatural history of an immunologically mediated liver disease. Gastroenterology 120:1009-1022. [DOI] [PubMed] [Google Scholar]
- 31.Rehermann, B., K. M. Chang, J. McHutchinson, R. Kokka, M. Houghton, C. M. Rice, and F. V. Chisari. 1996. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. J. Virol. 70:7092-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehermann, B., P. Fowler, J. Sidney, J. Person, A. Redeker, M. Brown, B. Moss, A. Sette, and F. V. Chisari. 1995. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 181:1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehermann, B., D. Lau, J. H. Hoofnagle, and F. V. Chisari. 1996. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Investig. 97:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reignat, S., G. J. Webster, D. Brown, G. S. Ogg, A. King, S. L. Seneviratne, G. Dusheiko, R. Williams, M. K. Maini, and A. Bertoletti. 2002. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J. Exp. Med. 195:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha, B., A. Grandien, and A. A. Freitas. 1995. Anergy and exhaustion are independent mechanisms of peripheral T cell tolerance. J. Exp. Med. 181:993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossol, S., G. Marinos, P. Carucci, M. V. Singer, R. Williams, and N. V. Naoumov. 1997. Interleukin-12 induction of Th1 cytokines is important for viral clearance in chronic hepatitis B. J. Clin. Investig. 99:3025-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sells, M. A., M. L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sing, G. K., A. Ladhams, S. Arnold, H. Parmar, X. Chen, J. Cooper, L. Butterworth, K. Stuart, D. D'Arcy, and W. G. Cooksley. 2001. A longitudinal analysis of cytotoxic T lymphocyte precursor frequencies to the hepatitis B virus in chronically infected patients. J. Viral Hepatol. 8:19-29. [DOI] [PubMed] [Google Scholar]
- 39.Sobao, Y., K. Sugi, H. Tomiyama, S. Saito, S. Fujiyama, M. Morimoto, S. Hasuike, H. Tsubouchi, K. Tanaka, and M. Takiguchi. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 34:922-929. [DOI] [PubMed] [Google Scholar]
- 40.Sobao, Y., H. Tomiyama, K. Sugi, M. Tokunaga, T. Ueno, S. Saito, S. Fujiyama, M. Morimoto, K. Tanaka, and M. Takiguchi. 2002. The role of hepatitis B virus-specific memory CD8 T cells in the control of viral replication. J. Hepatol. 36:105-115. [DOI] [PubMed] [Google Scholar]
- 41.Thimme, R., S. Wieland, C. Steiger, J. Ghrayeb, K. A. Reimann, R. H. Purcell, and F. V. Chisari. 2003. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, S., M. Chen, and P. Yang. 1992. Acute exacerbation of chronic type B hepatitis are accompanied by increased T cell responses to hepatitis B core and e antigens. Implications for hepatitis B e antigen seroconversion. J. Clin. Investig. 98:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webster, G., S. Reignat, M. Maini, S. Whalley, G. Ogg, A. King, D. Brown, P. Amlot, R. Williams, D. Vergani, G. Dusheiko, and A. Bertoletti. 2000. Incubation phase of acute hepatitis B in man: dynamic of cellular immune mechanisms. Hepatology 32:1117-1124. [DOI] [PubMed] [Google Scholar]