Abstract
Oral and oropharyngeal cancers are characterized by relatively low 5- year survival rates due to many factors, including local recurrence. The identification of new molecular markers may serve for the estimation of prognosis and thus augment treatment decisions and affect therapy outcome. The aim of this study was to describe the morphological characteristics and the DNA methylation status of the CDKN2A,CDH1, ATM, FHIT and RAR- genes in the central and peripheral part of the tumor and the surgical margin and evaluate their prognostic significance. 53 patients with oral and oropharyngeal cancer were enrolled to the prospective study, and had been primarily treated surgically. Correlations between morphological data, hypermethylation status and clinicopathological data, as well as prognosis, were assessed. Nuclei polymorphism highly correlated with T stage (p < 0.0001), N stage (p < 0.046), and metastases to the lymph nodes pN (p < 0.004 ). Also, the number of cells in irregular mitosis correlated with T stage (p < 0.004), and highly with pN (p < 0.009). The significance of CDKN2A hypermethylation as a good prognostic factor was also established in the Kaplan-Meir test. The ultrastructural analysis showed that none of the examined tumors had homogenous texture and that resection margin specimens clean in HE stained tissue samples frequently contained single tumor cells or few cells in groups surrounded by connective tissue. This indicates the superiority of electron microscopy over standard histopathological analysis. Thus, a combination of such morphological examination with epigenetic parameters described herein could result in the discovery of promising new prognostic markers of the disease.
Keywords: Oral cancer, Ultrastructural examination, Methylation, Prognostic factors
Introduction
Oral cancer is a serious and growing medical problem. It is the sixth most common cancer in the world and occurs in people aged 50 and over [1, 2]. However, about 6 % of patients suffering from this type of disease are under 45 years old, or even under 40 in countries with a high incidence [3–6]. The clinical data regarding the course of the disease are not clear-cut [7]. The risk factors are well known (tobacco smoking, alcohol abusing), but there are currently many reports regarding non-smoking and non-drinking populations with HPV infection [8–11]. Treatment failure is still high; thus, the search for prognostic markers which could be useful in early detection of recurrence is ongoing.
Among many prognostic factors, the assessment of surgical margin seems to be crucial. Routine histopathological examination using hematoxylin–eosin staining of paraffin-embedded tissue samples is a golden standard in evaluation of the resection field by pathologists. However, this technique does not allow to describe the ultrastructure of the tumor and surgical margin. For the last-mentioned purpose, the epoxy resin-embedded samples are prepared. Then, before electron microscopy studies are done toluidine blue-stained semi-thin sections are examined. Using this technique, the pathologist may evaluate more details including nuclear polymorphism, cells in mitosis, as well the cellular characteristic (size, shapes, size of nuclei, number of nucleoli per nucleus, presence of polykaryocytes, typical cytoplasmic ultrastructural characteristic for squamous epithelium). This technique allows also to find more precisely suspected changes, as dysplastic or neoplastic cell “traveling” through normal tissue. Besides this, immunohistochemistry and molecular analysis could bring new information. It has to be underlined that genetic and epigenetic changes are also detected in histopathologically clean resection fields. It could cause local relapse in mucosa primarily free of cancer cells. This has been explained by the Slaughter’s model of “field cancerization” [12]. In turn, the Califano model is based on the increased number of genetic alterations in the field, with a division into early and late ones [13]. Finally, there is the “patch-field” model proposed by Braakhuis [14]. All these theories assume that the whole mucosa is predisposed to carcinogenesis due to exposure to exogenous genotoxins [15, 16].
Among the molecular alterations found in the early stage of carcinogenesis are epigenetic changes. Silencing of tumor suppressor genes associated with promoter hypermethylation is a common feature in human cancers and serves as a mechanism for the loss of their function. [17–19]. The hypermethylation status of different genes was stated to be important for prognosis in cancer patients. Methylation analyses are performed frequently using tumor tissue, but some results showed that the estimation of DNA methylation changes in the resection margin has an even greater prognostic value [20].
The epigenetic alterations of CDKN2A (p16; located at 9p21), CDH1 (16q22.1), FHIT (3p14.2), RAR-β (3p24.2), and ATM (11q22.3) have been found to be related to clinical prognosis in cancer of the head and neck [18]. The analysis of methylation of CDKN2A gene has been suggested to be a useful method in the molecular diagnostics of the resection field [21]. The hypermethylation of CDH1 gene was considered to be a bad prognostic factor in tongue cancer [22] and ATM gene hypermethylation was connected with poorer prognosis in head and neck cancers [23]. Methylated RAR-β gene is completely suppressed in immortal dysplasia and oral carcinoma [24]. However, the results are still inconsistent; thus, the search for epigenetic markers useful as prognostic factors remains still an open question.
The aim of this study was to establish the morphological characteristics of various parts of the tumor including comparisons between the central, peripheral part and the surgical margin of the cancer of the oral cavity and oropharynx. We also assessed the methylation status of five genes (CDKN2A, CDH1, ATM, FHIT, and RAR-β) in the same samples. In the following study, the correlations between morphological data, genes hypermethylation status, clinicopathological data, and survival were assessed.
Material and methods
Patients
Patients with oral and oropharyngeal cancer who had been primarily treated surgically in the Department of Otolaryngology and Clinical Oncology, University of Medical Sciences in Poznań, Poland, between July 2009 and April 2012 were enrolled to the prospective study. The study group consisted of 53 patients, 4 women and 49 men, aged 24–82. The clinical data including sex, age, stage of the tumor, histological grading, risk factors, and localization of the tumor are listed in Table 1. The tissue samples were taken from the central (A) and peripheral part of the tumor (B) and from the surgical margin (C). All samples underwent routine histopathological examination (HE-stained specimens) and ultrastructural assessment using toluidine blue staining in a light microscope and in transmission electron microscopy (EM). The resection margin was considered clean in histopathological examination, if the distance from the invasive cancer to the margin was more than 5 mm—according to the UK Royal College of Pathologists. DNA methylation status for the chosen genes was established in the same samples (A, B, and C).
Table 1.
The clinical characterization of the study group
| Number of patients | 53 |
| Male | 49 |
| Female | 4 |
| Age (years) | |
| Range | 29–82 |
| Mean | 57 |
| Habits | |
| Smoking | 35 |
| Smoking and alcohol | 13 |
| No drinking and no smoking | 5 |
| TNM | |
| T1 | 6 |
| T2 | 24 |
| T3 | 12 |
| T4 | 11 |
| N0 | 25 |
| N1 | 15 |
| N2 | 12 |
| N3 | 1 |
| M0 | 0 |
| G stage | |
| G1 | 7 |
| G2 | 41 |
| G3 | 5 |
| Tumor localization | |
| Tonsils (no infiltration of the base of the tongue) | 13 |
| Tongue (2/3 anterior, no infiltration of the floor of the mouth) | 16 |
| Tongue and floor of the mouth | 11 |
| Palate | 3 |
| Lips | 1 |
| Base on the tongue and tonsils | 8 |
| Cheek | 1 |
Ultrastructural assessment
Tissue samples were pre-fixed in Karnovsky solution (glutaraldehyde 2 % and paraformaldehyde 3.4 % in 0.1 M cacodylate buffer pH 7.4) overnight (1 h at room temperature and cooled down to 4 °C). After pre-fixation, samples were washed several times in 0.1 M phosphate buffer and post-fixed in 1 % OsO4 in 0.1 M phosphate buffer with K4Fe(CN)6 · 3H2O; 0.16 % for 3 h at +4 °C. After post-fixation, dehydration in gradually increasing concentrations of ethanol and then acetone was carried out. Embedding in Epon 812 and polymerization were the next steps of the procedure. Semi-thin sections were cut from all blocks, stained with toluidine blue of basic pH, and checked with the use of an Olympus BX 41. After selection of the proper slide area ultrathin sections were cut and doubly stained with uranium acetate and lead citrate for EM. Selection of ultrathin section areas and micrographs was done by JEM 1011.
Since the aims of the study also included a careful check of the margins of tumors and their periphery (oncologic sterility), the control performed on the base of semi-thin sections was very accurate. In case of any doubts concerning presence of single tumor cells in surrounding tissue, EM checks were done.
In this series, we compared groups with a clean resection margin in HE-stained specimens and a positive margin in morphological assessment, versus a group with a clean resection margin in both examinations with survival, TNM status, G stage, and pN.
DNA extraction and gene methylation analysis
DNA was isolated from all samples according to standard procedures (proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation). For DNA methylation analysis, samples with more than 80 % of cancer cells in the field were enrolled. The methylation status of RAR-β, CDH1, FHIT, CDKN2A, and ATM was assessed using the methylation-specific polymerase chain reaction (MSP). DNA was converted in the presence of sodium bisulfite using the EZ DNA Methylation Kit from ZymoResearch (Orange, CA, USA). TrueStart Hot Start Taq DNA Polymerase from Fermentas (Burlington, Canada) was used for the amplification of RAR-β and ATM, whereas FastStart Taq DNA Polymerase (Roche Diagnostics, Germany) was used for the amplification of CDH1, FHIT, and CDKN2A. The primers and reaction conditions for MSP were as previously reported in other studies [25–29]. All the primers were obtained from Oligo.pl (Warsaw, Poland). DNA extracted from the lymphocytes of healthy blood donors was used as a negative control, and completely methylated human DNA (New England Biolabs, Ipswich, MA, USA) was used as a positive control in the MSP reactions. Amplification products were separated on 2.5 % agarose gels and visualized in UV light after ethidium bromide staining.
First, we compared the results obtained for groups A, B, and C: samples with methylation vs those without it in relation to TNM, pN, G stage, and survival. Then, groups A and B were coupled (tumor zone) and these results were correlated with clinicopathological data.
Statistical analysis
The correlation between clinicopathological features and results of morphological examination, as well as gene methylation, was assessed using Chi-square, Mann–Whitney, and Kaplan–Meier tests (p < 0.05). The Statistica program was used for analysis.
Results
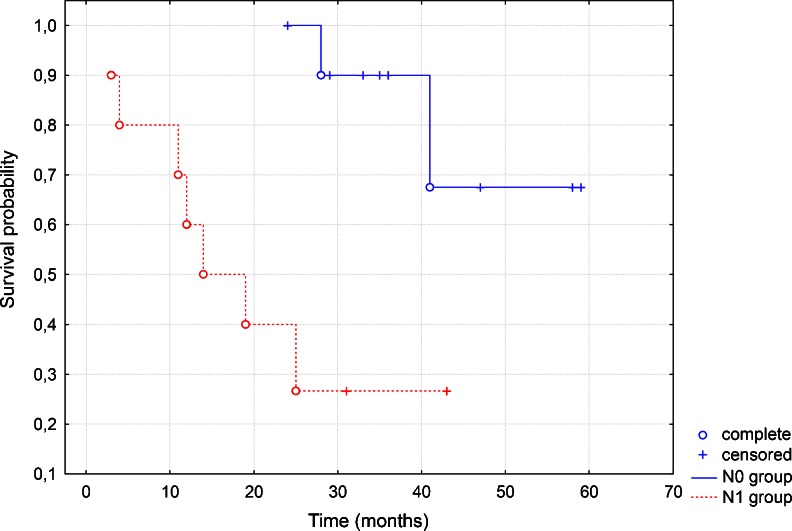
In the study group, the T2 stage of the tumor, N0 nodal involvement, and G2 malignancy grading prevailed. Of the 53 patients, 12 had a positive surgical margin in histopathological examination. In the series with a clean resection margin, we did not find histopathologically confirmed node metastases (pN) and the result was statistically significant (p = 0.0393). The second very important point was the finding that the group with pN0 and clean margin in HE staining, but having cancer cells in toluidine blue staining, present longer survival in the Kaplan–Meier test (Fig. 1).
Fig. 1.
The chart is showing differences between two groups: without neck metastases (pN0) and clean margin, and with neck metastases (pN1) and positive margin in HE staining. The first one presents longer survival in the Kaplan–Meier test
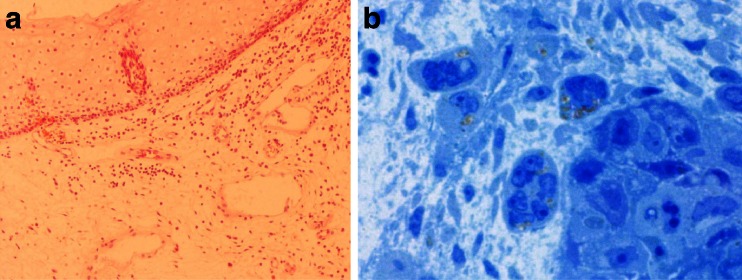
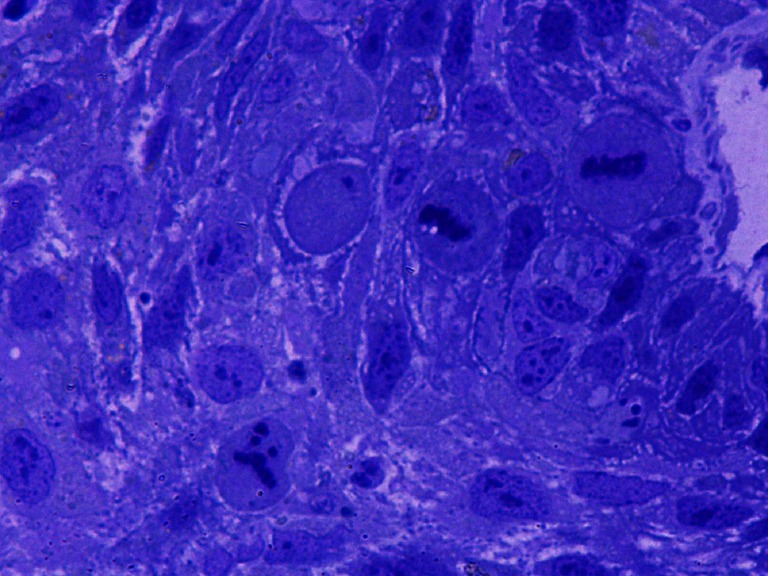
None of the tumors collected for research had a homogeneous texture (Fig. 2). In all cancers, fields of differentiated cells maturating to fully keratinized—or in the form of keratinizing—horny pearls (Fig. 3), or small horny plates lying on the free surface of the tumor occurred. Apart from maturating cells, others which had cytoplasmic features typical for spinous cells, i.e., cytokeratin bunches and irregular cytoplasmic bridges with irregular desmosomes or other forms of intercellular junctions, were found. Cells differed in size and shape of nuclei and cytoplasm (Fig. 4). Nuclear polymorphism was distinct and considerable, especially in the invasive front of the tumor. The ultrastructural characteristics of nuclei were very different for these cells compared to the nuclei of maturating cells. They were large, with an irregular contour, often with deep invagination of the nuclear envelope into the nucleoplasm (Fig. 5). Many nuclei had large, active, and numerous nucleoli. Parts of the nuclei were euchromatinized and had active nucleoli and very numerous perichromatin granules, with different diameters and areas of interchromatin granules. This is typical for cells which enter into the cell cycle. In the whole collected material, cells occurred in mitosis—and some of them were atypical or endomitotic, what could be interpreted as a loss of proper cell cycle control (Fig. 6). In some of the assessed tumors, disorders of the late telophase were found as a partial loss of the nuclear envelope. All of the studies carried out show that cancer formed heterogenous cellular populations, with differences of texture classified as different histological gradings: G1 or G2 or G3. Most commonly, the cells with lower G were found in the invasive front of the tumor. There was a statistically significant correlation between nuclei polymorphism and T stage (p < 0.0001), N stage (p < 0.046), and metastases to the lymph nodes confirmed in histopathological examination—pN (p < 0.004). The number of cells in irregular mitosis correlated with T stage (p < 0.004), and highly with pN (p < 0.009). In the examined group, there were differences between the clean resection margin in HE-stained specimens and those assessed using toluidine blue (Table 2). In the specimens prepared for the assessment of the ultrastructure of the tumor, even under a light microscope, it was possible to find cancer cells between normal glands, skeletal muscle cells, and connective tissue stroma (Fig. 7).
Fig. 2.
The picture shows non-homogenous structure of the tumor
Fig. 3.
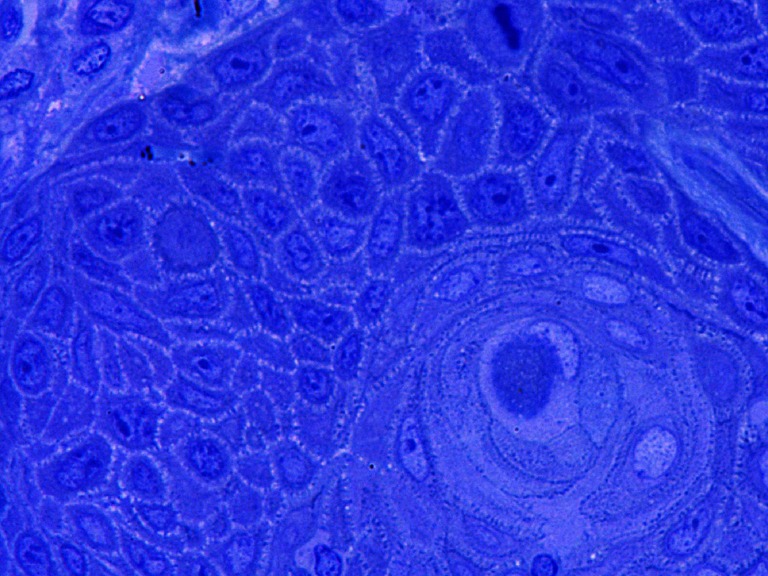
This figure shows horny pearl in G1 texture; other cells are modestly monomorphic in G1-2 carcinoma
Fig. 4.
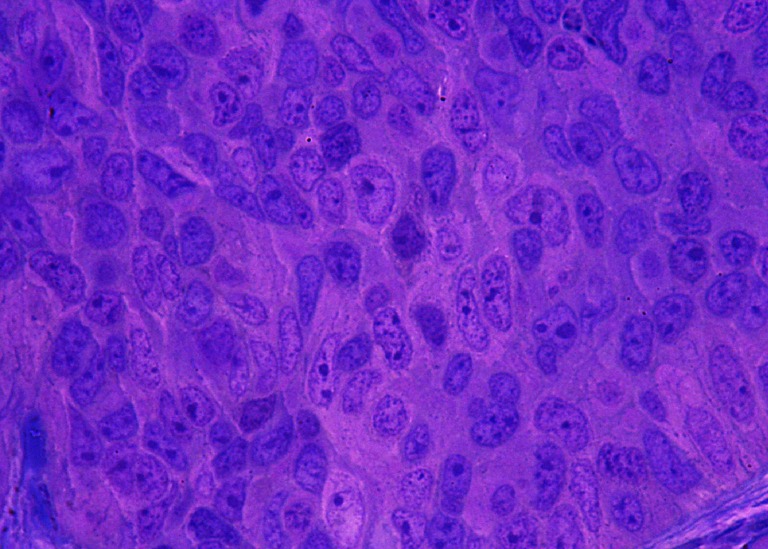
Solid area of the G2-G3 stage of the tumor. Significant polymorphism of nuclei with numerous nucleoli dominates in the general picture of this tumor. The area occupied by nuclei evidently prevail over the area of cytoplasm
Fig. 5.
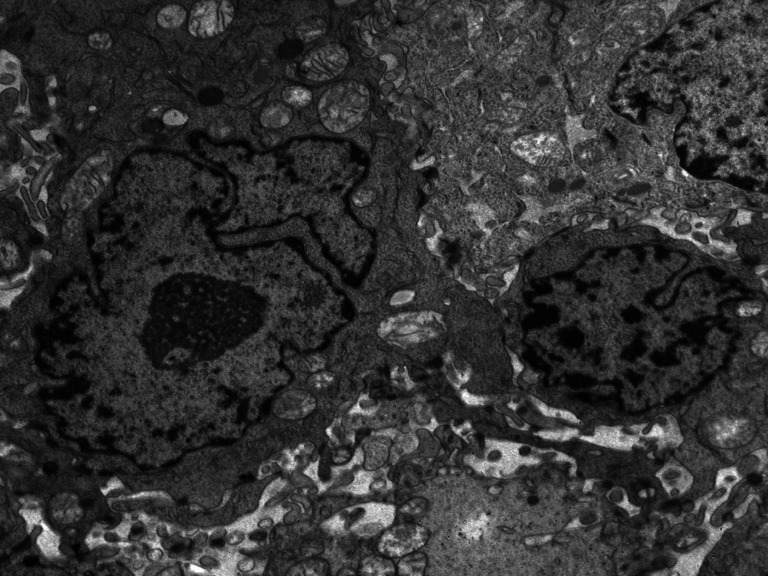
The micrograph presents differences between the nuclei of tumor cells. One is much larger, with a deeply invaginated nuclear membrane, enlarged active nucleolus, a field of interchromatin granules, and small groups of heterochromatin located in the vicinity of the nuclear membrane
Fig. 6.
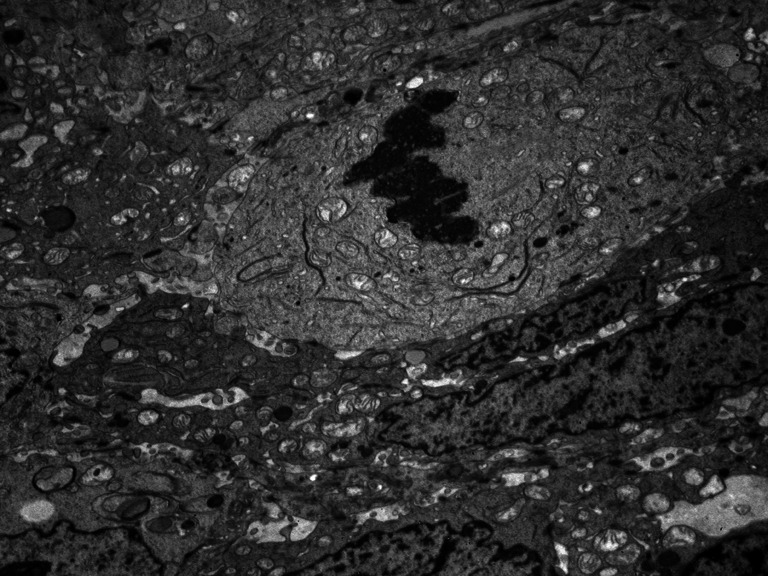
One of the tumor cells forms chromosomes and removes the nuclear membrane. Display of organelles is typical for the prophase. Neighboring cells do not divide
Table 2.
The number and percentage of patients with clean and positive resection margin in two types of examinations
| Margin U | Margin HE 0 | Margin HE 1 | Total |
|---|---|---|---|
| 0 | 20 (37.74 %) | 4 (7.55 %) | 24 (45.28 %) |
| 1 | 21 (39.62 %) | 8 (15.09 %) | 29 (54.72 %) |
| Total | 41 (77.36 %) | 12 (22.64 %) | 53 (100 %) |
U ultrastructural, HE hematoxylin and eosin-stained, 0 clean, 1 involved
Fig. 7.
Comparison between HE- (a) and toluidine blue- (b) stained specimens of the same patient. First one were classified as a clean resection margin; however, in the second one single cancer cells are present
We also correlated the group with a clean resection margin in HE-stained specimens but having cancer cells in ultrastructural examination with clinicopathological features. This group did not present a poor prognosis and did not correlate with TNM status or G stage. Apart from ultrastructural analysis, the methylation status of selected tumor suppressors was also assessed.
The methylation was found most often for CDH1 and CDKN2A genes in the A and B groups. However, there was no statistically significant correlation between gene hypermethylation and stage of the tumor, nodal involvement, and G stage, except for a correlation between CDH1 in the B and N1 stage (p = 0.031). For the next two genes, RAR β and FHIT, hypermethylation was found less frequently and also did not correlate with clinicopathological data. We did not detect methylation of ATM gene in any of the samples under analysis. Gene methylation was also found in the clean resection margin, and the percentage ranged between 1.8 and 26.4 (Tables 3 and 4). Altogether, the molecular changes in the resection margin did not correlate with TNM, malignancy grading, or survival. Likewise, combining the results of groups A and B did not provide any correlation between hypermethylation and clinicopathological data.
Table 3.
The presence of gene hypermethylation in the central (A) and peripheral parts (B) of the tumor, as well in the surgical margin (C)
| A | B | C | |
|---|---|---|---|
| CDH1 | 20/53 | 24/53 | 14/53 |
| (37.7 %) | (45.3 %) | (26.4 %) | |
| p16 | 14/53 | 18/53 | 10/53 |
| (26.4 %) | (33.9 %) | (18.8 %) | |
| RARbeta | 4/53 | 0 | 3/53 |
| (7.5 %) | (5.6 %) | ||
| FHIT | 1/53 | 1/53 | 1/53 |
| (1.8 %) | (1.8 %) | (1.8 %) | |
| ATM | 0 | 0 | 0 |
Table 4.
The analysis of gene methylation in clean and involve resection margin in two types of examinations: U—ultrastructural and HE—hematoxylin and eosin-stained, “−”—clean, “+”—involved; A—central part of tumor, B—peripheral part of tumor and C—resection margin
| ATM | CDH1 | CDKN2A | FHIT | RAR-β | |
|---|---|---|---|---|---|
| HE+ | 0 | A:5 (9.43 %) | A:3 (5.66 %) | 0 | A:1 (1.88 %) |
| B:5 (9.43 %) | B:5 (9.43 %) | B:0 | |||
| C:3 (5.66 %) | C:4 (7.54 %) | C:0 | |||
| HE− | 0 | A:13 (24.52 %) | A:10 (18.86 %) | A:1 (1.88 %) | A:2 (3.77 %) |
| B:16 (30.18 %) | B:12 (22.64 %) | B:1 (1.88 %) | B:0 | ||
| C:9 (16.98 %) | C:6 (11.32 %) | C:1 (1.88 %) | C:2 (3.77 %) | ||
| U+ | 0 | A:15 (28.30 %) | A:5 (9.43 %) | A:1 (1.88 %) | A:3 (5.66 %) |
| B:15 (28.30 %) | B:7 (13.20 %) | B:1 (1.88 %) | B:0 | ||
| C:10 (18.86 %) | C:6 (11.32 %) | C:1 (1.88 %) | C:1 (1.88 %) | ||
| U− | 0 | A:5 (9.43 %) | A:7 (13.20 %) | 0 | A:1 (1.88 %) |
| B:7 (13.20 %) | B:9 (16.98 %) | B: 0 | |||
| C:3 (5.66 %) | C:3 (5.66 %) | C:1 (1.88 %) |
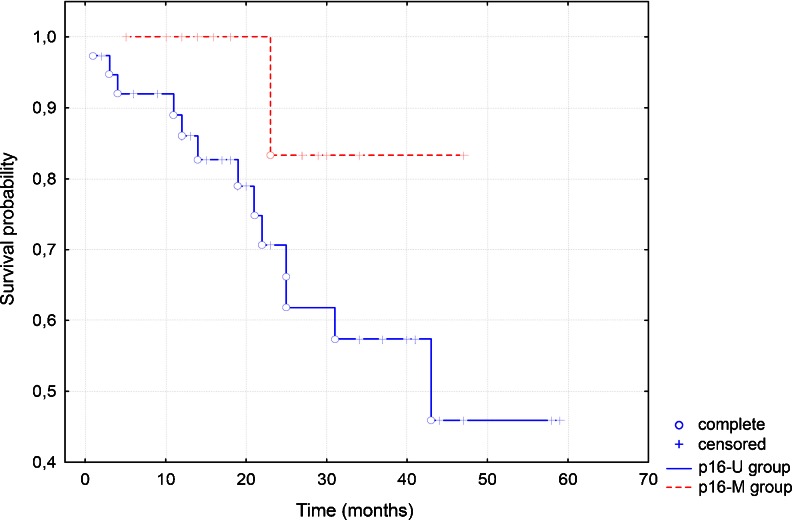
In the Kaplan–Meier test, there was no statistically significant correlation between gene methylation and survival, except methylation of CDKN2A. The group with CDKN2A hypermethylation presented longer survival (Fig. 8).
Fig. 8.
The chart shows that the group with p16 methylation (p16-M) presents better survival than the group without hypermethylation (p16-U) in the Kaplan–Meier test
Discussion
Five-year survival rates in oral cancer are still unsatisfactory. The epidemiological data show some differences between Western and Eastern European countries, as well as the USA. However, they never cross the 50 % threshold [1, 7] that points at a deep need to find new prognostic factors.
Carcinogenesis is a multistep process [15, 16]. Cancer development is the result of the accumulation of molecular alterations. Epigenetic and genetic changes lead to the deregulation of cell cycle through (epi)mutations of tumor suppressor genes and proto-oncogenes. In some of the cells, mutated genes conform with a proliferative activity and are responsible for the progression of cells to invasive carcinoma. Environmental carcinogens can initiate this process in the area of epithelial cells. The same carcinogens have often a global effect on the whole oral epithelium. This fact can be responsible for the multifocal progression of mutated cells toward invasive carcinoma by the same oral mucosa and thus can be represented by the morphological heterogeneity of cancer texture in the analyzed material. Our study shows that none of the examined tumors had the homogenous structure. This tissue texture characteristic was represented in small, medium, and large tumors, and was not dependent on tumor primary location. The observations gathered in our study are in line with the ideas formulated by Braakhuis [14]. It can also be thought that progression of certain tumor cells to the invasive stage does not take place simultaneously [30].
The gold standard in the assessment of cancer tissue is histological examination using HE-stained samples. Evaluation of the same tissue using a transmission electron microscope improves the accuracy of the morphological characteristics of examined cells. In papers describing carcinomas of oral mucosa, attention was paid to the ultrastructural characteristics of the architecture of tumors, formation and quality of the epithelial basement membrane, junctional complexes, including hemidesmosomes, cytoplasmic structures and the size, and structure and chromatin characteristics of nuclei and nucleoli. In the assessment of our specimens, we used previously described data of ultrastructural estimation [31–33]. In our study, special attention was paid to the characteristics of cell nuclei. Polymorphism of these structures includes differences in size, nuclear envelope contour (regular, oval without deep invagination, to deep indentation into the chromatin to polykaryocytes, with a lack of nuclear envelope on some small areas of nuclei), and structure of chromatin (heterochromatin beneath the nuclear envelope to highly euchromatic nuclei with prominent and also multiple, nucleoli). Additionally, in some tumor areas, regular as well as irregular mitotic figures were present. In our study group, there were statistically significant correlations between nuclei polymorphism of A and B with T stage (p < 0.0001), N stage (p < 0.046), and metastases to the lymph nodes confirmed in histopathological examination (pN p < 0.004). At the same time, the second examined parameter, i.e., the number of cells in irregular mitosis for the same area, correlated with the T stage (p < 0.004) and highly with pN (p < 0.009). These findings could explain the dynamics of cancer biology and could also be very useful as a prognostic factor in oral cancer.
We also analyzed ultrastructure of the resection margin because data regarding the prognostic importance of HE-stained specimens are not clear-cut. Some authors conclude that it is a robust prognosticator, whereas others contradict this thesis [34–39]. Going to the details, the first group supports the thesis that the presence of single cell or groups of cells in stroma surrounding the main tumor texture in the resection margin is a separate, negative predicting factor. In the present study, this situation did not correlate with worse prognosis. This may be at least partly explained by the fact that patients after surgery routinely undergo additional therapy, i.e., irradiation and/or chemotherapy which may be sufficient to eliminate cancer cells and prevent local relapse.
The ultrastructural characteristics of cell nuclei, including their heterogeneity encouraged us to confront the morphologic data with epigenetic profile. Solid tumors, like many other human diseases, show aberrant epigenetic alterations [19, 40–42]. Methylation of CpG islands associated with tumor suppressor genes is found to be crucial in the multistep mechanism of carcinogenesis. In our study, gene hypermethylation was detected most commonly in CDH1 and CDKN2A genes in central and peripheral parts of the tumor. These alterations prevailed in the peripheral part of the tumor, what may be associated with the presence of more rapidly proliferating cells that are more susceptible to molecular changes. However, only one statistically significant correlation was established between CDH1 in the B zone and N stage. Data regarding the hypermethylation process and its influence on clinicopathological features and prognosis are still unclear. Ha and Califano [18] noticed that methylation of CDKN2A ranged from 0 to 85 % in oral cancer. Nagata et al. found that methylation of four statistically selected genes including E-cadherin and RAR-β allows to detect oral cancer with 100 % sensitivity and 87.5 % specificity [43].
In our study, CDKN2A hypermethylation did not correlate with clinicopathological data, except survival, which was observed in the Kaplan–Meier curve. Hence, our observations for only this one parameter are in agreement with other authors, claiming the usefulness of the determination of the methylation status of CDKN2A as prognosis marker [44–47]. Dong et al. [44] found a statistically significant correlation between methylation of CDKN2A and nodal involvement and survival in buccal carcinoma, however, without association with gender, age, T stage, and G stage. In this study, the overall survival was poorer for the group with hypermethylation in the Kaplan–Meier test, but in multivariable analysis using the Cox regression model methylation was not statistically correlated with survival. In a multicentre study covering 353 cases of head and neck cancer, Roh et al. found that CDKN2A methylation correlated with a decreased survival [45]. However, in tumors with disruptive TP53 mutation, methylation of CDKN2A was protective. In our study, TP53 mutations were not tested. Veganzones-de-Castro et al. working on colorectal cancer made the same observation as we did [48]. They found that patients with poorly differentiated tumors having CDKN2A promoter methylated presented longer free survival than those without CDKN2A methylation. We cannot rule out that only one allele was methylated and the other one still remained and performed its function.
The methylation of RAR-β and FHIT was found less frequently in the central and peripheral parts of the tumor. In our study, methylation of these two genes did not correlate with age, stage of the tumor, malignancy grading, or survival. However, the methylation in tumor zone was more prevalent than in the resection margin. Tanaka et al. [49] found that methylation of the 5′ CpG island is an important mechanism for the inactivation of the FHIT gene in esophageal carcinoma. Hypermethylation of FHIT was relatively often found in oral cancer (27 %) [18] and in laryngeal carcinoma (26 %) [50]. However, it did not correlate with clinicopathological data and survival. RAR-β is completely suppressed in immortal dysplasia and oral carcinoma [24]. Hypermethylation was often found in oral cancer (47–100 %) [18], and relatively often in the laryngeal cell lines (64.7 %) [51]. Paluszczak et al. found the methylation of this gene in 59 % of laryngeal carcinoma cases, but it did not correlate with prognosis [50]. We found the same statistical results in our study. In our group of patients, we did not find aberrant methylation of ATM. Hypermethylation of this gene in HNSCC was described as connected with poorer prognosis [23, 52] but we did not confirm these observations.
The molecular assessment of the surgical margin was an important part of our research. In our study, we did not find statistically significant correlation between methylation of the resection margin and prognosis. Methylation of CDKN2A in the surgical margin was found as a prognostic factor for tongue cancer by Sinha et al. [20]. Other authors, however, found molecular changes in the resection margin, but without correlation with survival [21, 53]. Some authors propose criteria which determine the reliability of a molecular marker in the resection field [54, 55]. After the precise analysis Bradley et al. [54] concluded that the TP53 mutations fulfil the proposed criteria. Finally, we should keep in mind that cancer recurrence could have many reasons, which was precisely described by Braakhuis et al. [56].
In conclusion, we would like to stress that ultrastructural assessment in our patient group brought important information about cancer biology. Parameters like nuclei polymorphism and number of cells in irregular mitosis are strong prognosticators of patient survival. The epigenetic changes found in the tumor area did not correlate with clinicopathological features except CDKN2A methylation, which was correlated with better prognosis. The molecular assessment of the resection margin did not bring new prognostic information. However, a longer observation time and a larger number of patients are required to draw final conclusions.
Overall, we report frequent changes in tissue ultrastructure and gene hypermethylation of CDKN2A and CDH1 in oral cancer. The inclusion to our study of the estimation of other molecular parameters may result in the discovery of novel, prognostic markers of this disease.
Acknowledgments
This study was supported by Polish National Science Centre scientific grant NN 403 290736.
Conflicts of interest
None
Footnotes
The precise scientific description of each specimen (HE-stained, semi-thin sections, and EMs) are available from the main author.
References
- 1.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–16. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Warnakulasuriya S. Causes of oral cancer—an appraisal of controversies. Br Dent J. 2009;207(10):471–5. doi: 10.1038/sj.bdj.2009.1009. [DOI] [PubMed] [Google Scholar]
- 3.Soudry E, Preis M, Hod R, Hamzany Y, Hadar T, et al. Squamous cell carcinoma of the oral tongue in patients younger than 30 years: clinicopathologic features and outcome. Clin Otolaryngol. 2010;35(4):307–12. doi: 10.1111/j.1749-4486.2010.02164.x. [DOI] [PubMed] [Google Scholar]
- 4.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Curr Opin Oncol. 2009;21(3):194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 5.Gawecki W, Szyfter K, Szyfter W. Clinical and histopathological analysis of squamous cell carcinoma of the head and neck in young adults. Otolaryngol Pol. 2007;61(1):11–6. doi: 10.1016/S0030-6657(07)70375-X. [DOI] [PubMed] [Google Scholar]
- 6.Gawecki W, Kostrzewska-Poczekaj M, Gajecka M, Milecki P, Szyfter K, et al. The role of genetic factor in etiopathogenesis of squamous cell carcinoma of the head and neck in young adults. Eur Arch Otorhinolaryngol. 2007;264(12):1459–65. doi: 10.1007/s00405-007-0386-x. [DOI] [PubMed] [Google Scholar]
- 7.Mielcarek-Kuchta D, Wierzbicka M, Mietkiewska D, Kopec T, Banaszewski J, et al. Retrospective analysis of the treatment failure of the oral cancer in Department of Otolaryngology and Clinical Oncology University of Medical Sciences in Poznan in the year 2000–2005. Otolaryngol Pol. 2011;65(1):26–32. doi: 10.1016/S0030-6657(11)70624-2. [DOI] [PubMed] [Google Scholar]
- 8.Klussmann JP, Preuss SF, Speel EJ. Human papillomavirus and cancer of the oropharynx. Molecular interaction and clinical implications. HNO. 2009;57(2):113–22. doi: 10.1007/s00106-008-1867-y. [DOI] [PubMed] [Google Scholar]
- 9.Klussmann JP, Mooren JJ, Lehnen M, Claessen SM, Stenner M, et al. Genetic signatures of HPV-related and unrelated oropharyngeal carcinoma and their prognostic implications. Clin Cancer Res. 2009;15(5):1779–86. doi: 10.1158/1078-0432.CCR-08-1463. [DOI] [PubMed] [Google Scholar]
- 10.Hafkamp HC, Manni JJ, Haesevoets A, Voogd AC, Schepers M, et al. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122(12):2656–64. doi: 10.1002/ijc.23458. [DOI] [PubMed] [Google Scholar]
- 11.Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121(8):1813–20. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- 12.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6(5):963–8. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56(11):2488–92. [PubMed] [Google Scholar]
- 14.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63(8):1727–30. [PubMed] [Google Scholar]
- 15.Hecht SS. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol. 2002;3(8):461–9. doi: 10.1016/S1470-2045(02)00815-X. [DOI] [PubMed] [Google Scholar]
- 16.Wiencke JK. DNA adduct burden and tobacco carcinogenesis. Oncogene. 2002;21(48):7376–91. doi: 10.1038/sj.onc.1205799. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(1):R50–9. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 18.Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7(1):77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 20.Sinha P, Bahadur S, Thakar A, Matta A, Macha M, et al. Significance of promoter hypermethylation of p16 gene for margin assessment in carcinoma tongue. Head Neck. 2009;31(11):1423–30. doi: 10.1002/hed.21122. [DOI] [PubMed] [Google Scholar]
- 21.Shaw RJ, Hall GL, Woolgar JA, Lowe D, Rogers SN, et al. Quantitative methylation analysis of resection margins and lymph nodes in oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 2007;45(8):617–22. doi: 10.1016/j.bjoms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Chang HW, Chow V, Lam KY, Wei WI, Yuen A. Loss of E-cadherin expression resulting from promoter hypermethylation in oral tongue carcinoma and its prognostic significance. Cancer. 2002;94(2):386–92. doi: 10.1002/cncr.10211. [DOI] [PubMed] [Google Scholar]
- 23.Ai L, Vo QN, Zuo C, Li L, Ling W, et al. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol Biomarkers Prev. 2004;13(1):150–6. doi: 10.1158/1055-9965.EPI-082-3. [DOI] [PubMed] [Google Scholar]
- 24.McGregor F, Muntoni A, Fleming J, Brown J, Felix DH, et al. Molecular changes associated with oral dysplasia progression and acquisition of immortality: potential for its reversal by 5-azacytidine. Cancer Res. 2002;62(16):4757–66. [PubMed] [Google Scholar]
- 25.Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110(9):2019–26. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- 26.Gronbaek K, Worm J, Ralfkiaer E, Ahrenkiel V, Hokland P, et al. ATM mutations are associated with inactivation of the ARF-TP53 tumor suppressor pathway in diffuse large B-cell lymphoma. Blood. 2002;100(4):1430–7. doi: 10.1182/blood-2002-02-0382. [DOI] [PubMed] [Google Scholar]
- 27.King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49(1):36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 28.Smith JA, Fan CY, Zou C, Bodenner D, Kokoska MS. Methylation status of genes in papillary thyroid carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133(10):1006–11. doi: 10.1001/archotol.133.10.1006. [DOI] [PubMed] [Google Scholar]
- 29.Huang Q. A.L., Zhang ZY, Fan CY, Weiss LM., Promoter hypermethylation and protein expression of the p16 gene: analysis of 43 cases of B-cell primary gastric lymphomas from China. Mod Pathol. 2004;17(4):416–22. doi: 10.1038/modpathol.3800072. [DOI] [PubMed] [Google Scholar]
- 30.Yokota J, Kohno T. Molecular footprints of human lung cancer progression. Cancer Sci. 2004;95(3):197–204. doi: 10.1111/j.1349-7006.2004.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannan S, Kartha CC, Chandran GJ, Pillai MR, Sudha L, et al. Ultrastructure of oral squamous cell carcinoma: a comparative analysis of different histological types. Eur J Cancer B Oral Oncol. 1994;30B(1):32–42. doi: 10.1016/0964-1955(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 32.Cheng LH, Hudson J. Ultrastructural changes in malignant transformation of oral mucosa. Br J Oral Maxillofac Surg. 2002;40(3):207–12. doi: 10.1054/bjom.2001.0778. [DOI] [PubMed] [Google Scholar]
- 33.Kannan S, Kartha CC, Balaram P, Chandran GJ, Pillai MR, et al. Ultrastructural analysis of the adjacent epithelium of oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 1996;34(1):51–7. doi: 10.1016/S0266-4356(96)90136-9. [DOI] [PubMed] [Google Scholar]
- 34.Kurita H, Nakanishi Y, Nishizawa R, Xiao T, Kamata T, et al. Impact of different surgical margin conditions on local recurrence of oral squamous cell carcinoma. Oral Oncol. 2010;46(11):814–7. doi: 10.1016/j.oraloncology.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Spiro RH, Guillamondegui O, Jr, Paulino AF, Huvos AG. Pattern of invasion and margin assessment in patients with oral tongue cancer. Head Neck. 1999;21(5):408–13. doi: 10.1002/(SICI)1097-0347(199908)21:5<408::AID-HED5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.McMahon J, O'Brien CJ, Pathak I, Hamill R, McNeil E, et al. Influence of condition of surgical margins on local recurrence and disease-specific survival in oral and oropharyngeal cancer. Br J Oral Maxillofac Surg. 2003;41(4):224–31. doi: 10.1016/S0266-4356(03)00119-0. [DOI] [PubMed] [Google Scholar]
- 37.Upile T, Fisher C, Jerjes W, El Maaytah M, Searle A, et al. The uncertainty of the surgical margin in the treatment of head and neck cancer. Oral Oncol. 2007;43(4):321–6. doi: 10.1016/j.oraloncology.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Binahmed A, Nason RW, Abdoh AA. The clinical significance of the positive surgical margin in oral cancer. Oral Oncol. 2007;43(8):780–4. doi: 10.1016/j.oraloncology.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Woolgar JA. Histopathological prognosticators in oral and oropharyngeal squamous cell carcinoma. Oral Oncol. 2006;42(3):229–39. doi: 10.1016/j.oraloncology.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 41.Feinberg AP. Cancer epigenetics is no Mickey Mouse. Cancer Cell. 2005;8(4):267–8. doi: 10.1016/j.ccr.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Weiss D, Stockmann C, Schrodter K, Rudack C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 2013;36(3):213–24. doi: 10.1007/s13402-013-0129-5. [DOI] [PubMed] [Google Scholar]
- 43.Nagata S, Hamada T, Yamada N, Yokoyama S, Kitamoto S, et al. Aberrant DNA methylation of tumor-related genes in oral rinse: a noninvasive method for detection of oral squamous cell carcinoma. Cancer. 2012;118(17):4298–308. doi: 10.1002/cncr.27417. [DOI] [PubMed] [Google Scholar]
- 44.Dong Y, Wang J, Dong F, Wang X, Zhang Y. The correlations between alteration of p16 gene and clinicopathological factors and prognosis in squamous cell carcinomas of the buccal mucosa. J Oral Pathol Med. 2012;41(6):463–9. doi: 10.1111/j.1600-0714.2012.01132.x. [DOI] [PubMed] [Google Scholar]
- 45.Roh JL, Wang XV, Manola J, Sidransky D, Forastiere AA, et al. Clinical correlates of promoter hypermethylation of four target genes in head and neck cancer: a cooperative group correlative study. Clin Cancer Res. 2013;19(9):2528–40. doi: 10.1158/1078-0432.CCR-12-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Freitas Cordeiro-Silva M. E. Stur, L.P. Agostini, J.R. de Podesta, J.C. de Oliveira, et al., Promoter hypermethylation in primary squamous cell carcinoma of the oral cavity and oropharynx: a study of a Brazilian cohort. Mol Biol Rep. 2012;39(12):10111–9. doi: 10.1007/s11033-012-1885-4. [DOI] [PubMed] [Google Scholar]
- 47.Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, et al. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer. 2006;94(4):561–8. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veganzones-de-Castro S, Rafael-Fernandez S, Vidaurreta-Lazaro M. V. de-la-Orden, B. Mediero-Valeros, et al., p16 gene methylation in colorectal cancer patients with long-term follow-up. Rev Esp Enferm Dig. 2012;104(3):111–7. doi: 10.4321/S1130-01082012000300002. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka H, Shimada Y, Harada H, Shinoda M, Hatooka S, et al. Methylation of the 5' CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Cancer Res. 1998;58(15):3429–34. [PubMed] [Google Scholar]
- 50.Paluszczak J, Misiak P, Wierzbicka M, Wozniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011;47(2):104–7. doi: 10.1016/j.oraloncology.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Paluszczak J, Krajka-Kuzniak V, Malecka Z, Jarmuz M, Kostrzewska-Poczekaj M, et al. Frequent gene hypermethylation in laryngeal cancer cell lines and the resistance to demethylation induction by plant polyphenols. Toxicol In Vitro. 2011;25(1):213–21. doi: 10.1016/j.tiv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 52.He Y, Chen Q, Li B. ATM in oral carcinogenesis: association with clinicopathological features. J Cancer Res Clin Oncol. 2008;134(9):1013–20. doi: 10.1007/s00432-008-0365-7. [DOI] [PubMed] [Google Scholar]
- 53.Shaw RJ, Hobkirk AJ, Nikolaidis G, Woolgar JA, Triantafyllou A, et al. Molecular staging of surgical margins in oral squamous cell carcinoma using promoter methylation of p16(INK4A), cytoglobin, E-cadherin, and TMEFF2. Ann Surg Oncol. 2013;20(8):2796–802. doi: 10.1245/s10434-012-2713-8. [DOI] [PubMed] [Google Scholar]
- 54.Bradley PJ, MacLennan K, Brakenhoff RH, Leemans CR. Status of primary tumour surgical margins in squamous head and neck cancer: prognostic implications. Curr Opin Otolaryngol Head Neck Surg. 2007;15(2):74–81. doi: 10.1097/MOO.0b013e328058670f. [DOI] [PubMed] [Google Scholar]
- 55.Rodrigo JP, Ferlito A, Suarez C, Shaha AR, Silver CE, et al. New molecular diagnostic methods in head and neck cancer. Head Neck. 2005;27(11):995–1003. doi: 10.1002/hed.20257. [DOI] [PubMed] [Google Scholar]
- 56.Braakhuis BJ, Bloemena E, Leemans CR, Brakenhoff RH. Molecular analysis of surgical margins in head and neck cancer: more than a marginal issue. Oral Oncol. 2010;46(7):485–91. doi: 10.1016/j.oraloncology.2010.01.019. [DOI] [PubMed] [Google Scholar]