Abstract
Retroviral tropism is determined in part by cellular restriction factors that block infection by targeting the incoming viral capsid. Indeed, human immunodeficiency virus type 1 (HIV-1) infection of many nonhuman primate cells is inhibited by one such factor, termed Lv1. In contrast, a restriction factor in humans, termed Ref1, does not inhibit HIV-1 infection unless nonnatural mutations are introduced into the HIV-1 capsid protein (CA). Here, we examined the infectivity of a panel of mutant HIV-1 strains carrying substitutions in the N-terminal CA domain in cells that exhibit restriction attributable to Lv1 or Ref1. Manipulation of HIV-1 CA could alter HIV-1 tropism, and several mutations were identified that increased or decreased HIV-1 infectivity in a target-cell-specific manner. Many residues that affected HIV-1 tropism were located in the three variable loops that lie on the outer surface of the modeled HIV-1 conical capsid. Some tropism determinants, including the CypA binding site, coincided with residues whose mutation conferred on HIV-1 CA the ability to saturate Ref1 in human cells. Notably, a mutation that reverses the infectivity defect in human cells induced by CypA binding site mutation inhibits recognition by Ref1. Overall, these findings demonstrate that exposed variable loops in CA and a partial CypA “coat” can modulate restriction and HIV-1 tropism and suggest a model in which the exposed surface of the incoming retroviral capsid is the target for inhibition by host cell-specific restriction factors.
The cellular tropism of a given retrovirus is determined by its ability to exploit required host cell factors and to avoid cellular antiviral activities. Many mammalian cells express specific inhibitors, termed restriction factors, that confer resistance to infection by specific retroviruses (5, 38). Of these, the best characterized is Friend virus susceptibility factor 1 (Fv1), a mouse gene product of endogenous retroviral origin (4) whose allelic variants can specifically block infection by either N-tropic murine leukemia virus (N-MLV) or B-tropic MLV (5, 22, 26, 37). An analogous activity exists in human cells that inhibits infection by N-MLV and equine infectious anemia virus (23, 41). This as yet unidentified restriction factor is termed restriction factor 1 (Ref1). A number of nonhuman primate cells can restrict infection by various divergent retroviruses, including human immunodeficiency virus type 1 (HIV-1), and the factor(s) that are assumed to be responsible are termed lentivirus susceptibility factor 1 (Lv1) (3, 10, 23, 33). Although the mechanisms by which Fv1, Ref1, and Lv1 inhibit infection are unknown, certain characteristics are common to the resistance phenotype conferred by each of these factors. Most notably, the viral determinant of restriction is the capsid protein (CA) (10, 12, 13, 23, 30, 34, 41), and high levels of incoming restricted virions can saturate Fv1, Ref1, or Lv1, thereby abrogating infection resistance (2, 3, 6, 10, 11, 14, 33, 35, 40, 42). Recently, we have shown that saturation of human or African green monkey cells with one retrovirus is capable of abolishing resistance to another retrovirus, provided that both viruses are restricted but irrespective of whether they are closely or distantly related (23). Thus, Ref1 and certain forms of Lv1 appear capable of recognizing retroviral capsids with very little amino acid sequence homology. These findings suggest that perhaps a structurally conserved feature of the retroviral capsid constitutes the target for restriction by these factors. This notion is plausible because a comparison of the three-dimensional structure of widely divergent retroviral CA proteins reveals that the overall protein folds are well conserved (9, 18, 21, 25, 27).
In the case of MLV, a single amino acid residue at CA residue 110 determines sensitivity to restriction by Ref1 as well as the N and B alleles of Fv1 (30, 41). Similarly, we have recently shown that mutation of a single CA residue (G89) in HIV-1 abolishes sensitivity to Lv1 in Owl monkey cells but confers sensitivity to Ref1 in human cells (43). G89 lies in the center of the cyclophilin A (CypA) binding site on HIV-1 CA (17), suggesting that this interaction is important for restriction. Indeed, elimination of the CypA-CA interaction using cyclosporine A (CsA) had similar effects on sensitivity to Lv1 in Owl monkey and Ref1 in human cells (43). Thus, CypA and its binding site are important determinants of restriction factor recognition of HIV-1 CA, at least in some species. However, neither CsA treatment nor the G89 mutation had major effects on restriction of HIV-1 in Rhesus or African green monkey cells (43). These findings indicate that additional determinants of restriction factor recognition should exist and that sequence requirements for recognition of the HIV-1 capsid by restriction factors varies with target cell species. Therefore, to gain further insight into the mechanism of HIV-1 restriction and to map additional determinants of restriction factor sensitivity, we introduced a number of mutations into surface exposed residues in the N-terminal domain of HIV-1 CA. We determined their effects on infectivity by using target cell lines from various primate species. This analysis revealed that HIV-1 species tropism is determined in a complex manner by residues that are predicted to lie on the outer exposed surface of the modeled conical HIV-1 capsid (20, 31). These data are consistent with a model in which primate lentivirus species tropism is strongly influenced by restriction factors that recognize the surface of the incoming capsid, perhaps by contacting multiple CA monomers within the intact capsid structure.
MATERIALS AND METHODS
Plasmid construction.
The envelope-defective SIVMAC/GFP reporter virus, a simian immunodeficiency virus from macaques that encodes green fluorescent protein (GFP) in place of Nef, has been described previously (10). A similar HXB-derived HIV-1/GFP reporter virus was modified by replacing the BssHII-SalI fragment extending from the 5′ untranslated sequence to the vpr gene with the corresponding fragment from the NL4-3 HIV-1 clone and is referred to as wild-type HIV-1/GFP. All subsequent mutations were introduced into this plasmid.
To generate HIV-1 clones encoding chimeric CA domains in which loop sequences were derived from SIVMAC, two overlapping primers were designed that spanned loop sequences but encoded the corresponding loop sequence from SIVMAC. These were used in PCRs together with a primer either spanning the BssHII site in the HIV-1 5′ untranslated sequence or a primer spanning the ApaI site in the HIV-1 nucleocapsid-encoding sequence. The two PCR products were then mixed and subjected to PCR amplifications using the BssHII and ApaI primers. The final PCR products were digested by BssHII and ApaI and inserted into the NL4-3 clone, sequenced, and then transferred to the wild-type HIV-1/GFP construct as BssHII-SalI fragments. A similar strategy using overlapping primers introducing amino acid mutations was used to generate the CA mutants: R82A/L83A, V86A/H87A, P90A, A92E, P90A/A92E, I91A/P93A, G94A, and P122A.
All the remaining mutants were obtained from a previously described panel, constructed in the context of an HIV-1 NL4-3-based clone (44). Each was transferred to the HIV-1/GFP construct (described above) as a BssHII-SalI fragment. In all cases, the presence of the CA mutation in the final HIV-1/GFP construct was verified by DNA sequencing.
To generate HIV-1 GagPol expression plasmids, wild-type or mutant GagPol-encoding sequences were amplified by PCR, using primers that incorporated 5′ EcoRI and 3′ NotI sites and inserted into the previously described expression vector, pCRV1 (23).
Cell lines and viruses.
The human TE671, Owl monkey (OMK), Rhesus monkey (FrhK-4), and African green monkey (CV-1) fibroblast cell lines were obtained from the American Tissue Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with fetal calf serum and antibiotics.
Wild-type or mutant GFP-reporter virus stocks were made by cotransfecting 293T cells with a proviral plasmid and a vesicular stomatitis virus glycoprotein (VSV-G) expression plasmid as previously described (10, 23). Virus-like particles (VLPs) were made in the same way except that GagPol expression plasmids were used in place of the proviral plasmid. All reporter virus and VLP stocks were quantified using a colorimetric reverse transcription assay (Cavidi Tech).
N-MLV reporter virus stocks were generated by cotransfecting 293T cells with an N-MLV GagPol, a GFP-expressing MLV retroviral vector, and a VSV-G expression plasmid, as previously described (23). N-MLV stocks were quantified by titration on Fv1-null Mus dunni tail fibroblasts.
Infection assays.
TE671, OMK, FrhK-4, or CV-1 cells were seeded at 2 × 104 cells/well in 24-well trays the day before infection. Cells were inoculated with fivefold serially diluted GFP reporter virus stocks in the presence of 5 μg of polybrene/ml. Infected cells were enumerated 48 h postinfection using a FACSCalibur cell sorter and CellQuest software (Becton-Dickinson).
Abrogation-of-restriction assays.
TE671 cells were seeded at 2 × 104 cells/well in 24-well trays the day before infection. Cells were inoculated with a fixed dose of N-MLV GFP reporter virus selected so that infection with this virus in the absence of VLPs gave low, but accurately measurable, levels of infection (about 0.3% GFP-positive cells). Restriction-abrogating VLPs were serially diluted twofold and added to the target cells simultaneously with the fixed dose of N-MLV in the presence of polybrene (5 μg/ml). Infection by the GFP reporter virus was measured 48 h later as described above.
RESULTS
Exposed loops in the N-terminal domain of HIV-1 CA affect species tropism.
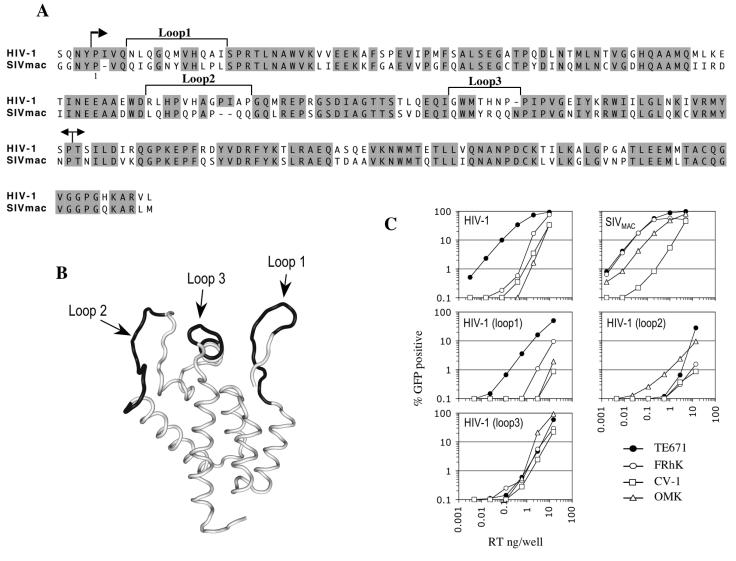
We and others have previously shown that VSV-G-pseudotyped HIV-1 and SIVMAC exhibit distinct tropisms for nonhuman primate cells that are governed, in large part, by the CA protein (10, 23, 24, 34). Even though the amino acid sequences of HIV-1 and SIVMAC CA proteins are quite well conserved (about 65% identical residues), an alignment, shown in Fig. 1A, reveals that nonconserved residues cluster in segments. These include the CypA binding loop, which we have previously demonstrated to be an important determinant of Lv1 and Ref1 recognition (43). Based on sequence variation and their position in the tertiary CA structure proximal to the CypA binding loop, we first focused our analysis on three such segments, reasoning that they might contain further determinants of restriction factor recognition. As shown in Fig. 1B, these segments are all surface exposed and encompass three loops within the N-terminal CA domain. Hereafter, we refer to these three segments as loop1, loop2, and loop3. Loop1 comprises the N-terminal β-hairpin, loop2 spans the CypA binding site between helices 4 and 5, and loop3 includes the C-terminal portion of helix 6 and a mini-β-hairpin that links helices 6 and 7.
FIG. 1.
Exposed loops in the HIV-1 CA N-terminal domain are tropism determinants. (A) Amino-acid alignment of HIV-1(NL4-3) and SIVMAC CA proteins. Sequence identities are shaded. The first arrow indicates the protease cleavage site between matrix and CA proteins. Numbering starts with the N-terminal residue of CA. The three regions defined as loop1, loop2 and loop3 in this study are indicated. The boundaries of the N- and C-terminal domains are marked with arrows. (B) Worm diagram representation of the HIV-1 CA N-terminal domain Cα-backbone structure indicating the locations of loop1, loop2, and loop3 in black. (C) Titration of HIV-1-, SIVMAC-, and HIV-1-based mutant viruses where CA loop1, loop2, or loop3 sequence was replaced by that of SIVMAC. The percentages of infected (GFP-positive) human (TE671), Rhesus monkey (FRhK-4), African green monkey (CV-1), and Owl monkey (OMK) cells are plotted as a function of the VSV-G-pseudotyped GFP reporter virus inoculum (in nanograms of reverse transcriptase [RT] per well).
Initially, we precisely replaced each loop in HIV-1 capsid by that of SIVMAC. GFP reporter viruses bearing these chimeric CA proteins were titrated on human (TE671), Owl monkey (OMK), Rhesus monkey (FrhK-4), and African green monkey (CV-1) target cells, and their infectivities were compared with those of the parental HIV-1 and SIVMAC reporter viruses (Fig. 1C). All three chimeras were less infectious than wild-type HIV-1 on human TE671 cells. The HIV-1(loop1) chimera was otherwise quite similar to HIV-1 in that it was about 50- to 100-fold more infectious on human cells than on the nonhuman primate cell lines. However, the loss of infectivity resulting from loop1 substitution was slightly more pronounced in human and AGM cells than in Rhesus or Owl monkey cells.
In contrast, the HIV-1(loop2) chimera was actually more infectious than wild-type HIV-1 in OMK cells but less infectious in each of the other cell lines, dramatically so (almost 1,000-fold) in human TE671 cells (Fig. 1C). This finding is consistent with our previous observation that the CypA binding properties of HIV-1 CA can have large effects on restriction in human and Owl monkey cells (43). Finally, the HIV-1(loop3) chimera was also more infectious than wild type HIV-1 in OMK cells and less infectious in TE671 cells and displayed infectivity similar to that of wild type HIV-1 in Rhesus FrhK-4 and AGM CV1 cells. In fact, the loop3 chimera exhibited an almost complete loss of the species-specific infectivity phenotype that is evident in wild-type HIV-1 and was similarly infectious in each of the human and nonhuman primate cell lines (Fig. 1C). Overall, the data in Fig. 1 indicate that sequences residing in the variable loops of the N-terminal domain of capsid can have very dramatic effects on the species tropism of HIV-1.
Mutational analysis of the CA N-terminal domain exposed loops.
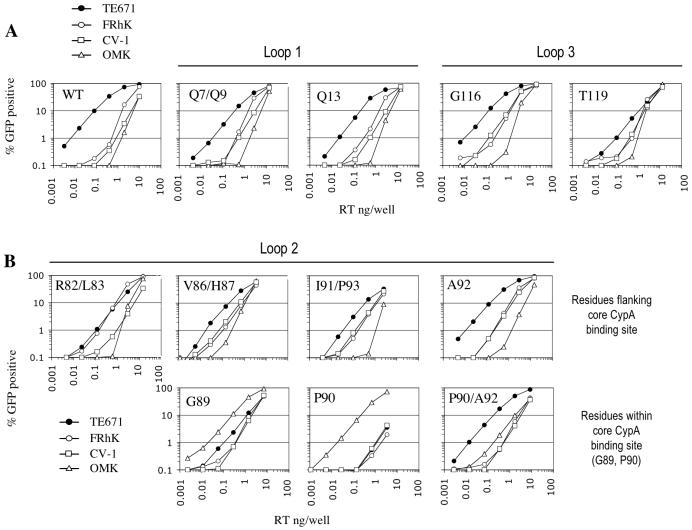
In order to map the residues within the CA loops responsible for the species-specific infectivity phenotypes more precisely, we determined the infectivity profiles of mutant viruses containing single- or double-amino-acid mutations within these segments. Residues were replaced by alanine, unless otherwise stated. A number of CA mutations, some of which have been characterized previously (44), either had no effect or caused a decrease in infectivity that was not specific to any particular cell line (data not shown). In contrast, mutation of residues Q7A/Q9A, which lie at the apex of the β-hairpin in loop1, or Q13A reduced infectivity specifically in human cells (Fig. 2A). While these results suggest that residues Q7/Q9 and Q13 in loop1 contribute to human cell-specific HIV-1 tropism, the effect of these mutations was quite modest (three to fivefold) and was similar to the modest species-specific effect of replacement of the entire loop 1 sequence with that of SIVMAC (Fig. 1C).
FIG. 2.
Tropism determinants within CA N-terminal domain variable loops. The same panel of cell lines as used in Fig. 1 was infected with HIV-1 GFP reporter viruses bearing the indicated CA mutations either within either loop1 or loop3 (A) or the cyclophilin binding loop2 (B). The percentage of infected (GFP-positive) cells is plotted as a function of the VSV-G-pseudotyped GFP reporter virus inoculum, as in Fig. 1. RT, reverse transcriptase.
Loop3 substitution had a more dramatic effect on HIV-1 tropism (Fig. 1C), and the effects of four individual mutations in this segment were examined (Fig. 2A; also data not shown). Of these, the G116A mutant exhibited infectivity similar to that of wild type HIV-1 in human cells but was slightly more infectious (up to fivefold) than wild-type HIV-1 in AGM, Rhesus, and OMK cells. Another loop3 mutant (T119A) had a different effect and partly reproduced the phenotype of the loop3 chimera (Fig. 1). Specifically the T119A mutant exhibited substantially reduced infectivity (>10-fold), selectively in human TE671 cells but not in other primate cell lines (Fig. 2A). Other mutations introduced into loop3 were lethal or caused decreases in infectivity that were not target cell specific (data not shown).
Our previous studies have demonstrated the importance of the CypA binding site within loop2 in determining restriction in both Owl monkey and human cells (43). Specifically, substitution of a single residue within loop2 (G89V) abolished restriction in OMK cells but conferred sensitivity to restriction by Ref1 in human cells. To determine whether other proximal residues are important for species-specific restriction, we introduced mutations at every position within HIV-1 loop 2 that differs in SIVMAC. As is shown in Fig. 2B, several of these mutations decreased infectivity in human cells but increased infectivity in nonhuman primate cells. Specifically, the R82A/L83A mutant was about four- to fivefold more infectious in Rhesus cells than wild-type HIV-1, such that its infectivity was equivalent in human and Rhesus cells. The V86A/H87A and I91A/P93A mutations also led to increased infectivity compared to wild-type HIV-1 in both Rhesus and AGM cells, and V86A/H87A was also more infectious in OMK cells.
Mutations within the core CypA binding site were the only single-residue substitutions that led to complete or almost complete loss of restriction in any nonhuman primate cell line. As was previously reported for a G89V mutation (43), a substitution at the neighboring position (P90A) led to a very dramatically increased titer (about 100-fold), specifically in OMK cells (Fig. 2B). Like G89V, this mutation has been shown to eliminate CA-CypA binding (7, 16, 46). This mutation also strongly inhibited (about 100-fold) infection in human cells, even more dramatically than G89V (Fig. 2B).
Previously, it has been shown that an A92E substitution can confer CsA resistance and restore replication competence in human cells to an HIV-1 strain that is rendered defective by CypA binding site mutation (1, 7). Our single-cycle infectivity analysis (Fig. 2B) revealed that combining the A92E mutation with P90A increased infectivity over that with the single P90A mutant by almost 100-fold in human cells and also increased infectivity significantly in Rhesus and AGM cells. Conversely, the P90A/A92E double mutant was about 10-fold less infectious than the P90A single mutant in OMK cells, although it was clearly more infectious than wild-type HIV-1 therein (Fig. 2B). Thus, in the context of a preexisting P90A substitution, the A92E mutation had dramatic and opposing effects in human and Owl monkey cells that were quite different from its modest effect on tropism in the absence of P90A substitution. Overall, many of the residues in loop2 and other CA loops had clear effects on infectivity that were frequently opposing and highly dependent on the target cell, with residues within, close to, and distal to the core CypA binding site apparently contributing to the overall species tropism of HIV-1.
Mutational analysis of residues outside the variable loops.
We also examined a panel of virus mutants containing substitutions in CA regions other than the exposed loops. In general, several of these mutations, for example, P38A, E45A (helix 2), T54A (helix 3), Q63A/Q67A, K70A, E71A (helix 4) and R143A (helix 7), either had negligible effects on infectivity or decreased infectivity in a manner that was not clearly specific to any species (data not shown) (44). However, as is shown in Fig. 3, other mutations, including N74A (helix 4) and N139A (helix 7), selectively decreased infectivity in human cells by three- to fivefold. A more pronounced loss of infectivity (six- to sevenfold), specifically in human cells, was observed in a virus containing an A78D/E79A/R82A triple mutation in helix 4 at the base of loop2 (Fig. 3). However, of the mutations that targeted residues outside the loop sequences, the E128A/R132A double mutant, which affects exposed residues in helix 7, had the clearest species-specific effects; this substitution reduced infectivity by about 12-fold in human cells but also increased infectivity in Rhesus and AGM cells by 4- to 6-fold, such that equivalent infectivity was observed in each of these three cell lines (Fig. 3). Overall, however, with the exception of residues in helices 4 and 7, close to the base of loops 2 and 3, the impact of mutations outside the loops on species-specific tropism was relatively modest and infrequent.
FIG. 3.
Effects of mutations outside CA variable loops on HIV-1 tropism. The panel of cell lines, as in Fig. 1, was infected with HIV-1 GFP reporter viruses bearing the indicated CA mutations. A78* refers to a triple mutant (A78D/E79A/R82A). The percentage of infected (GFP-positive) cells is plotted as a function of the VSV-G-pseudotyped GFP reporter virus inoculum, as in Fig. 1. RT, reverse transcriptase.
Abrogation of Ref1 restriction in human cells by mutant HIV-1 capsids.
A number of CA mutations led to reduced HIV-1 infectivity, specifically in human cells. To determine whether this was due to recognition of the mutant viral capsids by Ref1, we examined whether saturation of TE671 cells with mutant virus-like particles could inhibit Ref1-mediated restriction of N-MLV infection. We have previously used this approach to demonstrate that saturation of TE671 cells with HIV-1 VLPs carrying the G89V mutation can completely abolish N-MLV restriction (43), implying that this mutant capsid is recognized by, and can therefore saturate, Ref1. Thus, selected CA mutations that caused human cell-specific losses in infectivity were introduced into an HIV-1 GagPol expression plasmid that was coexpressed with VSV-G to generate VLPs. These VLPs were then tested for their ability to saturate Ref1 and restore restricted N-MLV infectivity in human cells.
As previously shown (23, 43), saturation of TE671 cells with wild-type HIV-1 particles had only a modest threefold inhibiting effect on N-MLV restriction (Fig. 4A and B). However, VLPs containing a mutation in loop1 (Q7A/Q9A) that conferred decreased infectivity in the context of virions were about fivefold better able than wild-type VLPs to abrogate Ref1-mediated restriction, increasing N-MLV titers by more than 16-fold (Fig. 4A). This result suggests that the modest but specific loss of infectivity in human cells induced by the Q7A/Q9A mutation is at least partly a consequence of recognition by Ref1 and that loop1 contains determinants of recognition by this restriction factor. In contrast, mutation of residues in loop3 (T119A) and in helix 7 (E128A/R132A) that caused more substantial human cell-specific losses in infectivity did not affect the ability of VLPs to saturate Ref1. Therefore, the human cell-specific infectivity loss resulting from some HIV-1 CA mutations may be independent of Ref1.
FIG. 4.
Abolishment of N-MLV restriction in human cells by mutant HIV-1 VLPs. TE671 cells were inoculated with a fixed dose of N-MLV GFP in the presence of increasing amounts of HIV-1 wild-type or Q7A/Q9A, T119A, or E128A/R132A mutant VLPs (A). Alternatively, P90A, A92E, or P90A/A92E mutant VLPs were used (B). The percentage of infected (GFP-positive) cells is plotted as a function of restriction-abolishing particle dose, given in nanograms of reverse transcriptase (RT) per well.
We have previously shown that the CypA binding site within loop2 is also an important determinant of Ref1 susceptibility or resistance (43), and as was the case for G89V, the P90A substitution resulted in VLPs that were potent suppressors of Ref1 restriction in that they were capable of completely rescuing N-MLV infectivity in human cells (Fig. 4B). Of note, the A92E substitution alone had no effect on the ability of HIV-1 VLPs to saturate Ref1 (Fig. 4B). However, combining the P90A mutation with an A92E substitution resulted in VLPs that could not saturate Ref1 and enhance N-MLV infectivity. Thus, the A92E substitution reversed both the infectivity defect (Fig. 2B) and the Ref1 saturating phenotype (Fig. 4B) induced by the P90A mutation, suggesting that it eliminates the interaction between Ref1 and the P90A mutant HIV-1 capsid.
DISCUSSION
We have identified a number of mutations in the N-terminal domain of the HIV-1 CA protein that have clear species-specific effects on infectivity. In principle, mutations within CA that reduce viral infectivity could do so as a consequence of a number of effects, including the following: (i) a generalized loss of viral fitness due to structural alteration, (ii) gain of interaction with an inhibitory restriction factor, or (iii) loss of interaction with hypothetical host factors that facilitate infection. In the absence of identified host factors (with the exception of CypA) that modulate HIV-1 infectivity by binding to CA, it is somewhat difficult to determine which effect governs the phenotype displayed by a particular mutant. Moreover, it is possible that a single mutation might have more than one functional consequence: for example, a nonspecific reduction in fitness combined with a loss of interaction with a restriction factor might be phenotypically neutral. However, because we analyzed infectivity in a number of cell lines with distinct restriction phenotypes for wild-type HIV-1 infection, we can interpret target-cell-specific effects of CA mutations as indicating the involvement of target-cell-specific molecules in modulating infection via effects on CA.
Some of these mutants have been previously assessed for effects on infectivity in human cells only. While there is generally good agreement between the findings reported herein and previous analyses (15, 44), some minor discrepancies may result from the use of different human target cells (TE671 in this study, HeLa-CD4 in previous studies) and viral envelopes (VSV-G in this study, HIV-1 Env in previous studies). Indeed, we have found that certain loop2 mutations have different effects on infectivity in TE671 versus HeLa cells (T. Hatziioannou and S. Cowan, data not shown). We hypothesize that this is due to polymorphism in, or differential expression of, human gene products, such as Ref1, that modulate infectivity via effects on CA.
A number of CA mutations clearly and selectively inhibited HIV-1 infectivity in human cells. In many cases, the same mutations had no effect, or even enhanced infectivity, in nonhuman primate cells. These residues either could be directly involved in capsid recognition by restriction factors or, alternatively, they could affect the overall structure of capsid and thus affect restriction in an indirect manner. In some cases, reduced infectivity in human cells was accompanied by an enhanced ability of VLPs carrying the same mutations to saturate Ref1. These data suggest that both the N-terminal β-hairpin (loop1) and the CypA binding site (loop2) contain determinants of Ref1 sensitivity or resistance. However, for other mutations, a loss of infectivity specifically in human TE671 cells did not always coincide with an enhanced ability to saturate Ref1. It is possible that these mutants are inhibited by a different restriction factor or that the loss of infectivity in human cells is mediated through other species or cell-type-specific mechanisms.
In general, it proved easier to generate HIV-1 mutants that have specifically reduced infectivity in human cells than mutants that have lost restriction in nonhuman primate cells. Based on these findings, it seems likely that restriction in most nonhuman primate cells is influenced by a number of determinants in CA and that several mutations will be required to completely abolish HIV-1 restriction in those cells. The only exception to this is the Owl monkey cell line. With this cell line, restriction can be completely bypassed by single-amino-acid mutations that also prevent CypA binding. These data are in agreement with our previously published observations that the CA-CypA interaction in OMK cells facilitates Lv1 restriction. Conversely, this interaction inhibits Ref1 restriction in human cells (43). HIV-1 CA mutants (G89V and P90A) that are unable to bind CypA are less infectious in human cells and are able to interact with Ref1. Importantly, we extend this finding to show that a mutation (A92E) that compensates for the loss of infectivity in human cells induced by a CypA binding site mutation (P90A) does so by abolishing recognition of the CypA-free capsid by Ref1.
Although the mutations that clearly had species-specific effects on infectivity did not always lie in proximal positions in the linear amino acid sequence, they were found to cluster when mapped on the three-dimensional structure of HIV-1 CA monomer (Fig. 5). Moreover, the three loop segments examined in our study can undergo concerted conformational shifts (39), suggesting that they may function as a single cooperative structural unit.
FIG. 5.
Localization of HIV-1 tropism determinants to the surface of the HIV-1 CA. (A) Representation of a CA N-terminal domain monomer, with loops and tropism determinants indicated. Residues whose mutation had pronounced species-specific effects on infectivity are indicated in red (A78/E79/R82, R82/L83, V86/H87, G89, P90/A92, I91/P93, G116, T119, and E128/R132). Mutations that induced modest but clearly species-specific effects on infectivity are indicated in orange (Q7/Q9, Q13, N74, and N139), and mutations that induced little or no species-specific effects are in yellow (Q4, P38, E45, T54, Q63/Q67, K70, E71, G94, P122, and R143). The approximate position of the C-terminal CA domain is also indicated, as is the orientation of the molecule with respect to the “outside” (top) and “inside” (bottom) faces of the capsid lattice. (B) A single N-terminal domain hexamer with residues colored in the same way as in panel A is shown as viewed from the outside of the capsid lattice. The positions of loop1, loop2, and loop3 are indicated. (C) View of part of the hexameric lattice (N-terminal CA domain only) showing the juxtaposition of tropism determinants in several adjacent CA hexamers. (D) Representation of the intact HIV-1 capsid. The position of the pentameric “defects” that determine capsid shape are shown in red, and a group of CA hexamers similar to that shown in panel C with N-terminal domains colored pink and C-terminal domains colored blue is shown schematically on part of the hexagonal lattice.
As illustrated in Fig. 5A and B, the N-terminal domains of HIV-1 CA are thought to assemble into hexameric rings, with each contacting neighboring rings via C-terminal CA domain interactions (8, 20, 31). Ultimately, a conical lattice of hexameric rings is formed that constitutes the complete viral capsid (Fig. 5C and D). Interestingly, the mutations that affected tropism were located primarily in capsid regions predicted to project outward on the surface of the hexameric rings in models for the intact viral capsid (Fig. 5). Thus, tropism determinants were located in regions that are not predicted to contact other capsid molecules but would rather be exposed on the outer surface of the intact capsid lattice (20, 31). Presumably, therefore, these residues would be free to interact with other proteins, such as capsid-targeting restriction factors. In the context of a hexameric lattice, the presence of CypA bound to loop2 on a proportion of the CA molecules would likely affect the accessibility of some, but not all, residues identified as species-specific tropism determinants. This might explain why CypA affects HIV-1 tropism for some, but not all, primate species.
Based on the surface distribution of the tropism determinants in the CA lattice (Fig. 5C), it is entirely possible that a single entity, for example, a restriction factor, could simultaneously contact multiple hexameric rings. Two striking features of retroviral capsids are the following: (i) they can adopt very different overall shapes, including cones, spheres and cylinders, despite being comprised of very similar hexameric units, and (ii) they also generally lack any formal symmetry element (19, 20, 28, 31, 45). The differential distribution of pentameric “defects” in the otherwise hexameric CA lattice (e.g., Fig. 5D) is thought to govern this structural polymorphism. A consequence of intrinsic capsid asymmetry is that each hexameric building block is positioned in a slightly different local environment relative to its neighbors. Thus, any restriction factor that binds simultaneously to neighboring hexamers would be likely to recognize only restricted areas on the surface of the capsid lattice. It is plausible, therefore, that retroviral capsids might have evolved highly heterogeneous and unusual shapes, such as cones, in response to host restriction factors that target these structures.
The findings described herein might have practical consequences. Specifically, we identified several mutations that increased HIV-1 infectivity in nonhuman primate cells, although none were able to completely restore infectivity in cells from the rhesus macaque, the most commonly used primate species in AIDS research. Other studies have also shown that alteration of residues within loop2 can increase titers of HIV-1 in several nonhuman primate cells and at least partially bypass Lv1 restriction (29). However, neither our analysis nor previous analyses have been exhaustive, and it seems likely that further mutational analyses of capsid might lead to the generation of HIV-1 strains that are completely resistant to Lv1-mediated inhibition in macaques. Other adaptations, particularly with respect to the Vif/APOBEC3G axis (32, 36), may also be required, but these and other recent findings suggest that the adaptation of HIV-1 to full replication competence in a practically useful nonhuman species may be possible.
Acknowledgments
We thank Greg Towers for reagents and helpful discussions.
This work was supported in part by NIH grants AI50111 (to P.D.B.) and AI45405 (to W.I.S.). T.H. is the recipient of an AmFAR Scholar Award. P.D.B. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassin, R. H., G. Duran-Troise, B. I. Gerwin, and A. Rein. 1978. Abrogation of Fv-1b restriction with murine leukemia viruses inactivated by heat or by gamma irradiation. J. Virol. 26:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 6.Boone, L. R., C. L. Innes, and C. K. Heitman. 1990. Abrogation of Fv-1 restriction by genome-deficient virions produced by a retrovirus packaging cell line. J. Virol. 64:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, J. A., T. Wilk, R. Welker, H. G. Krausslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. Embo J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 10.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decleve, A., O. Niwa, E. Gelmann, and H. S. Kaplan. 1975. Replication kinetics of N- and B-tropic murine leukemia viruses on permissive and nonpermissive cells in vitro. Virology 65:320-332. [DOI] [PubMed] [Google Scholar]
- 12.DesGroseillers, L., and P. Jolicoeur. 1983. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol. 48:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfman, T., and H. G. Gottlinger. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duran-Troise, G., R. H. Bassin, A. Rein, and B. I. Gerwin. 1977. Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell. 10:479-488. [DOI] [PubMed] [Google Scholar]
- 15.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 18.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 19.Ganser, B. K., A. Cheng, W. I. Sundquist, and M. Yeager. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 21.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 22.Goff, S. P. 1996. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell. 86:691-693. [DOI] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, Z., L. Jin, D. L. Peterson, and C. L. Lawson. 1999. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286:83-93. [DOI] [PubMed] [Google Scholar]
- 26.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67-122. [DOI] [PubMed] [Google Scholar]
- 27.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 28.Kingston, R. L., N. H. Olson, and V. M. Vogt. 2001. The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy. J. Struct. Biol. 136:67-80. [DOI] [PubMed] [Google Scholar]
- 29.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak, C. A., and A. Chakraborti. 1996. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225:300-305. [DOI] [PubMed] [Google Scholar]
- 31.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 32.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 33.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pincus, T., J. W. Hartley, and W. P. Rowe. 1975. A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1-sensitive and resistant cell cultures. Virology 65:333-342. [DOI] [PubMed] [Google Scholar]
- 36.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 37.Stoye, J. P. 1998. Fv1, the mouse retrovirus resistance gene. Rev. Sci. Tech. 17:269-277. [DOI] [PubMed] [Google Scholar]
- 38.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 40.Tennant, R. W., J. A. Otten, A. Brown, W. K. Yang, and S. J. Kennel. 1979. Characterization of Fv-1 host range strains of murine retroviruses by titration and p30 protein characteristics. Virology 99:349-357. [DOI] [PubMed] [Google Scholar]
- 41.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 retrovirus restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 44.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]