Abstract
Recently, APOBEC3G has been identified as a host factor that blocks retroviral replication. It introduces G to A hypermutations in newly synthesized minus strand viral cDNA at the step of reverse transcription in target cells. Here, we identified the human APOBEC3F protein as another host factor that blocks human immunodeficiency virus type 1 (HIV-1) replication. Similar to APOBEC3G, APOBEC3F also induced G to A hypermutations in HIV genomic DNA, and the viral Vif protein counteracted its activity. Thus, APOBEC family members might have evolved as a general defense mechanism of the body against retroviruses, retrotransposons, and other mobile genetic elements.
In the human immunodeficiency virus type 1 (HIV-1), there exists an accessory gene between the pol and env open reading frames which encodes the viral infectivity factor Vif (6). This gene was originally called sor (short open reading frame), orfQ, orf1, orfP′, and orfA (22, 24-26), and it produces a 23-kDa protein (13, 17, 31). The same gene has been found in all lentiviruses except equine infectious anemia virus (23). It is expressed from the 4-kb HIV transcripts in a Rev-dependent manner (7, 27). Initial studies demonstrated that Vif increases HIV infectivity up to 1,000 times (4, 33).
The effect of Vif on viral infectivity is highly cell type dependent. Cells can be divided into groups that are permissive and nonpermissive for HIV infection (1). In permissive cells, including HeLa, COS, C8166, Jurkat, U937, SupT1, or 293T, Vif is not required for HIV infection. However, in nonpermissive cells, including H9, CEM, Hut68, peripheral blood mononuclear cells, and macrophages, Vif is required for HIV replication (5, 34). It was demonstrated that the nonpermissive cells express a dominant inhibitor to inactivate HIV-1 in the absence of Vif (18, 30). Since Vif is an RNA-binding protein (3, 16, 36), it seems that Vif might protect the HIV genome from the attack of this molecule. Recently, this inhibitor was identified as APOBEC3G (originally called CEM-15) (28), which belongs to the APOBEC (apolipoprotein B mRNA-editing enzyme-catalytic polypeptide) family. These proteins catalyze the cytosine deamination reaction that changes nucleotides from cytosine to uracil. Although the first member of this family, APOBEC1, is an RNA-editing enzyme, APOBEC3G was shown to use DNA as a template (11). APOBEC3G causes cytidine deamination of HIV-1 negative-strand DNA during the reverse transcription step (10, 19, 37). Hypermutations induced by APOBEC3G either introduce stop codons in viral protein open reading frames or trigger the degradation of viral DNA by uracil-DNA glycosylase, therefore blocking HIV replication (8, 9, 15). Until now, nine APOBEC family members have been identified in humans. APOBEC2 is found on chromosome 6; all the others (APOBEC1, AID [activation-induced deaminase], and APOBEC3A, -3B, -3C, -3D, -3F, and -3G) are found on chromosome 22 (12). However, their antiviral activities have not been determined. Here, we report that like APOBEC3G, APOBEC3F also has similar antiviral activity, which is blocked by Vif.
Cloning and expression of APOBEC proteins.
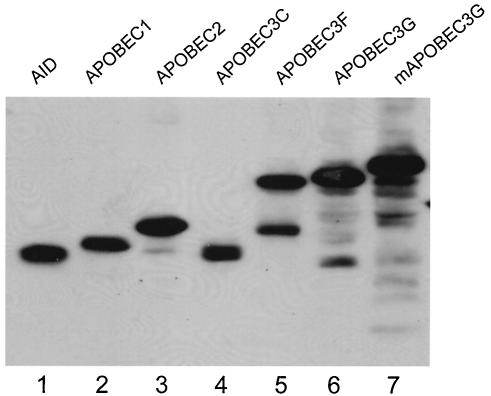
To determine the effect of APOBEC family members on HIV replication, we obtained the cDNAs coding for human AID, APOBEC2, APOBEC3C, APOBEC3F, and murine APOBEC3G(mAPOBEC3G) from Open Biosystems (Huntsville, Ala.), human APOBEC3G from K. Streble (National Institute of Allergy and Infectious Diseases), and APOBEC1 from N. O. Davidson (Washington University, St. Louis, Mo.). They were subsequently cloned into a mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, Calif.) with a V5 epitope tag from SV5 paramyxovirus and a polyhistidine epitope at the C terminus. These plasmids were transfected into 293T cells, and their expression was detected with a monoclonal anti-V5 antibody (Fig. 1). All these proteins were expressed abundantly with the expected mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As previously observed in human and murine APOBEC3G (20), we also observed degradation products in human APOBEC3F (Fig. 1, lane 5).
FIG. 1.
Expression of APOBEC proteins. Human APOBEC proteins AID, APOBEC1, APOBEC2, APOBEC3C, APOBEC3F, APOBEC3G, and mAPOBEC3G were cloned into pcDNA3 with a V5 tag. These proteins were expressed in 293T cells and detected with a monoclonal anti-V5 antibody.
APOBEC3F turns HIV-permissive cells into nonpermissive cells.
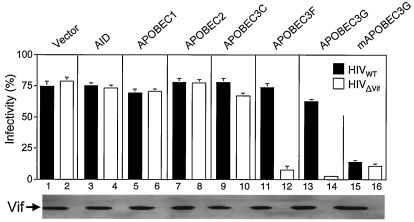
Next, we determined if these proteins could inhibit HIV replication by a single-round infectivity assay. The wild-type virus was from pNL4-3. The pNLΔVif clone was generated by inserting a NaeI linker at the PfiMI site of pNL4-3. The wild-type or Vif-defective HIV viruses were produced from 293T cells in the presence of different APOBEC proteins via CaPO4 transfection. Viral levels were then quantified by an enzyme-linked immunosorbent assay for p24Gag content. Expression levels of Vif from these viruses were confirmed by Western blotting with an anti-Vif antibody (Fig. 2, lower panel). Equal amounts of viruses were used to infect GHOST-R3/X4/R5 indicator cells, which were stably transfected with the HIV-2 long terminal repeat linked to a green fluorescence protein expression construct. Once HIV particles infect these cells, the viral transcriptional transactivator (Tat) activates the HIV-2 long terminal repeat and induces the expression of green fluorescence protein. After 48 h, the infection can then be detected and quantified by determining the green fluorescence intensity by flow cytometry. As presented in Fig. 2, AID, APOBEC1, APOBEC2, and APOBEC3C had no effect on the replication of both wild-type and Vif-defective HIV (compare lanes 3 to 10 with the control lanes 1 and 2). As previously reported (20), human APOBEC3G only inhibited the replication of ΔVif-HIV (lanes 13 and 14), and the murine APOBEC3G inhibited the replication of both wild-type and ΔVif-HIV (lanes 15 and 16). Interestingly, we found that APOBEC3F inhibited the replication of the Vif-defective HIV but not the wild-type HIV to an extent similar to that of APOBEC3G (Fig. 2, lanes 11 to 12). Thus, we conclude that, similar to human APOBEC3G, human APOBEC3F inhibits Vif-defective HIV replication, and the block can be overcome by Vif.
FIG. 2.
Function of APOBEC proteins in viral replication. APOBEC proteins were coexpressed with wild-type or Vif-defective HIV in 293T cells. After normalization by p24Gag enzyme-linked immunosorbent assay, equal amounts of viruses were used to infect GHOST-R3/X4/R5 cells. The expression of Vif in cell lysates from wild-type or Vif-defective viruses was demonstrated by Western blotting (bottom).
Sequence comparison between human APOBEC3F and -3G.
APOBEC3G and APOBEC3F are both located on the chromosome 22q13.1 separated by 24,667 bp (12). In studies by Jarmuz et al. (12), the expression of APOBEC3F was also detected in colorectal adenocarcinoma, chronic myelogenous leukemia, and epithelial cells. Their N termini share almost 100% similarity. APOBEC3F has seven exons, and APOBEC3G has eight exons. Both of them contain duplicated active sites, inserts, and linker peptides in exons 6 and 7 (12). Sequence alignments demonstrate conserved zinc fingers, glutamates involved in proton shuttling in the active site (C/HXE, PCXXC), and two critical aromatic residues (F/Y) involved in RNA binding (12) (Fig. 3). All these results suggest that APOBEC3F could have a similar cytidine deaminase activity as APOBEC3G to block HIV replication.
FIG. 3.
Sequence alignment between human APOBEC3G (3G) and APOBEC3F (3F). Two zinc-finger domains are underlined; two active sites for cytidine deaminase (C/HXE, PCXXC), two linker peptides, and two inserts are boxed, where X represents any amino acid residues; critical aromatic residues (F/Y) for RNA binding are presented in bold. For APOBEC3F, only residues different from APOBEC3G are presented. Dots indicate identity, and dashes represent deleted residues.
APOBEC3F induces G to A hypermutations in HIV minus strand cDNA during reverse transcription.
To prove this hypothesis, GHOST-R3/X4/R5 cells were infected with various HIV viruses produced from 293T cells. After 5 h, cells were trypsinized and collected. Viral DNAs were then extracted by a DNeasy tissue kit (QIAGEN Inc., Valencia, Calif.). A primer pair that targeted the HIV Vif/Vpr region was used to amplify a 420-bp fragment. Fragments were further purified by agarose gel, cloned into pCR4-TOPO vector (Invitrogen), and sequenced by flanking T3 and T7 primers. As presented in Fig. 4, sequences from the wild-type and Vif-defective viruses did not contain any mutations when these viruses were produced in the absence of APOBEC proteins (Fig. 4, see WT and ΔVif panels). In the presence of APOBEC3F or APOBEC3G, sequences from the wild-type HIV-infected cells contained very few mutations (Fig. 4, WT/A3F and WT/A3G panels). However, in sharp contrast, sequences from Vif-defective HIV-infected cells contained a very high frequency of G to A mutations (Fig. 4, ΔVif/A3F and ΔVif/A3G panels). Of note, APOBEC3G induced more dramatic hypermutations than did APOBEC3F (Fig. 4, ΔVif/A3F and ΔVif/A3G panels). In the presence of mAPOBEC3G, sequences from the wild-type and Vif-defective HIV-infected cells contained a high frequency of G to A mutations (Fig. 4, WT/mA3G and ΔVif/mA3G panels). Since the G to A mutation in viral double-strand DNA corresponds to the C to U mutation in the single minus strand DNA, we conclude that, like human and murine APOBEC3G, APOBEC3F catalyzes the deamination of deoxycytidine (dC) to deoxyuridine (dU) during HIV minus strand synthesis. These changes are responsible for the disruption of HIV replication. Interestingly, APOBEC3G targeted more GGGG repeats and APOBEC3F targeted single G molecules.
FIG. 4.
APOBEC3F induces G to A mutations in viral genome. Wild-type (WT) and Vif-defective viruses were produced in the presence and absence of APOBEC3G (A3G), APOBEC3F (A3F), and mAPOBEC3G (mA3G). These viruses were used to infect GHOST-R3/X4/R5 cells. Six hours later, cellular DNAs were extracted from these infected cells, and viral DNAs were amplified by PCR. After cloning into TA-cloning vector, these viral DNAs were then sequenced. The pNL4-3 sequence from nucleotides 5698 to 5787 is presented at the top. Five sequences from each infection are presented, and only the mutated nucleotides are shown. Dots indicate sequence identity.
Vif excludes APOBEC3F and APOBEC3G from incorporation into HIV particles.
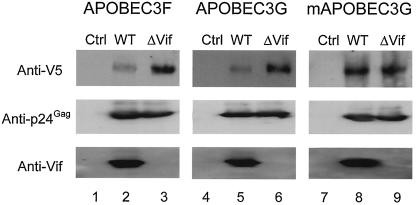
Our results indicate that APOBEC3F blocks HIV replication, and its activity is counteracted by Vif. For its effects, Vif excludes APOBEC3G from virions, therefore blocking the function of APOBEC3G (2, 14, 20, 21, 29, 32, 35). To determine if Vif could also exclude APOBEC3F from incorporation into virions, wild-type and Vif-defective HIV were produced in the presence of APOBEC3F, APOBEC3G, or mAPOBEC3G, respectively. Viruses were further purified by loading onto a 20% sucrose cushion and spun at 27,000 rpm for 2 h in a Beckman SW28 rotor. Supernatants and sucrose layers were carefully removed, and viral particles were resuspended in STE buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA). Equal levels of viral particles were loaded to compare levels of APOBEC proteins (Fig. 5, see anti-p24Gag blotting). As expected, in the presence of Vif, levels of APOBEC3F and APOBEC3G were significantly reduced in virions (Fig. 5, compare lanes 2 and 5 with lanes 3 and 6). As previously reported (20), Vif did not reduce the levels of mAPOBEC3G in HIV particles as previously reported (Fig. 5, lanes 8 and 9). Thus, we conclude that Vif inhibits the activity of these human, but not murine, APOBEC family members by blocking their incorporation into HIV particles.
FIG. 5.
Levels of APOBEC proteins in viral particles. 293T cells were cotransfected with wild-type (WT) or Vif-defective proviruses and different APOBEC expression vectors. Viral particles were collected 48 h later and purified by ultracentrifugation through a 20% sucrose cushion. Equal amounts of viral proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the expressions of APOBEC proteins, p24Gag, and Vif were determined by Western blotting.
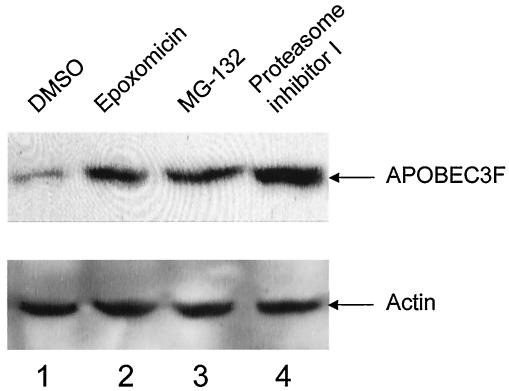
To further define how Vif excludes APOBEC3F from virions, we measured intracellular levels of APOBEC3F in the presence of Vif. Indeed, in cells expressing Vif, APOBEC3F was barely detected (Fig. 6, lane 1). Since it was reported that Vif degrades APOBEC3G via the proteasome (2, 21, 28, 32, 35), we also determined if this pathway was involved in the removal of APOBEC3F. Indeed, levels of APOBEC3F were increased when cells were treated with proteasomal inhibitors epoxomicin, MG-132, or proteasome inhibitor I (Fig. 6, lanes 2 to 4). Thus, the ubiquitylation and proteasomal degradation play the same role in the removal of APOBEC3F and APOBEC3G by Vif in cells.
FIG. 6.
The degradation of APOBEC3F by Vif is blocked by proteasomal inhibitors in cells. 293T cells were cotransfected with Vif and APOBEC3F expression vectors. Twenty hours later, cells were treated with proteasomal inhibitors epoxomicin, MG-132, and proteasome inhibitor I at 10 μM for 12 h. Levels of APOBEC3F were then analyzed by Western blotting. DMSO, dimethyl sulfoxide.
In summary, we presented data that another APOBEC family member, the human APOBEC3F, inhibits HIV replication by catalyzing the deamination of dC to dU during HIV minus strand synthesis and causes G to A hypermutations in the viral genome. Similar to the human APOBEC3G, Vif overcomes the antiviral activity of APOBEC3F by excluding its incorporation into viral particles. These results suggest that APOBEC family members might have evolved as a general defense mechanism of the body against retroviruses, retrotransposons, and other mobile genetic elements.
Acknowledgments
We thank K. Strebel for pcDNA-APOBEC3G, N. O. Davidson for APOBEC1 cDNA, and the NIH AIDS Research and Reference Reagent Program for various reagents.
This work was supported by research grants from the National Institutes of Health (B.M.P.), California Universitywide AIDS Research Program (B.M.P., Y.-H.Z.), and a training grant from the National Institutes of Health (Y.-H.Z.).
REFERENCES
- 1.Bour, S., and K. Strebel. 2000. HIV accessory proteins: multifunctional components of a complex system. Adv. Pharmacol. 48:75-120. [DOI] [PubMed] [Google Scholar]
- 2.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 3.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 5.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo, R., F. Wong-Staal, L. Montagnier, W. A. Haseltine, and M. Yoshida. 1988. HIV/HTLV gene nomenclature. Nature 333:504. [DOI] [PubMed] [Google Scholar]
- 7.Garrett, E. D., L. S. Tiley, and B. R. Cullen. 1991. Rev activates expression of the human immunodeficiency virus type 1 vif and vpr gene products. J. Virol. 65:1653-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goff, S. P. 2003. Death by deamination: a novel host restriction system for HIV-1. Cell 114:281-283. [DOI] [PubMed] [Google Scholar]
- 9.Gu, Y., and W. I. Sundquist. 2003. Good to CU. Nature 424:21-22. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 11.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247-1253. [DOI] [PubMed] [Google Scholar]
- 12.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 13.Kan, N. C., G. Franchini, F. Wong-Staal, G. C. DuBois, W. G. Robey, J. A. Lautenberger, and T. S. Papas. 1986. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science 231:1553-1555. [DOI] [PubMed] [Google Scholar]
- 14.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.KewalRamani, V. N., and J. M. Coffin. 2003. Virology. Weapons of mutational destruction. Science 301:923-925. [DOI] [PubMed] [Google Scholar]
- 16.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, T. H., J. E. Coligan, J. S. Allan, M. F. McLane, J. E. Groopman, and M. Essex. 1986. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science 231:1546-1549. [DOI] [PubMed] [Google Scholar]
- 18.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 20.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 21.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 22.Muesing, M. A., D. H. Smith, C. D. Cabradilla, C. V. Benton, L. A. Lasky, and D. J. Capon. 1985. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313:450-458. [DOI] [PubMed] [Google Scholar]
- 23.Oberste, M. S., and M. A. Gonda. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 6:95-102. [DOI] [PubMed] [Google Scholar]
- 24.Rabson, A. B., and M. A. Martin. 1985. Molecular organization of the AIDS retrovirus. Cell 40:477-480. [DOI] [PubMed] [Google Scholar]
- 25.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Pescador, R., M. D. Power, P. J. Barr, K. S. Steimer, M. M. Stempien, S. L. Brown-Shimer, W. W. Gee, A. Renard, A. Randolph, J. A. Levy, et al. 1985. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science 227:484-492. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1991. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is Rev-dependent and regulated by splicing. Virology 183:677-686. [DOI] [PubMed] [Google Scholar]
- 28.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 29.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 30.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 31.Sodroski, J., W. C. Goh, C. Rosen, A. Tartar, D. Portetelle, A. Burny, and W. Haseltine. 1986. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science 231:1549-1553. [DOI] [PubMed] [Google Scholar]
- 32.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 33.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ′A' (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 34.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H., R. J. Pomerantz, G. Dornadula, and Y. Sun. 2000. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74:8252-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]