Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV), a newly identified group 2 coronavirus, is the causative agent of severe acute respiratory syndrome, a life-threatening form of pneumonia in humans. Coronavirus replication and transcription are highly specialized processes of cytoplasmic RNA synthesis that localize to virus-induced membrane structures and were recently proposed to involve a complex enzymatic machinery that, besides RNA-dependent RNA polymerase, helicase, and protease activities, also involves a series of RNA-processing enzymes that are not found in most other RNA virus families. Here, we characterized the enzymatic activities of a recombinant form of the SARS-CoV helicase (nonstructural protein [nsp] 13), a superfamily 1 helicase with an N-terminal zinc-binding domain. We report that nsp13 has both RNA and DNA duplex-unwinding activities. SARS-CoV nsp13 unwinds its substrates in a 5′-to-3′ direction and features a remarkable processivity, allowing efficient strand separation of extended regions of double-stranded RNA and DNA. Characterization of the nsp13-associated (deoxy)nucleoside triphosphatase ([dNTPase) activities revealed that all natural nucleotides and deoxynucleotides are substrates of nsp13, with ATP, dATP, and GTP being hydrolyzed slightly more efficiently than other nucleotides. Furthermore, we established an RNA 5′-triphosphatase activity for the SARS-CoV nsp13 helicase which may be involved in the formation of the 5′ cap structure of viral RNAs. The data suggest that the (d)NTPase and RNA 5′-triphosphatase activities of nsp13 have a common active site. Finally, we established that, in SARS-CoV-infected Vero E6 cells, nsp13 localizes to membranes that appear to be derived from the endoplasmic reticulum and are the likely site of SARS-CoV RNA synthesis.
Severe acute respiratory syndrome (SARS) is a life-threatening form of pneumonia characterized by high fever, nonproductive cough, chills, myalgia, lymphopenia, and progressing infiltrates on chest radiography (48). Between November 2002 and June 2003, an epidemic emerged that, facilitated by international air travel, spread within few weeks from its origin in Guangdong Province, China, to many other countries. There is now clear evidence that SARS is caused by a previously unknown coronavirus, termed the SARS coronavirus (SARS-CoV) (14, 16, 32, 49). The SARS-CoV genome encompasses 29,727 nucleotides [excluding the 3′-terminal poly(A) tail] (43, 52, 53), and there is phylogenetic evidence to suggest that SARS-CoV represents an early split-off from the coronavirus group 2 lineage (65). By analogy with other coronaviruses and other members of the order Nidovirales (17, 34, 37), SARS-CoV gene expression involves a set of complex transcriptional, translational, and posttranslational regulatory mechanisms (72). It starts with the translation of two large replicative polyproteins, pp1a (≈490 kDa) and pp1ab (≈790 kDa), which are encoded by the viral replicase gene (21,221 nucleotides), which comprises open reading frames (ORFs) 1a and 1b (Fig. 1). Expression of the ORF1b-encoded region of pp1ab involves ribosomal frameshifting into the −1 frame just upstream of the ORF1a translation termination codon (72).
FIG.1.
Expression and primary structure of the SARS-CoV nsp13 helicase. (A) Overview of the domain organization and (predicted) proteolytic processing of the SARS-CoV replicase polyproteins pp1a and pp1ab. Nsp13 is encoded by ORF 1b and is processed from pp1ab by the 3C-like proteinase. The processing end products of pp1a are designated nsp1 to nsp11, and those of pp1ab are designated nsp1 to nsp10 and nsp12 to nsp16. Note that nsp1 to nsp10 may be released by proteolytic processing of either pp1a or pp1ab, whereas nsp11 is processed from pp1a and nsp12 to nsp16 are processed from pp1ab. Nsp11 and nsp12 have a number of common residues at their N termini. Cleavage sites that are (predicted to be) processed by the viral main proteinase are indicated by grey arrowheads, and sites that are processed by the papain-like proteinase 2 are indicated by black arrowheads. Ac, acidic domain (92); X, X domain (21), which is predicted to have ADP-ribose 1"-phosphatase activity (65); SUD, SARS-CoV unique domain (65); PL2, papain-like cysteine proteinase 2 (72); Y, Y domain containing a transmembrane domain and a putative metal-binding domain (65, 72, 92); TM1, TM2, and TM3, putative transmembrane domains 1 to 3, respectively; 3CL, 3C-like main proteinase (3, 72); RdRp, putative RNA-dependent RNA polymerase domain (19, 29, 43, 52); HEL, superfamily 1 helicase domain (72); ExoN, putative 3′-to-5′ exonuclease (65); XendoU, putative poly(U)-specific endoribonuclease (65); MT, putative S-adenosylmethionine-dependent ribose 2′-O-methyltransferase (65, 81); C/H, domains containing conserved Cys and His residues and predicted to bind metal ions. (B) Sequence comparison of coronavirus helicases. The alignment was generated with the ClustalW program (version 1.82) (http://www.ebi.ac.uk/clustalw/) and used as the input for the ESPript program, version 2.1 (http://prodes.toulouse.inra.fr/ESPript/cgi-bin/ESPript.cgi). The nsp13 sequences of SARS-CoV (isolate Frankfurt 1; accession no. AY291315), mouse hepatitis virus (MHV, strain A59; NC_001846), bovine coronavirus (BCoV, isolate LUN; AF391542), human coronavirus 229E (HCoV-229E; X69721), porcine epidemic diarrhea virus (PEDV, strain CV777; AF353511); transmissible gastroenteritis virus (TGEV, strain Purdue 46; AJ271965), and avian infectious peritonitis virus (IBV, strain Beaudette; M95169) were derived from the replicative polyproteins of these viruses, whose sequences were obtained from the DDBJ, EMBL, and GenBank databases. Conserved helicase motifs I to VI (18) are indicated. Near the N terminus, the 12 conserved Cys and His residues predicted to form a binuclear zinc-binding cluster (77) are indicated by @. Also indicated is the conserved Lys288 residue (corresponding to Lys5589 in pp1ab), which, in the MBP-nsp13_KA control protein, was replaced with Ala. Lys288 is part of conserved helicase motif I (18), which is also called the Walker A box (82). Highlighted in grey is the C-terminal nsp13 sequence against which the rabbit antiserum, α-nsp13, used in this study was raised.
The production of the functional replication-transcription complex involves extensive proteolytic processing by two viral cysteine proteinases (52, 65, 72, 91). The activities of these proteinases, a papain-like (accessory) cysteine proteinase (PLpro) residing in nsp3 and a 3C-like (main) proteinase (termed 3CLpro or Mpro) residing in nsp5, were recently established and characterized (3, 15, 72, 85). The membrane-bound replicase complex synthesizes both genome-length RNAs (replication) and as many as eight subgenomic mRNAs (transcription) (65, 72). As in other coronaviruses, all SARS-CoV subgenomic mRNAs have a 5′-terminal leader sequence which is acquired from the 5′ end of the genome through a unique discontinuous RNA synthesis mechanism (72). mRNAs 2, 4, 5, and 9 encode the four structural proteins S (ORF2), E (ORF4), M (ORF5), and N (ORF9a). Furthermore, no fewer than eight other proteins (with unknown functions) are predicted to be expressed from mRNAs 3 (ORFs 3a and 3b), 6 (ORF 6), 7 (ORFs 7a and 7b), 8 (ORFs 8a and 8b), and 9 (ORF9b) (65, 72).
In cells infected with mouse hepatitis virus (MHV), the best-studied coronavirus in terms of molecular biology, the majority of the viral replicase subunits were found to be associated with intracellular membranes, a feature encountered in many positive-stranded RNA viruses. During the peak of MHV RNA synthesis, key nonstructural proteins such as RNA-dependent RNA polymerase (RdRp) and helicase, de novo RNA synthesis, and also the viral nucleocapsid protein were found to colocalize in punctate cytoplasmic foci (13, 61, 74). The results of ultrastructural studies of the replicase complex in MHV-infected cells have remained equivocal thus far. While the results of one study pointed mainly to membranes of (late) endosomal origin as the site of viral replication (74), a more recent study (22) described virus-induced double-membrane structures resembling those implicated in the RNA synthesis of another nidovirus, the arterivirus prototype equine arteritis virus (47, 66).
Our knowledge of the molecular mechanisms controlling coronavirus RNA synthesis is still rather limited. However, the available information strongly suggests that the enzyme machinery of coronaviruses may differ fundamentally from that of other positive-stranded RNA viruses. First, at more than 20 kb, the coronavirus replicase genes are unparalleled in size and complexity among those of all positive-stranded RNA viruses. Second, coronaviruses seem to employ a set of RNA-processing activities that are rare or even unique among positive-stranded RNA viruses (65). Third, the conserved sequential order of the functional domains in pp1a and pp1ab of members of the order Nidovirales differs significantly from that of other positive-stranded RNA virus polyproteins, and the functional subunits themselves are only distantly related to their viral and cellular homologs (17, 30). The last observation also applies to the coronavirus (nidovirus) superfamily 1 helicase encoded by the central region of ORF1b (19).
Coronaviral and other nidoviral helicase domains are unique among their known positive-stranded RNA virus homologs in that they are linked in a single protein to an N-terminal binuclear zinc-binding domain consisting of 12 conserved Cys/His residues (19, 25). The zinc-binding domain of the arterivirus nsp10 helicase was shown previously to be involved in diverse processes of the viral life cycle, such as genome replication, mRNA transcription, and virion biogenesis (77). Nidovirus helicases also differ from other positive-stranded RNA viral helicases in that they occupy a position in the viral polyprotein downstream of the RNA-dependent RNA polymerase (RdRp). This arrangement is unique among positive-stranded RNA viruses, in which the helicase protein generally precedes the RdRp in the viral polyprotein (30). Also, biochemical data revealed functional differences between the nidovirus enzymes (4, 58, 59) and the well-characterized superfamily 2 helicases of pesti- and flaviviruses, supporting the idea that coronavirus (and nidovirus) helicases serve functions in the viral life cycle that are distinct from those of other RNA viral helicases.
The SARS-CoV helicase domain is believed to be part of nsp13, a 601-amino-acid cleavage product of pp1ab that is flanked by two 3CLpro cleavage sites, 5301Gln|Ala5302 and 5902Gln|Ala5903 (65, 72). Recently, we were able to show that the corresponding protein sequence (residues 5302 to 5902) mediates ATPase and DNA duplex-unwinding activities (72), indicating that the domain borders were predicted correctly. In the present study, we purified a recombinant form of nsp13 from Escherichia coli and characterized the enzymatic activities of this protein in detail. Nsp13 was revealed to be highly promiscuous with respect to its substrates. Thus, nsp13 was able to separate RNA and DNA duplexes, both of which were unwound with high processivity in a 5′-to-3′ direction. Furthermore, nsp13 was shown to hydrolyze all standard nucleotides and ribonucleotides and, by use of a common active site, also cleaves the γ-phosphate moiety of 5′-triphosphorylated RNA substrates, an activity not reported previously for coronavirus helicases. It is tempting to suggest that the 5′ RNA triphosphatase activity is involved in the synthesis of the 5′ cap structure of viral mRNAs. Using an nsp13-specific antiserum, we established that in SARS-CoV-infected Vero E6 cells, nsp13 localizes to membranes that may be derived from the endoplasmic reticulum and are the likely site of SARS-CoV RNA synthesis.
MATERIALS AND METHODS
Cells, virus, and antisera.
Vero E6 cells (kindly provided by P. Kaukinen, University of Helsinki, Finland) were infected with SARS-CoV strain Frankfurt 1 (kindly provided by H. F. Rabenau and H. W. Doerr, Johann-Wolfgang-Goethe-Universität, Frankfurt am Main, Germany). A preliminary characterization of SARS-CoV replication in Vero E6 cells demonstrated that viral RNA synthesis peaked at around 9 to 10 h postinfection and that a maximum virus titer of about 108 PFU per ml was released at approximately 12 h postinfection (data not shown).
An anti-SARS-CoV nsp13 rabbit antiserum was raised essentially as described by Snijder et al. (67). A peptide representing the nsp13 C terminus (NH2-KKLQFTSLEIPRRNVATLQ-COOH) was coupled to bovine serum albumin and used for repeated immunization of a New Zealand White rabbit. The specificity of the immune response was confirmed by Western blot analysis with an E. coli-expressed, histidine-tagged form of nsp13 (data not shown). Furthermore, immunofluorescence double-labeling studies (see below) with a serum from a convalescent SARS patient (kindly provided by L. L. M. Poon, Hong Kong University, Hong Kong) confirmed the reactivity of the nsp13 serum with SARS-CoV-infected cells, while mock-infected cells were not labeled (data not shown).
Immunofluorescence microscopy.
SARS-CoV-infected Vero E6 cells on glass coverslips were fixed with paraformaldehyde at various time points after infection and processed for immunofluorescence microscopy essentially as described by van der Meer et al. (75). Following permeabilization, indirect immunofluorescence assays were carried out with the nsp13 rabbit antiserum at a dilution of 1:800 in phosphate-buffered saline containing 5% fetal bovine serum. As the secondary antibody, an indocarbocyanine-conjugated donkey anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories) was used.
For double-labeling studies with marker proteins targeted to specific cellular compartments, Vero E6 cells were transfected with plasmid pECFP-ER or pEGFP-Golgi with Lipofectamine Plus (Invitrogen) 16 h prior to infection with SARS-CoV. Plasmid pECFP-ER (Clontech) contains a cytomegalovirus promoter that directs the expression of a variant of the enhanced cyan fluorescent protein (ECFP, a green fluorescent protein [GFP] derivative) that is targeted to the lumen of the endoplasmic reticulum by an N-terminal signal sequence and a C-terminal KDEL endoplasmic reticulum retrieval sequence. Construct pEGFP-Golgi was engineered by transferring the EGFP coding region from vector pEGFP-ER (Clontech) into expression vector pEYFP-Golgi (Clontech). Construct pEGFP-Golgi expresses an EGFP variant that is targeted to the transmedial region of the Golgi complex due to the presence of the N-terminal 81 amino acids of human β-1,4-galactosyltransferase. A mouse monoclonal antibody (1D3) (79) and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories) were used to visualize the localization of the cellular enzyme protein disulfide isomerase (PDI), a resident protein of the endoplasmic reticulum and intermediate compartment.
Samples were examined with a Zeiss Axioskop 2 fluorescence microscope (equipped with the appropriate filter sets, a digital Axiocam HRc camera, and Zeiss Axiovision 3 software) or with a Zeiss confocal laser scanning microscope. Images were optimized with Adobe Photoshop 6.0.
Protein expression and purification.
SARS-CoV nsp13 is encoded by nucleotides 16167 to 17969 of the SARS-CoV (strain Frankfurt 1) genome (72) (GenBank accession no. AY291315). For protein expression, the pMal-SARS-CoV-nsp13 and pMal-SARS-CoV-nsp13_KA plasmid DNAs (72) were used to transform E. coli TB1 cells (New England Biolabs). The maltose binding protein (MBP)-SARS-CoV nsp13 and MBP-SARS-CoV nsp13_KA fusion proteins were expressed and purified by amylose affinity chromatography with protocols described previously for the purification of coronavirus proteases (23, 24, 89, 90). Thereafter, the partially purified fusion proteins were concentrated and loaded onto HiLoad 16/60 Superdex 200 columns (Pharmacia Biotech) run under isocratic conditions with 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM dithiothreitol, 0.1 mM EDTA, and 5% glycerol. Fractions containing the desired protein were concentrated and stored at −80°C.
NTPase assay.
MBP-SARS-CoV nsp13 (10 nM) was incubated at 20°C with various concentrations of a nucleoside triphosphate (NTP) or deoxynucleoside triphosphate (dNTP) (0.5 to 10 μM) in buffer consisting of 20 mM HEPES-KOH (pH 7.4), 10% glycerol, 5 mM magnesium acetate, 2 mM dithiothreitol, 0.1 mg of bovine serum albumin per ml, and the appropriate α-32P-labeled (d)NTP (400 Ci/mmol; Amersham). In control reactions, MBP-SARS-CoV nsp13_KA was used. Following incubation for 2 to 15 min, the reactions were stopped by adding EDTA (pH 8.0) to a final concentration of 100 mM. The reaction products were analyzed by thin-layer chromatography with polyethyleneimine-cellulose F plates (Merck) with potassium phosphate (pH 4.0) as the liquid phase. The potassium concentration varied between 0.13 and 0.4 M, depending on the type of (d)NTP to be analyzed. Substrate hydrolysis was measured by phosphorimaging with ImageQuant software as described previously (58), and kinetic parameters were determined with Hofstee plots (26).
Preparation of RNA and DNA substrates.
The methods used to produce partial-duplex RNA and DNA substrates were described previously (58). To produce 5′,3′-DNA-T30, oligonucleotide D2 (5′-GGTGCAGCCGCAGCGGTGCTCG-[T]30-3′) and the 5′-32P-labeled oligonucleotide D3 (5′-[T]30-CGAGCACCGCTGCGGCTGCACC-3′) were annealed. To produce 3′-DNA-T30, the 5′-32P-labeled oligonucleotide D1 (5′-CGAGCACCGCTGCGGCTGCACC-3′), and oligonucleotide D2 were annealed. To produce 5′-DNA-T30, the 5′-32P-labeled oligonucleotide DR (5′-GGTGCAGCCGCAGCGGTGCTCG-3′) and oligonucleotide D3 were annealed. To produce DNA-0, the 5′-32P-labeled oligonucleotide D1 and oligonucleotide DR were annealed.
To produce 5′-RNA4, two RNAs, RNA-Eco and RNA-Xho, were transcribed in vitro and annealed. RNA-Eco was transcribed in the presence of [α-32P]CTP with T3 RNA polymerase and EcoRV-linearized pBluescript II KS(+) DNA (Stratagene) as a template. RNA-Xho was transcribed with T7 RNA polymerase and XhoI-linearized pBluescript II KS(+) DNA as a template. The partial-duplex 5′-RNA4 contains a 27-bp duplex region and 5′ single-stranded regions of 24 and 75 nucleotides, respectively. The production of 3′-RNA2 has been described previously (58). This partial-duplex RNA contains a 22-bp duplex region and a 3′ single-stranded region of 15 nucleotides.
To produce DNA3, single-stranded DNAs 8 and 19 were annealed. These single-stranded DNAs were produced as follows. First, two PCRs, PCR-8 and PCR-19, were performed. In both reactions, one of the primers was 5′-biotinylated. In PCR-8, oligonucleotides 5′-TAATACGACTCACTATAGGGACTTAAGTACCTTATCTATCTACA-3′ (forward primer) and 5′-biotin-TTTAGTAAAGGCCTCTAGGATGTT-3′ (reverse primer) were used to amplify a 1,021-bp fragment from full-length human coronavirus HCoV-229E cDNA as the template (70). In PCR-19, oligonucleotides 5′-biotin-ACTTAAGTACCTTATCTATCTACA-3′ (forward primer) and 5′-AAAGATGCCGGCCATAGCAAAAAT-3′ (reverse primer) were used to amplify a 100-bp fragment from full-length HCoV-229E cDNA. Second, by virtue of the 5′-biotin, PCR-8 and PCR-19 were bound to streptavidin Dynabeads (Dynal) according to the manufacturer's instructions. Third, the DNA duplexes were melted with 0.15 M sodium hydroxide, and the nonbiotinylated DNA strands were isolated from the supernatant. Finally, single-stranded DNA19 was 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase and annealed with single-stranded DNA 8. This partial-duplex DNA3 contained a 100-bp duplex region and 5′ and 3′ single-stranded regions of 21 and 900 nucleotides, respectively.
To produce RNA3, RNA76 and RNA77 were annealed. RNA76 was transcribed with T7 RNA polymerase and a 320-bp DNA template amplified by PCR from a full-length cDNA copy of the HCoV-229E genome RNA (70) with oligonucleotides 5′-TAATACGACTCACTATAGGGACTTAAGTACCTTATCTATCTACA-3′ (forward primer) and 5′-CAGGCCATTAGGAACAGTTACTGG-3′ (reverse primer). RNA77 was transcribed in the presence of [α-32P]CTP with T7 RNA polymerase and a 220-bp DNA template amplified by PCR from a full-length cDNA copy of the HCoV-229E genome RNA with oligonucleotides 5′-GATGCTGGAGTCGTAGTGTAATTG-3′ (forward primer) and 5′-AATAATACGACTCACTATAGGGCAGGCCATTAGGAACAGTTACTGG-3′ (reverse primer). The partial-duplex RNA contains a 200-bp duplex and a 5′ single-stranded region of 100 nucleotides.
For the RNA 5′-triphosphatase assays, an RNA substrate, RNA53, with the sequence 5′-GGGAAAAAAA-3′ was transcribed in vitro with T7 RNA polymerase. In two separate transcription reactions, which were performed in the presence of either [α-32P]GTP or [γ-32P]GTP, [α-32P]-labeled and 5′-γ-32P-labeled forms, respectively, of RNA53 were produced. As a template for these in vitro transcription reactions, a double-stranded DNA containing a T7 promoter was produced by annealing two complementary oligonucleotides, 5′-AATAATACGACTCACTATAGGGAAAAAAA-3′ and 5′-TTTTTTTCCCTATAGTGAGTCGTATTATT-3′.
Duplex-unwinding assay.
Typical reactions contained 14 nM MBP-nsp13 or 28 nM MBP-nsp13_KA and 10 nM RNA or DNA substrate. The reactions were performed in buffer containing 20 mM HEPES-KOH (pH 7.4), 10% glycerol, 5 mM magnesium acetate, 2 mM dithiothreitol, 0.1 mg of bovine serum albumin per ml, and 2 mM ATP. Following incubation for 30 min at 30°C, the samples were mixed with an equal volume of loading buffer (20% glycerol, 0.2% sodium dodecyl sulfate) containing bromphenol blue dye and analyzed by electrophoresis in 6 to 10% polyacrylamide gels (acrylamide/bisacrylamide ratio, 19:1) buffered with 0.5× Tris-borate-EDTA containing 0.1% sodium dodecyl sulfate.
RNA 5′-triphosphatase assay.
MBP-nsp13 and MBP-nsp13_KA (each at 40 nM) were incubated for 5 to 60 min at 30°C with 500 nM RNA53 in buffer consisting of 20 mM HEPES-KOH (pH 7.4), 10% glycerol, 5 mM magnesium acetate, 2 mM dithiothreitol, and 0.1 mg of bovine serum albumin per ml. The reaction products were analyzed by thin-layer chromatography on polyethyleneimine-cellulose F plates (Merck) with 0.15 M lithium chloride-0.15 M formic acid (pH adjusted to 3.1 with LiOH) as the liquid phase. Alternatively, the reaction samples were separated by electrophoresis in 8% polyacrylamide gels (acrylamide/bisacrylamide ratio, 19:1) buffered with 0.5× Tris-borate-EDTA containing 0.1% sodium dodecyl sulfate.
RESULTS
Subcellular localization of nsp13 in SARS-CoV-infected cells.
A preliminary characterization of SARS-CoV replication in Vero E6 cells indicated that maximum progeny titers were reached at approximately 12 h postinfection and that viral RNA synthesis peaked at 9 to 10 h postinfection (data not shown). Consequently, we chose to study the subcellular localization of nsp13 at the early and late stages of infection at 6 and 9 h postinfection, respectively. To this end, we used immunofluorescence microscopy and a rabbit antiserum raised against a peptide representing the nsp13 C terminus (see Fig. 1B and Materials and Methods). Staining of the cells for nsp13 (Fig. 2) revealed strictly cytoplasmic labeling, suggestive of localization to intracellular membrane compartments, as described for other nidoviruses. At 6 h postinfection, the labeling was mostly punctate, often with a higher labeling density at one side of the nucleus (Fig. 2A, top panel). Part of the signal was close to the nucleus, but there was also a significant amount of labeling in other regions of the cytoplasm and even close to the cell periphery. Three hours later, nsp13 was much more intensely labeled and largely concentrated in the perinuclear region, again often at one side of the nucleus but sometimes also in the form of two large patches at either side of the nucleus (Fig. 2A, bottom panel).
FIG. 2.
Immunofluorescence microscopy analysis showing the intracellular distribution of the SARS-CoV nsp13 helicase in infected Vero E6 cells. Cells were fixed at 6 h postinfection (A, top row) or 9 h postinfection (A [bottom row], B, and C) and analyzed with a conventional fluorescence microscope (A) or laser scanning confocal microscope (B and C). The SARS-CoV nsp13 staining developed from a punctatedispersed pattern at 6 h postinfection to a large, mainly perinuclear staining at 9 h postinfection. Part of the nsp13 signal overlapped the labeling for the ECFP-ER and PDI marker proteins used in this study, whereas no colocalization with a Golgi marker protein (EGFP-Golgi) was observed. Bar, 20 μm. (A) Prior to infection, cell cultures were transfected with an expression plasmid encoding endoplasmic reticulum-targeted ECFP. Part of the cells remained untransfected or uninfected, explaining the cells positive for nsp13 or the marker protein only. (B) Nontransfected cells were infected with SARS-CoV at a multiplicity of infection of 1 and used for double labeling with antisera recognizing nsp13 and the cellular protein PDI, a resident protein of the endoplasmic reticulum and intermediate compartment. (C) Prior to infection, cell cultures were transfected with an expression plasmid encoding Golgi complex-targeted EGFP.
To obtain more insight into the localization of SARS-CoV nsp13, cells were transfected prior to infection with plasmids directing the expression of EGFP or ECFP marker proteins that were specifically targeted to the Golgi complex or the endoplasmic reticulum, respectively (Materials and Methods). In addition, a monoclonal antibody against PDI, a resident protein of the endoplasmic reticulum and intermediate compartment, was used in double-labeling studies in conjunction with the anti-nsp13 rabbit serum. Specimens were analyzed with both a conventional fluorescence microscope and a confocal laser scanning microscope. At 6 h postinfection, the punctate labeling pattern of SARS-CoV nsp13 partially overlapped the signal for ECFP-ER (Fig. 2A) and PDI (data not shown). Also at 9 h postinfection, there was an overlap between the labeling for nsp13 and ECFP-ER (Fig. 2A), which was corroborated by confocal microscopy (data not shown). Essentially similar results were obtained by using confocal microscopy to analyze specimens double labeled for nsp13 and PDI (Fig. 2B). However, with both ECFP-ER and PDI, a significant part of the marker protein labeling did not overlap the nsp13 staining (Fig. 2A and 2B). At this time of infection, cell morphology began to change due to the cytopathogenic effects of the SARS-CoV infection. A clear difference in shape between infected and noninfected cells was observed (data not shown). Confocal microscopy revealed that at 6 h (data not shown) and 9 h (Fig. 2C) postinfection, the signals of the EGFP-Golgi marker protein and SARS-CoV nsp13 were completely separate. Taken together, our data suggested that nsp13, and therefore likely the core of the SARS-CoV replication machinery, localizes to a subdomain of the endoplasmic reticulum and/or an endoplasmic reticulum-derived membrane compartment.
Heterologous expression and purification of SARS-CoV nsp13.
To obtain sufficient amounts of SARS-CoV nsp13 for biochemical studies, we used an E. coli expression system. SARS-CoV pp1ab residues 5302 to 5902 were fused at the N terminus to the E. coli MBP. As Fig. 3A (lane 5) shows, fusion protein MBP-nsp13 of sufficient purity was obtained with a two-step purification protocol involving amylose affinity and size exclusion chromatography. The same approach was used to express and purify an MBP-nsp13 control protein, MBP-nsp13_KA, in which the conserved lysine residue (5589 in pp1ab) of the Walker A box (82) (helicase motif I in Fig. 1) was replaced with Ala (Fig. 3A, lane 9). The identities of the proteins were confirmed by Western blot analysis with the nsp13-specific rabbit antiserum described above.
FIG. 3.
Purification of recombinant MBP-nsp13 and MBP-nsp13_KA fusion proteins from E. coli cells. (A) Aliquots taken at each step of the purification protocol were analyzed on a sodium dodecyl sulfate-12% polyacrylamide gel, and the proteins were stained with Coomassie brilliant blue dye. Lanes: 1, protein molecular mass markers, with masses indicated on the left (in kilodaltons); 2, cleared lysate of IPTG-induced E. coli TB1 bacteria transformed with the expression plasmid pMal-SARS-CoV-nsp13; 3 and 4, peak fractions 1 and 2, respectively, from the amylose-agarose chromatography column; 5, pooled peak fractions from the Superdex 200 column; 6, cleared lysate of IPTG-induced E. coli TB1 bacteria transformed with the expression plasmid pMal-SARS-CoV-nsp13_KA; 7 and 8, peak fractions 1 and 2, respectively, from the amylose-agarose chromatography column; 9, pooled peak fractions from the Superdex 200 column. The fusion proteins are indicated by an arrowhead. (B) Western immunoblot analysis with SARS-CoV nsp13-specific rabbit antiserum. Lanes: 1, cleared lysate of E. coli TB1 bacteria transformed with the expression plasmid pMal-SARS-CoV-nsp13; 2, cleared lysate of IPTG-induced E. coli TB1 bacteria transformed with the expression plasmid pMal-SARS-CoV-nsp13; 3, purified MBP-nsp13; 4, purified MBP-nsp13_KA. The positions of protein molecular mass markers are indicated on the left (in kilodaltons).
The antiserum raised against SARS-CoV pp1ab residues 5885 to 5902 detected a protein of approximately 107 kDa in lysates obtained from isopropylthiogalactopyranoside (IPTG)-induced E. coli TB1 cells transformed with pMal-SARS-CoV-nsp13 plasmid DNA (Fig. 3B). The size of the IPTG-induced protein corresponds to the calculated molecular mass of the MBP-nsp13 fusion protein, and, as expected, the protein was not detected in noninduced cells. The purified MBP-nsp13 and MBP-nsp13_KA proteins (Fig. 3B, lanes 3 and 4) were also stained by the nsp13-specific antiserum and comigrated with the IPTG-induced protein in lane 2 (Fig. 3B), which confirmed the identities of the purified proteins. In repeated experiments, we failed to remove the amino-terminal MBP from the purified proteins by factor Xa treatment. Even after prolonged incubation with factor Xa endopeptidase, only a minor proportion of the recombinant proteins were cleaved (data not shown). Since both the SARS-CoV nsp13 helicase and other nidovirus helicases have been shown previously to tolerate N-terminal fusions (25, 58, 59, 72) and histidine-tagged forms of SARS-CoV nsp13 proved to be less soluble and active (K. A. Ivanov and J. Ziebuhr, unpublished data), we decided to use the MBP-nsp13 fusion proteins in the experiments reported here.
NTPase and dNTPase activities of SARS-CoV nsp13.
NTP hydrolysis is known to provide the energy required for translocation of RNA helicases along single-stranded RNA and duplex RNA unwinding. In a first set of experiments, we characterized the specificity of SARS-CoV nsp13 with respect to the nucleotide cofactors used. Previous studies had revealed that coronavirus and arterivirus helicases are able to hydrolyze ATP, GTP, and, in the case of porcine reproductive and respiratory syndrome virus helicase, CTP and UTP also (4, 58, 60). We now found that the MBP-nsp13 fusion protein but not the MBP-nsp13_KA control protein was able to hydrolyze all common ribonucleotides and nucleotides (Table 1). The Km values determined in the absence of nucleic acids were consistently found to be in the low micromolar range, with ATP, GTP, and dATP being hydrolyzed slightly more efficiently than other (ribo)nucleotides (Table 1). Taken together, the data suggest that SARS-CoV nsp13 displays no marked selectivity for the sugar or the nucleobase of the NTP substrate.
TABLE 1.
Analysis of SARS-CoV MBP-nsp13 NTPase substrate specificitya
| Nucleotide | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| ATP | 1.23 ± 0.12 | 2.3 ± 0.2 | 1.87 |
| UTP | 3.40 ± 0.07 | 1.5 ± 0.3 | 0.44 |
| GTP | 0.82 ± 0.05 | 1.5 ± 0.2 | 1.83 |
| CTP | 1.37 ± 0.05 | 1.3 ± 0.3 | 0.95 |
| dATP | 1.14 ± 0.04 | 1.5 ± 0.1 | 1.32 |
| dTTP | 3.01 ± 0.70 | 0.6 ± 0.1 | 0.20 |
| dGTP | 0.88 ± 0.12 | 0.6 ± 0.1 | 0.68 |
| dCTP | 2.86 ± 0.45 | 0.9 ± 0.1 | 0.31 |
The kinetic constants for NTP hydrolysis were determined from Hofstee plots (26) by the assay described in Materials and Methods. The reactions were performed with 10 nM enzyme in the presence of NTP and dNTP concentrations ranging from 0.5 to 10 μM.
5′-to-3′ RNA and DNA duplex-unwinding activities of SARS-CoV nsp13.
In a previous report, we established that SARS-CoV MBP-nsp13 separates “forked” substrates with 5′ and 3′ single-stranded regions on the same side of a partial-duplex DNA substrate (72). From a technical point of view, the nsp13-associated DNA helicase activity is advantageous because it allows potential enzyme inhibitors and mutant forms of nsp13 to be tested in DNA-based (rather than RNA-based) unwinding assays. We do not believe that this activity is of biological significance to the life cycle of coronaviruses (see Discussion) and, therefore, went on to characterize the predicted RNA duplex-unwinding activity of nsp13, mainly with respect to its polarity and processivity. To this end, a standard helicase assay with partially double-stranded RNA molecules was used (58).
MBP-nsp13 proved to be inactive on partial-duplex RNA substrates containing 3′ single-stranded tails, whereas partial-duplex substrates containing 5′ single-stranded tails were readily unwound by MBP-nsp13 (but not the MBP-nsp13_KA control protein) (Fig. 4A). The data conclusively show the 5′-to-3′ polarity of the dsRNA unwinding reaction; that is, nsp13 binds to 5′ single-stranded regions of partial-duplex RNAs and unwinds the duplex region in a 5′-to-3′ direction with respect to the RNA strand to which nsp13 initially binds. Consistent with the nsp13 RNA helicase activity and our previous data obtained for other coronavirus and arterivirus helicases (58, 59), the separation of partial-duplex DNA by SARS-CoV nsp13 was also critically dependent on the presence of 5′ single-stranded regions on the DNA substrate, demonstrating the 5′-to-3′ polarity of the double-stranded DNA-unwinding reaction. The RNA and DNA duplex-unwinding activities of nsp13 were found to depend on the presence of a nucleotide cofactor (data not shown). Furthermore, the NTPase-deficient protein MBP-nsp13_KA lacked helicase activity (Fig. 4 and 5), suggesting that NTP hydrolysis provides the energy for duplex unwinding.
FIG. 4.
RNA and DNA duplex-unwinding activities of SARS-CoV nsp13 have 5′-to-3′ polarity. The reaction conditions were as described in Materials and Methods. The structures of the substrates are shown schematically, with the radiolabeled strands marked by asterisks. (A) Helicase assay with RNA substrates 5′-RNA4 (lanes 1 to 4) and 3′-RNA2 (lanes 5 to 9), containing 5′ and 3′ single-stranded regions, respectively. Both RNA substrates contained a 22-bp duplex region. The reaction products were separated on a nondenaturing 10% polyacrylamide gel and visualized by autoradiography. Lanes 1 and 5, reactions without protein; lanes 2 and 6, heat-denatured RNA substrate; lanes 3 and 7, reactions containing MBP-nsp13; lanes 4 and 8, reactions containing MBP-nsp13_KA. (B) Helicase assay with DNA substrates. With the exception of DNA-0, which was entirely double stranded, the substrates consisted of identical 22-bp duplexes to which 30-nucleotide-long, single-stranded oligo(dT) tails were attached at different positions. Lanes 1, 5, 9, and 13, reactions without protein; lanes 2, 6, 10, and 14, heat-denatured DNA substrates; lanes 3, 7, 11, and 14, reactions containing MBP-nsp13_KA; lanes 4, 8, 12, and 16, reactions containing MBP-nsp13.
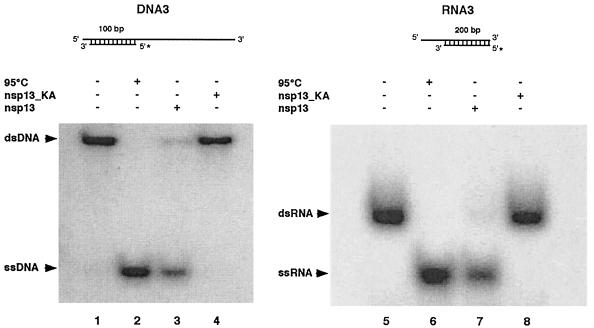
FIG. 5.
Effective unwinding of DNA and RNA substrates containing extended duplex regions by SARS-CoV nsp13. Reaction products were separated on 4.5% (left panel) and 5% (right panel) polyacrylamide gels. Lanes 1 and 5, reactions without protein; lanes 2 and 6, heat-denatured substrates; lanes 3 and 7, reactions containing MBP-nsp13; lanes 4 and 8, reactions containing MBP-nsp13_KA.
Interestingly, the SARS-CoV nsp13 helicase exhibited a remarkable processivity on both DNA and RNA substrates, allowing efficient separation of extended base-paired regions in both types of nucleic acids (Fig. 5). Thus, SARS-CoV nsp13 (this study) and other nidovirus helicases (58, 59) belong to a small group of helicases that are able to act on both DNA and RNA substrates. However, whereas the vast majority of the latter homologs exhibit a clear preference for either RNA or DNA (38, 51, 83), which generally corresponds to the physiological role of a given enzyme, no such preference for either RNA or DNA was observed with the coronavirus helicases. The hepatitis C virus NS3 helicase seems to be an equally interesting case among the RNA virus helicases characterized so far. Here, the established function of NS3 in cytoplasmic RNA synthesis stands in striking contrast to the finding that NS3 has robust DNA helicase but only poorly processive RNA helicase activities in vitro, which led to the proposal that, besides its role in viral RNA replication, NS3 might also act on host cell DNA (45).
RNA 5′-triphosphatase activity of SARS-CoV nsp13.
Based on early studies on the structure of mouse hepatitis virus RNAs (35, 36), coronavirus RNAs are generally accepted to carry a 5′ cap structure. Thus far, however, the enzymatic activities required for cap synthesis, that is, RNA 5′-triphosphatase, RNA guanylyltransferase, and RNA [guanine-7]methyltransferase (62, 63), have not been identified in the coronavirus proteome, suggesting that coronaviruses either depend on cellular activities or have evolved other strategies and/or proteins to accomplish 5′ cap synthesis.
As nsp13 was established to be capable of hydrolyzing all common nucleotides (see above), which already indicates that the substrate specificity is determined primarily by the 5′-phosphate groups rather than the nucleobase or sugar moieties, we reasoned that nsp13 may also act as an RNA 5′-triphosphatase. To explore this possibility, 5′-γ-32P-labeled RNA substrates were prepared by in vitro transcription and incubated with MBP-nsp13. The analysis of the reaction products by thin-layer chromatography (Fig. 6A) indeed revealed that MBP-nsp13 released radioactivity from 5′-γ-32P-labeled RNA, supporting our hypothesis. The radiolabel comigrated with that of the orthophosphate produced by [γ-32P]GTP hydrolysis, indicating that nsp13 cleaves the β-γ phosphate bond of the 5′-terminal nucleotide of the RNA substrate.
FIG. 6.
SARS-CoV nsp13 has RNA 5′-triphosphatase activity. 5′-γ-32P-labeled RNA was prepared as described in Materials and Methods and incubated with MBP-nsp13 or MBP-nsp13_KA. The reaction products were separated by thin-layer chromatography (A) and polyacrylamide gel electrophoresis (B) and visualized by autoradiography. (A) Lane 1, [γ-32P]GTP without protein; lane 2, [γ-32P]GTP and MBP-nsp13 (GTPase activity); lane 3, 5′-γ-32P-labeled RNA without protein; lane 4, 5′-γ-32P-labeled RNA and MBP-nsp13; lane 5, 5′-γ-32P-labeled RNA and MBP-nsp13_KA. (B) Lane 1, [γ-32P]GTP without protein; lane 2, [γ-32P]GTP and MBP-nsp13 (GTPase activity); lane 3, 5′-γ-32P-labeled RNA without protein; lanes 4, 5, and 6, 5′-γ-32P-labeled RNA and MBP-nsp13. Reactions were terminated by the addition of EDTA after 10 min (lane 4), 30 min (lane 5), and 60 min (lanes 2 and 6).
To provide additional evidence for a “true” RNA 5′-triphosphatase activity of nsp13, we sought to formally exclude the possibility that contaminating E. coli RNase activities degraded the RNA substrate and thereby released [γ-32P]GTP, which was then hydrolyzed by the GTPase activity of nsp13. Consequently, following incubation with nsp13, we analyzed the substrate RNAs by polyacrylamide gel electrophoresis, along with GTP as a control. As illustrated in Fig. 6B, no [γ-32P]GTP or [γ-32P]GTP-containing oligoribonucleotides smaller than the substrate RNAs were detectable, confirming that the radiolabel was indeed released from the intact RNA substrate rather than from [γ-32P]GTP generated by RNA degradation. When RNA transcribed in the presence of [α-32P]GTP was used as a substrate, no radioactivity was released, demonstrating that nsp13 does not have a general phosphohydrolase activity comparable to that of calf intestine phosphatase (Fig. 7).
FIG. 7.
Substrate specificity of the SARS-CoV nsp13-associated RNA 5′-triphosphatase activity. The substrate RNA, 5′-GGGAAAAA-3′, was synthesized by in vitro transcription in the presence of [α-32P]GTP (lanes 1 to 4) or [γ-32P]GTP (lanes 5 to 8). Reactions were performed as described in Materials and Methods. The RNA substrates were incubated without protein (lanes 1 and 5), with MBP-nsp13 (lanes 2 and 6), with MBP-nsp13_KA (lanes 3 and 7), or with alkaline phosphatase from calf intestine (CIP) (lanes 4 and 8). Reaction products were separated by thin-layer chromatography on polyethyleneimine cellulose-F plates and visualized by autoradiography. Reaction mixtures were incubated for 60 min (lanes 2, 3, 6, and 7) or 30 min (lanes 4 and 8).
The fact that the NTPase-deficient control protein MBP-nsp13_KA lacked RNA 5′-triphosphatase activity indicated to us that the NTPase and RNA 5′-triphosphatase activities may have a common active site. To corroborate this hypothesis, we performed competition experiments with ATP, adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), ADP, and AMP. As Fig. 8 shows, ATP indeed acted as an effective competitive inhibitor of the 5′ RNA triphosphatase activity of nsp13, whereas both ADP and AMP-PNP acted less efficiently and AMP had almost no inhibitory effect even at the high concentration used in these assays. These data led us to conclude that (i) the SARS-CoV nsp13 NTPase and RNA 5′ triphosphatase activities have a common (or largely overlapping) active site and (ii) consistent with the NTPase activity data reported above, the 5′-terminal γ- and β-phosphate groups rather than the nucleobase interact with the NTPase/RNA 5′-triphosphatase subdomain of nsp13.
FIG. 8.
Inhibition of the SARS-CoV nsp13-associated RNA 5′-triphosphatase activity by ATP. The plot illustrates the effect of including 2 mM ATP, ADP, AMP, or the ATP analog AMP-PNP on the RNA 5′-triphosphatase activity of MBP-nsp13. The average values of two experiments are plotted.
DISCUSSION
Helicases are a diverse class of enzymes that are involved in virtually every aspect of RNA and DNA metabolism. Helicases possess a common core structure which, depending on their specific function, is extended by accessory domains that confer both activity and specificity on a given enzyme (5, 64). The structure information available for a number of helicases indicates that members of the two largest helicase superfamilies, SF1 and SF2 (18, 20), have a similar core structure consisting of two RecA-like domains (5, 10, 12). Despite this structural similarity, the two superfamilies have been suggested to differ functionally (64). This conclusion is based on the available structure information showing that SF1 enzymes appear to bind to nucleic acids via stacking interactions of aromatic residues with the bases of the nucleic acid substrate (80), while SF2 enzymes bind via interactions with the phosphodiester backbone (31). Without doubt, this concept is intriguing and may explain the conservation of two superfamilies, although it needs to be further supported by additional structure information on SF1 and SF2 enzymes. The consequences of the different binding modes are not yet entirely clear but must be related to the function of a given enzyme.
On the basis of these considerations, it becomes clear that the functions of positive-stranded RNA virus helicases, which have representatives in all three main classes of helicases, SF1 to SF3, may vary considerably (27). Unfortunately, with the exception of the SF2 helicases of the Flaviviridae family, which have been characterized extensively (33, 39, 83), the information on positive-stranded RNA virus helicases is very limited, in terms of both structure and biochemical activity. However, despite this lack of information, it is reasonable to suggest that the functions of coronavirus (nidovirus) and flavivirus helicases must differ. Not only do flavivirus and nidovirus helicases belong to different helicase superfamilies (20, 27), they also display different polarities in their unwinding activities and feature a different subdomain structure. In an effort to provide first insights into the enzymatic reactions catalyzed by the SARS-CoV nsp13 helicase, we characterized a recombinant form of this protein.
Enzymatic activities of SARS-CoV nsp13.
The study shows conclusively that SARS-CoV nsp13 has a variety of enzymatic functions. These include NTPase, dNTPase, RNA 5′-triphosphatase, RNA helicase, and DNA helicase activities. In previous studies (58, 59, 72) we demonstrated that, consistent with many other helicase-associated NTPase activities (27, 41), nidovirus NTPase activities can be significantly stimulated by homopolynucleotides, most probably because of conformational changes that are triggered in the helicase's Mg2+- and NTP-binding sites upon binding of nucleic acids (68). Comparison of the kinetic constants of the SARS-CoV nsp13 (d)NTPase activities revealed little variation, suggesting that the nucleotide-binding site of nsp13 has a low specificity (see below). Similar data were also obtained for a recombinant form of SARS-CoV nsp13 carrying a 36-residue, N-terminal extension when (d)NTP hydrolysis was measured in the presence of polynucleotides (69). Other RNA virus helicase-associated NTPase activities were also reported to lack marked selectivities for specific nucleotides (4, 44, 50, 69, 83). It is thus tempting to suggest that the lack of selectivity for specific nucleotide cofactors is a general feature of RNA virus helicases. Given the high nucleotide consumption upon maximum viral RNA synthesis in the host cell, it may be advantageous for the helicase's duplex-unwinding activity not to depend strictly on a specific nucleotide. In this way, the risk of depletion of cellular ATP or GTP pools is also reduced, which would have detrimental effects on diverse metabolic pathways of the host cell and, thus, its viability in general.
Our data lead us to conclude that nsp13 is also able to bind to triphosphorylated RNA 5′ ends, allowing the enzyme to act as an RNA 5′-triphosphatase. Apparently, the NTPase and RNA 5′-triphosphatase activities have a common active site, and both the nucleotide and RNA substrates of the phosphohydrolase activity seem to be bound primarily via interactions to the γ- and β-phosphate groups. The conversion of 5′-triphosphorylated RNA substrates into RNA 5′-diphosphates has not previously been associated with coronavirus helicases. However, helicase-associated RNA 5′-triphosphatase activities have been reported in several other positive-stranded RNA viruses (6, 7, 40, 78, 84), indicating that positive-stranded RNA viruses may use helicases in pathways linked to both RNA synthesis and RNA modification. Helicase-associated RNA 5′-triphosphatase activities have been implicated in 5′ cap formation, and since coronaviruses are not known to encode a separate RNA 5′-triphosphatase, it seems reasonable to suggest that coronaviruses (and, possibly, other nidoviruses) employ the helicase to mediate the first step of 5′ cap synthesis. The increasing number of helicase-associated RNA 5′-triphosphatase activities identified in positive-stranded RNA viruses over the past decade may indicate that the dual use of helicases in both RNA synthesis and cap formation is a common theme in these viruses.
At present, it remains obscure which viral and/or cellular enzymes are involved in the guanylylation and (guanine-7) methylation reactions required to synthesize mature 5′ cap structures. For nsp16, which is the C-terminal pp1ab processing product, an RNA 2′-O-ribose methyltransferase has been predicted (65, 81) (Fig. 1). The conservation of the 2′-O-ribose methyltransferase in coronaviruses (and several other nidoviruses) indicates that specific nucleotides of coronavirus RNAs may be methylated at their ribose 2′-OH groups. Although the physiological (viral or cellular) substrates of the predicted RNA 2′-O-ribose methyltransferase remain to be determined, it is certainly reasonable to suggest that, by analogy to the conversion of cap0 to cap1/cap2 structures in cellular mRNAs, the 5′-terminal nucleotide(s) that is part of the 5′ m7GpppG( A)N cap structure of coronavirus mRNAs is among the most likely candidates for ribose 2′-OH methylations. Experiments to address this hypothesis are under way in our laboratory.
Functions of nsp13 in the coronavirus (nidovirus) life cycle.
The novel RNA 5′-triphosphatase activity established in this study for SARS-CoV nsp13 further extends the list of nidovirus helicase functions. Thus, both the equine arteritis virus reverse genetics data (77) and the biochemical data obtained for coronavirus helicases (58-60, 69, 72) demonstrate that nidovirus helicases are multifunctional proteins that are involved in diverse processes of the nidovirus replication cycle, including genome replication, subgenomic mRNA transcription, and 5′ cap formation. The previously proposed role of the nidovirus helicase in replication (77) is consistent with the remarkable processivity of the enzyme that was established in this study, as it enables nsp13 to effectively unwind double-stranded regions that the RdRp may encounter in the templates used for RNA synthesis.
As discussed above, both the structure of nidovirus helicases, which involves an N-terminal zinc-binding domain that is required for activity (A. Seybert, L. C. van Dinten, C. C. Posthuma, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr, unpublished data), and their 5′-to-3′ polarity clearly distinguish the nidovirus enzymes from the well-characterized helicases of the Flaviviridae family, all of which unwind their substrates in the opposite direction (27). Nidovirus helicases also differ from the positive-stranded plant RNA virus helicases involved in cell-to-cell movement, which again have deviant functional properties (28). It seems reasonable to believe that the divergent evolution of nidovirus helicases from the homologs of other positive-stranded RNA virus helicases was driven by the unique features of the nidovirus life cycle, of which the synthesis of an extensive set of subgenomic mRNAs encoding the structural (and accessory nonstructural) proteins is one of the most prominent hallmarks. Interestingly, the nidovirus helicase was proven to be critically involved in the latter process and appears to mediate an activity in subgenomic mRNA synthesis that is distinct from its replicative function(s) (76, 77).
Based on the ability of nidovirus helicases to translocate along single-stranded RNA in a 5′-to-3′ direction, we speculate that the helicase might release (the 3′ end of) the nascent negative strand from its template during negative-strand RNA synthesis. In other words, if bound to the plus-strand RNA, the 5′-to-3′ polarity of the nidovirus helicase activity may be used to release the nascent minus strand; for example, if attenuation signals that cause the polymerase to stall were encountered next to transcription regulatory sequence elements during minus-strand RNA synthesis (46, 56, 57). After the transcription regulatory sequence is copied and again detached by the helicase, the 3′ end of the nascent strand could be transferred to the 5′ end of the genomic RNA, where it binds to the complementary leader transcription regulatory sequence. The leader sequence is then copied to complete minus-strand synthesis. In this way, all the subgenomic negative strands acquire an antileader sequence at their 3′ end. The antileader-containing subgenomic minus strands are subsequently used as templates for the synthesis of 5′ leader-containing mRNAs.
Subcellular localization of the SARS-CoV replication complex.
Over the past decade, the association of the replication complex with intracellular membranes (of different origin) has emerged as a common feature of positive-strand RNA viruses that replicate in eukaryotic cells (1, 42, 54, 55). The association of viral replicases with (modified) cellular membranes is thought to be an important advantage in creating a suitable (micro)environment for viral RNA synthesis. Furthermore, the formation of membrane-bounded complexes may aid in shielding double-stranded RNA replication intermediates from the host defense mechanisms that may be triggered by double-stranded RNA, such as the RNA interference and interferon-induced pathways.
In the order Nidovirales, the subcellular localization of the replicase has only been studied in detail for the arterivirus equine arteritis virus and for the coronavirus MHV. In the case of equine arteritis virus, most replicase subunits (including RdRp and helicase) localize to virus-induced double-membrane structures that are derived from the endoplasmic reticulum and can be formed upon expression of two specific replicase cleavage products (47, 66, 75). In fact, the development and subcellular localization of the equine arteritis virus replication complex in Vero cells show some remarkable similarities to the images obtained for SARS-CoV nsp13 in this study (75). For the only coronavirus studied in detail thus far, MHV, the situation is less clear, which may be partially due to the use of (i) a variety of cell lines, (ii) different cellular marker proteins, and (iii) antisera recognizing different MHV replicase subunits (8, 9, 13, 22, 61, 74). Although all reports support the membrane association of MHV RNA synthesis, a variety of different cellular compartments (including the Golgi complex, endosomes, and the endoplasmic reticulum) have been implicated in viral RNA synthesis. Furthermore, the localization of the MHV helicase protein in particular was reported to change late in infection (9).
In the case of SARS-CoV, it is clear that the nsp13 helicase protein localizes exclusively to the cytoplasm. Despite its affinity for DNA substrates, this makes it unlikely that the protein can interact with host DNA, as was recently suggested for the hepatitis C virus helicase (45). Assuming that, as in other nidoviruses, the viral helicase is an appropriate marker, the images in Fig. 2 represent the first information on the localization of the SARS-CoV replication complex in the infected cell. Our data, although preliminary in nature, appear to be most consistent with the association of the complex with a subdomain of the endoplasmic reticulum or endoplasmic reticulum-derived membranes. However, in-depth ultrastructural studies will be required to confirm that this is indeed the case and to investigate whether this region contains double-membrane structures, as described for MHV by Gosert et al. (22), and may provide an answer to the question of whether these structures are derived from the endoplasmic reticulum, as postulated for equine arteritis virus.
Finally, to our knowledge, the nsp13 rabbit antiserum described in this study is the first reagent recognizing one of the subunits of the SARS-CoV replicase polyproteins, which are continuously expressed from the viral genome beginning at the earliest stages of infection. Consequently, this antiserum facilitates the early (4 to 6 h postinfection) and rapid detection of SARS-CoV replication in infected cells and may therefore be a useful tool in both diagnostics and fundamental research.
Conclusion.
Despite recent progress in the characterization of SARS-CoV replicative enzymes (3, 15, 65, 72, 85) and the increasing body of information available on homologs from other coronaviruses (nidoviruses), which, in some cases, can be used to predict the functional and structural properties of SARS-CoV proteins, there are still major gaps in our knowledge. The availability of full-length clones for SARS-CoV (87) and other coronaviruses that are amenable to analysis at a lower biosafety level provides an excellent basis for directed genetic analysis (2, 11, 70, 71, 73, 86, 88). These genetic approaches and the availability of recombinantly expressed, active enzymes and biochemical assays are anticipated to yield important new information on the molecular details of coronaviral replication and transcription and the complex interplay between the various enzymes involved in RNA synthesis and processing. We hope that, in the long term, the unique functional and structural properties of the coronavirus replicative enzymes may also lead to the development of selective enzyme inhibitors and even drugs suitable to combat SARS-CoV and other coronavirus (nidovirus) infections.
Acknowledgments
We thank H. F. Rabenau and H. W. Doerr (Johann-Wolfgang-Goethe-Universität, Frankfurt am Main, Germany) for providing the SARS-CoV Frankfurt-1 isolate and P. Kaukinen (University of Helsinki, Helsinki, Finland) for providing Vero E6 cells. We also thank L. L. M. Poon (Hong Kong University, Hong Kong) for sharing sera from SARS patients with us. We are grateful to Axel Rethwilm for the generous support of SARS-related research at the Institute of Virology and Immunology in Würzburg and acknowledge the assistance of Peter Bredenbeek, Fred Wassenaar, Clara Posthuma, Alexander Gorbalenya, and Willy Spaan in preparation of the nsp13 antiserum and SARS-CoV research at Leiden University Medical Center in general. We also thank Frans Prins (Leiden University Medical Center, Department of Pathology), for assistance with confocal microscopy.
The work of J.Z. and K.A.I. was supported by the Deutsche Forschungsgemeinschaft (ZI 618/2-3, EGK, SFB 479).
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 4.Bautista, E. M., K. S. Faaberg, D. Mickelson, and E. D. McGruder. 2002. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 298:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, L. E., H. S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. [DOI] [PubMed] [Google Scholar]
- 6.Bisaillon, M., J. Bergeron, and G. Lemay. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J. Biol. Chem. 272:18298-18303. [DOI] [PubMed] [Google Scholar]
- 7.Bisaillon, M., and G. Lemay. 1997. Characterization of the reovirus lambda1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 272:29954-29957. [DOI] [PubMed] [Google Scholar]
- 8.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 11.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delagoutte, E., and P. H. von Hippel. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines. I. Structures and properties of isolated helicases. Q. Rev. Biophys. 35:431-478. [DOI] [PubMed] [Google Scholar]
- 13.Denison, M. R., W. J. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten, C., S. Günther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguière, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Müller, V. Rickerts, M. Stürmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 15.Fan, K., P. Wei, Q. Feng, S. Chen, C. Huang, L. Ma, B. Lai, J. Pei, Y. Liu, J. Chen, and L. Lai. 2003. Biosynthesis, purification and substrate specificity of SARS coronavirus 3C-like proteinase. J. Biol. Chem. 279:1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stöhr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya, A. E. 2001. Big nidovirus genome. When count and order of domains matter. Adv. Exp. Med. Biol. 494:1-17. [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 19.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegyi, A., A. Friebe, A. E. Gorbalenya, and J. Ziebuhr. 2002. Mutational analysis of the active centre of coronavirus 3C-like proteases. J. Gen. Virol. 83:581-593. [DOI] [PubMed] [Google Scholar]
- 24.Herold, J., S. Siddell, and J. Ziebuhr. 1996. Characterization of coronavirus RNA polymerase gene products. Methods Enzymol. 275:68-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heusipp, G., U. Harms, S. G. Siddell, and J. Ziebuhr. 1997. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 71:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofstee, B. H., M. Dixon, and E. C. Webb. 1959. Non-inverted versus inverted plots in enzyme kinetics. Nature 184:1296-1298. [DOI] [PubMed] [Google Scholar]
- 27.Kadaré, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalinina, N. O., D. V. Rakitina, A. G. Solovyev, J. Schiemann, and S. Y. Morozov. 2002. RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296:321-329. [DOI] [PubMed] [Google Scholar]
- 29.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 30.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 31.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635-647. [DOI] [PubMed] [Google Scholar]
- 32.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 33.Kwong, A. D., J. L. Kim, and C. Lin. 2000. Structure and function of hepatitis C virus NS3 helicase. Curr. Top. Microbiol. Immunol. 242:171-196. [DOI] [PubMed] [Google Scholar]
- 34.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, M. M., C. D. Patton, and S. A. Stohlman. 1982. Further characterization of mRNAs of mouse hepatitis virus: presence of common 5′-end nucleotides. J. Virol. 41:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, M. M., and S. A. Stohlman. 1981. Genome structure of mouse hepatitis virus: comparative analysis by oligonucleotide mapping. Adv. Exp. Med. Biol. 142:69-82. [DOI] [PubMed] [Google Scholar]
- 37.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 38.Lee, C. G., and J. Hurwitz. 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 267:4398-4407. [PubMed] [Google Scholar]
- 39.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Y. I., T. W. Shih, Y. H. Hsu, Y. T. Han, Y. L. Huang, and M. Meng. 2001. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′-triphosphatase activity. J. Virol. 75:12114-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 42.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern, K. A., J. A. Landro, K. Hsiao, C. Lin, Y. Gu, M. S. Su, and J. A. Thomson. 1997. Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of a hepatitis C virus NS3-NS4A complex isolated from transfected COS cells. J. Virol. 71:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang, P. S., E. Jankowsky, P. J. Planet, and A. M. Pyle. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21:1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preugschat, F., D. R. Averett, B. E. Clarke, and D. J. Porter. 1996. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271:24449-24457. [DOI] [PubMed] [Google Scholar]
- 51.Rogers, G. W., Jr., W. F. Lima, and W. C. Merrick. 2001. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276:12598-12608. [DOI] [PubMed] [Google Scholar]
- 52.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Günther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 53.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kääriäinen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 57.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 58.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seybert, A., L. C. van Dinten, E. J. Snijder, and J. Ziebuhr. 2000. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 74:9586-9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seybert, A., and J. Ziebuhr. 2001. Guanosine triphosphatase activity of the human coronavirus helicase. Adv. Exp. Med. Biol. 494:255-260. [DOI] [PubMed] [Google Scholar]
- 61.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shuman, S. 1995. Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 50:101-129. [DOI] [PubMed] [Google Scholar]
- 63.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1-40. [DOI] [PubMed] [Google Scholar]
- 64.Singleton, M. R., and D. B. Wigley. 2002. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 184:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 67.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soultanas, P., M. S. Dillingham, S. S. Velankar, and D. B. Wigley. 1999. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 290:137-148. [DOI] [PubMed] [Google Scholar]
- 69.Tanner, J. A., R. M. Watt, Y. B. Chai, L. Y. Lu, M. C. Lin, J. S. Peiris, L. L. Poon, H. F. Kung, and J. D. Huang. 2003. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 278:39578-39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 71.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thiel, V., K. A. Ivanov, Á. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weiβbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 73.Thiel, V., N. Karl, B. Schelle, P. Disterer, I. Klagge, and S. G. Siddell. 2003. Multigene RNA vector based on coronavirus transcription. J. Virol. 77:9790-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. Krijnse Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Dinten, L. C., J. A. den Boon, A. L. Wassenaar, W. J. Spaan, and E. J. Snijder. 1997. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. USA 94:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Dinten, L. C., H. van Tol, A. E. Gorbalenya, and E. J. Snijder. 2000. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 74:5213-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kääriäinen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 79.Vaux, D., J. Tooze, and S. Fuller. 1990. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 345:495-502. [DOI] [PubMed] [Google Scholar]
- 80.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 81.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warrener, P., and M. S. Collett. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 69:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wengler, G. 1993. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 85.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 100:13190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ziebuhr, J., J. Herold, and S. G. Siddell. 1995. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ziebuhr, J., G. Heusipp, and S. G. Siddell. 1997. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 71:3992-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 92.Ziebuhr, J., V. Thiel, and A. E. Gorbalenya. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276:33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]